Abstract
Salmonella enterica serovar Enteritidis is frequently isolated from animal-source foods associated with human salmonellosis outbreaks. This serovar was spread to animal (mainly poultry) farms worldwide in the 1980s, and it is still detected in foods produced in many countries, including Brazil. The present study reports a retrospective genome-wide comparison of S. Enteritidis from foodborne outbreaks in Southern Brazil in the last two decades. Fifty-two S. Enteritidis isolates were obtained from foodborne outbreaks occurring in different cities of the Brazilian southernmost State, Rio Grande do Sul (RS), from 2003 to 2015. Whole-genome sequences (WGS) from these isolates were obtained and comparatively analyzed with 65 additional genomes from NCBI. Phylogenetic and Bayesian analyses were performed to study temporal evolution. Genes related to antibiotic resistance and virulence were also evaluated. The results demonstrated that all S. Enteritidis isolates from Southern Brazil clustered in the global epidemic clade disseminated worldwide originally in the 1980s. Temporal analysis demonstrated that all Brazilian isolates had a tMRCA (time to most recent common ancestor) in 1986 with an effective population size (Ne) increase soon after until 1992, then becoming constant up to now. In Southern Brazil, there was a significant decrease in the spreading of S. Enteritidis in the last decade. In addition, three antibiotic resistance genes were detected in all isolates: aac(6′)-Iaa, mdfA, and tet(34). These results demonstrate the high frequency of one only specific S. Enteritidis lineage (global epidemic clade) in foodborne outbreaks from Southern Brazil in the last two decades.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Salmonella enterica serovar Enteritidis is one of the leading pathogens causing human salmonellosis in the World [1]. S. Enteritidis has been associated with foodborne outbreaks related to the consumption of poultry products, such as eggs and undercooked chicken meat. The frequent occurrence in such foods is associated with the adaption of this serovar to commercial bird species (mainly Gallus gallus) and to the poultry farm environments [2, 3]. In general, high bacterial loads in the birds from the poultry flocks result in the contamination of the egg shells and/or the chicken carcasses at the slaughterhouses [4].
S. Enteritidis emerged as a major public health problem in the world in the mid-1980s. According to the World Health Organization (WHO), different countries reported an important increase in the isolation of S. Enteritidis in avian-source foods, such as eggs and chicken meat [5,6,7,8,9,10,11,12]. Phage typing was a useful epidemiologic tool for studying outbreaks of Salmonella serovars, including Enteritidis, for a long time [13, 14]. The first detected epidemic strains were from phage type 8 (PT8), but later there was a predominance of strains from phage type 4 (PT4), most of them detected on eggs and chicken meat in Europe [13,14,15,16].
There was an important reduction in the number of contaminated foods and human infections by foodborne S. Enteritidis since the beginning of the 2000s due to the implementation of preventive and control measures in animal production and a more strict bacteriological control in the whole poultry production processes. Live and inactivated vaccines against S. Enteritidis were also used in poultry flocks worldwide [4, 17, 18]. In Brazil, specific rules for Salmonella testing in broilers, laying hens, and foods were implemented and used by the whole poultry-producing chain [18]. Despite all these control measures, S. Enteritidis has remained as one of the most frequent bacterial foodborne pathogens with high impact in human health worldwide [19, 20].
S. Enteritidis strains have remarkable genetic diversity that allows them to adapt to hosts and to disseminate in different environmental conditions. It was previously classified into three different clades (Central/Eastern African, West African, Global Epidemic) plus the “outliers,” each one presenting strains with specific metabolic characteristics more adapted to some hosts and causing a wide range of human symptoms, including invasive disease [20]. S. Enteritidis strains from these different clades have some well-established genetic features, with the occurrence of specific pathogenicity islands and virulence genes. The two distinct African epidemic lineages emerged in the human population in the past century, and they are associated with a specific prophage profile, one virulence plasmid with several multidrug resistance (MDR) genes and patterns of genomic degradation similar to other host-restricted invasive Salmonella serovars (such as Typhi and Gallinarum) [20]. On the contrary, the global epidemic lineage is adapted to different hosts and transmission niches presenting a genome with less pseudogenes and without a plasmid with a MDR profile. Most S. Enteritidis strains from this lineage seem to have a low number of antimicrobial resistance genes and consequently high susceptibility to antibiotics [21,22,23].
Whole-genome sequencing (WGS) and bioinformatics analysis have been increasingly used to track foodborne pathogens. This technology has the power to differentiate isolates and to determine genetic relationships by applying the principles of evolutionary biology [24]. Moreover, data generated by the WGS technique can be submitted to in silico evaluation of genetic characteristics of each isolate [25]. Several studies have compared the S. Enteritidis genomes by single nucleotide polymorphisms (SNPs) to investigate the phylogenetic relationships, allowing to trace the source of contamination of a food [19, 20, 26, 27]. The present study aimed to perform a retrospective WGS comparison of S. Enteritidis isolates from foodborne outbreaks in Southern Brazil over a 13-year period. It was possible to assess the recent evolution and to define the main genetic characteristics of the main circulating S. Enteritidis lineage in Southern Brazil.
Materials and methods
Bacteria isolates
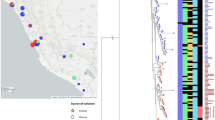
Fifty-two S. Enteritidis isolates were obtained from foodborne outbreaks (30.7% [16/52] representing isolates associate with poultry) occurring in different cities in Rio Grande do Sul (RS), the southernmost Brazilian state, from 2003 to 2015 (Fig. 1; Table S1). A single colony was removed from xylose lysine desoxycholate (XLD) agar plate and placed in a brain heart infusion (BHI) broth, following overnight incubation at 35 °C. Then, one aliquot of 0.1 mL was used to extract DNA with a commercial kit following the manufacturer’s instruction (NewGene, Simbios Biotecnologia, Cachoeirinha, Brazil).
Geographical distribution of 46 out of 52 S. Enteritidis isolates from foodborne outbreaks in thirty-three cities in the Brazilian state of Rio Grande do Sul (six isolates had no origin information). The circles represent the geographical location of the different cities in the study. The diameter is in accordance with the number of isolates in each city
One S. Enteritidis–specific PCR was used to test all isolates as previously described [28]. Amplification conditions followed an initial denaturation cycle of 3 min at 95 °C followed by 40 cycles of 15 s at 95 °C and 60 s at 60 °C performed on StepOne Plus equipment (Applied Biosystems, Carlsbad CA, USA). The evaluation of the result was performed directly on the equipment by the analysis of the presence of amplification curves and determination of the cycle threshold (Ct) values. Only DNAs with Ct value < 20 were used for the next steps.
Whole-genome sequencing
PureLink® Genomic DNA Mini Kit was used to extract the genomic DNA following the manufacturer’s instructions (Thermo Fisher Scientific, Waltham, MA, USA). DNA was visualized on 2% agarose gel stained with ethidium bromide. Sequencing library was prepared using Nextera XT kit (Illumina, Inc., San Diego, CA, USA). DNA concentration was adjusted to 0.5 ng/µL and sequenced on the Illumina NextSeq platform with 150 bp paired end reads (Wadsworth Center, New York State Dept. of Health, Albany, NY, USA).
Genome Data Acquisition, Multi-Locus Sequence Typing (MLST) and Salmonella In Silico Typing Resource (SISTR)
The raw sequence reads were trimmed and low-quality bases removed with Trimmomatic v0.33 [29]. Quality of trimmed reads was checked using FastQC v0.11.2 [30] prior to de novo assembly using SPAdes v3.6.0 [31]. The quality of draft genomes was evaluated using QUAST v4.0, and average coverage was estimated using BBmap v38.26 [32] and Samtools v1.8. Legacy MLST (7-gene MLST) sequence type and confirmation of serovar were determined using SISTR v0.3.1 [33].
SNP phylogeny
Single nucleotide polymorphisms were identified by kSNP3 with an optimal kmersize of 19 nucleotides [34] using a total of 117 S. Enteritidis assemblies, including the 52 isolates sequenced here and 65 WGS data downloaded from NCBI (Table S2), all from different sources (foods, animals, human, environment, organs). Additional NCBI data were defined according to S. Enteritidis previously sequenced in Brazil and other South American countries as well as reference genomes [20, 27]. A maximum parsimony tree was used to cluster the sequences based on the core SNPs identified.
The CFSAN SNP Pipeline v2.0.2 [35] was used to map high-quality SNPs (hqSNPs) in these 117 genomes. SF297-04 was used as reference. The resulting SNP matrix of preserved sites was then used to build a phylogeny using the maximum likelihood (ML) method with the IQtree program with 1000 bootstraps [36].
Tip-dated evolutionary analyses
Linear regression models implemented in TempEst v1.5 [37] were used to evaluate the temporal signal and clock likeness of the phylogenies based on associations between temporal sequence divergence and isolation dates. Tip-dated phylogenies were constructed using BEAUti v2.6.0 and BEAST v2.6.0 [38] with a combination of the GTR substitution model and lognormal relaxed clock and coalescent Bayesian skyline models [39]. The runs were carried out with a substitution rate prior set to 1.0 × 10−7/site/year (default of program).
Markov Chain Monte Carlo (MCMC) algorithm was run for 1 × 108 generations [20], and parameters were logged every 1 × 104 generations. The confirmation of model combination was identified based on marginal likelihood values and ESS (effective sample size) of run statistics (e.g., prior, posterior, tree likelihood, clock rate, and coalescent). The output statistics and traces were analyzed in Tracer v1.7.1, and the trees file was annotated in TreeAnnotator v2.6.0 (burn-in set at 10) and edited in FigTree v1.4.4. This unrooted maximum credibility tree was presented with height (i.e., ages relative to the youngest sequence), 95% highest posterior density (HPD) intervals on node bars, and posterior probabilities placed on the branch labels.
The investigation of the relationship among isolates was performed by comparing the median of SNPs among sequences. Isolates with less than 21 hqSNPs of difference were considered with strong epidemiological relationship [40].
Identification of gene for prediction of antimicrobial resistance
The Abricate v0.8 (https://github.com/tseemann/abricate) and BLASTN algorithm (http://blast.ncbi.nlm.nih.gov/Blast.cgi) were used to search the genome assemblies for antimicrobial resistance genes (using ResFinder database: https://bitbucket.org/account/user/genomicepidemiology/projects/DB). Matches with ≥ 75% sequence identity and ≥ 50% sequence length coverage were considered relevant, after the output.
Results
WGS data of S. Enteritidis and sequence type (ST)
Raw sequencing reads obtained in this study (n = 52) and downloaded from NCBI (n = 65) were assembled with a median of 25 contigs larger than 1 Kb (ranging from 18 to 163 Kb), a median N50 of 406,263 bp (ranging from 45,084 to 594,007 bp), and median average coverage of 114 × (ranging from 62 to 617 ×). The median length of the 117 assembled S. Enteritidis genomes (made of contigs ≥ 1000 bp) was 4.70 Mbp (ranging from 4.59 to 4.83 Mbp). In silico analysis showed that all isolates matched to serovar Enteritidis with SISTR. ST11 was detected in all S. Enteritidis genomes used in this study.
SNP-based phylogenetic analysis of S. Enteritidis
The phylogenetic relationship among the 52 genomes sequenced in the present study and 65 S. Enteritidis WGSs downloaded from NCBI identified 26,287 core SNPs, which were used to build an initial maximum parsimony phylogeny (Figure S1). All S. Enteritidis foodborne outbreak isolates sequenced in the present study (n = 52) clustered together with other strains of the global epidemic lineage from around the world in a monophyletic group. All these set of 117 isolates were used to further investigate the relationship of the isolates using a high-quality single nucleotide polymorphism (hqSNP) approach.
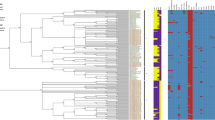
The CFSAN SNP Pipeline identified 0 to 129 pairwise hqSNPs (median 57). It was used the hqSNP alignment with the TVM e substitution model defined, of the previous analysis, to build a phylogeny that guided further analyses (Fig. 2). The topology of the ML tree clearly demonstrated that the isolates from food sources, humans, and poultry are phylogenetically similar. This monophyletic clade with S. Enteritidis isolated between 1990 and 2016 also contained all isolates from foodborne outbreaks of Rio Grande do Sul (n = 52) and other genomes data from Brazil (n = 52), Portugal (n = 1), UK (n = 1), USA (n = 1), Colombia (n = 2), Argentina (n = 5), and Uruguay (n = 2).
Maximum likelihood phylogeny constructed using high-quality SNPs (hqSNPs) identified among 117 S. Enteritidis genomes using the CFSAN SNP Pipeline. Red are isolates sequenced in this study of foodborne outbreaks (n = 52), black are Brazilian isolates downloaded from NCBI (n = 52), pink are isolates from other America countries (USA, Argentina, Colombia, and Uruguay [n = 10]), green are isolates from Europe (UK and Portugal [n = 2]), and brown is P125109. The phylogeny was constructed using IQ-TREE and is midpoint rooted, with branch lengths representing the number of substitutions per site
Bayesian phylogenetic analysis of S. Enteritidis
The 104 Brazilian S. Enteritidis genomes were used to perform the Bayesian temporal analysis. For this analysis, twelve genomes belonging to other countries and the genome P125109 were removed. In addition, two evolutionary analyses were performed: (i) with all Brazilian genomes (52 isolates sequenced in this study from foodborne outbreaks of Rio Grande do Sul State and 52 WGS data downloaded from NCBI from several Brazilian regions); (ii) with only the 52 isolates sequenced in the present study from foodborne outbreaks of Rio Grande do Sul State.
The results of the correlation coefficient of TempEst demonstrated that R = 0.80 (104 isolates) and R = 0.66 (52 isolates) indicated a correlation between isolation date and sequence divergence, which indicates that our dataset is suitable for temporal analysis.
One combination of lognormal relaxed clock model and coalescent Bayesian skyline was run in BEAST to reconstruct tip-dated Bayesian phylogenies. The model combination was run with a GTR substitution model. The 104 isolates included were estimated to evolve with a rate of 4,71 × 10−7 substitutions/site/year (95% HPD 4.14 × 10−7–5.28 × 10−7), while the temporal analysis of 52 isolates presented a rate of evolution of 5.40 × 10−7 substitutions/site/year (95% HPD 4.35 × 10−7–6.54 × 10−7).
All 104 analyzed isolates presented a common ancestor dated back to 1983 (95% HPD: 1946–2011). Interestingly, there were two main clusters with all more recent isolates in the branch located at the bottom of the tree (Fig. 3). The additional analysis of the 52 isolates from Southern Brazil presented a common ancestor 3 years later, dating back to 1986 (95% HPD: 1953–2009). Again there were two main clusters with all more recent isolates at the bottom of the tree (Fig. 4).
Maximum clade credibility tree constructed using high-quality SNPs (hqSNPs) identified among 104 S. Enteritidis genomes using the CFSAN SNP Pipeline, rooted using BEAST. Red are isolates of foodborne outbreaks from RS sequenced in this study (n = 52) and black are genomes downloaded from NCBI from several Brazilian regions (n = 52). Time in years is plotted along the X-axis. Branch labels denote posterior probabilities of branch support, and node bars correspond to 95% highest posterior density (HPD) intervals for node heights
Maximum clade credibility tree constructed using high-quality SNPs (hqSNPs) identified among 52 S. Enteritidis genomes (red) using the CFSAN SNP Pipeline, rooted using BEAST. Time in years is plotted along the X-axis. Branch labels denote posterior probabilities of branch support, and node bars correspond to 95% highest posterior density (HPD) intervals for node heights
Historical demographic trend of the S. Enteritidis population size over time was reconstructed using a Bayesian skyline. An important increase was observed in the population size in Brazil from late 1988 up to 1992 and afterward apparently remains constant until recently (Fig. 5). On the contrary, S. Enteritidis population size related to foodborne outbreaks of Rio Grande do Sul decreased after 2007 and stabilized in 2012 (Fig. 6). This situation may reflect the adoption of important preventive measures to control Salmonella in poultry farms as well as in food inspection in this period of time.
Effective population size over the time. Bayesian skyline plot, based on a “relaxed clock” coalescent framework analysis, was constructed using 104 sequences from foodborne outbreak isolates of Rio Grande do Sul State (n = 52, sequenced in this study) and genomes downloaded from NCBI from several Brazilian regions (n = 52). X-axis represents time in years, while Y-axis shows the effective population size. The blue band represents 95% highest posterior density (HPD) intervals
Effective population size and time for most recent common ancestor. Bayesian skyline plot, based on a “relaxed clock” coalescent framework analysis, was constructed using 52 sequences of foodborne outbreaks in Rio Grande do Sul sequenced in this study. X-axis represents time in years, while Y-axis shows the effective population size. The blue band represents 95% highest posterior density (HPD) intervals
Outbreak investigation
S. Enteritidis genomes from the same and different outbreaks (according information of city and year) were comparatively analyzed to assess molecular identity among the isolates. The median of SNPs among the forty-four isolates from the most disseminated cluster (Cluster I) ranged from 0 to 42 SNPs (Fig. 4). Interestingly there was a subcluster (A) that the median number of SNPs was 21 SNPs, indicating strong epidemiological relationship. This subcluster showed a range of twenty-nine isolates belonging to a period of 10 years (2006 to 2015) from different cities. There were even isolates from different cities that showed a difference value median of 0 hqSNPs. These values are within the range observed between isolates from the same source causing S. Enteritidis foodborne outbreaks [19, 40, 41].
Antimicrobial resistance profiling of S. Enteritidis isolates
Genomic analysis demonstrated that all 52 foodborne outbreak isolates from Southern Brazil (100%) had the antimicrobial resistance genes (ARGs) aac(6′)-Iaa, mdf(A), and tet(34). The aac(3)-Iva, aph(3′')-Ib, aph(4)-Ia, tet(A), and aph(6)-Id genes were present in 3.8% (2/52) isolates. No other antibiotic resistance gene was detected in the genomes from S. Enteritidis sequences (Figure S2).
Discussion
Foodborne diseases occur worldwide and are responsible for outbreaks in different countries. However, outbreak investigation is often limited because many episodes of foodborne diseases are not reported [42, 43]. Laboratorial data generated by the diagnostic and screening techniques help to reduce the incidence of outbreaks, supporting the adoption of preventive and control measures and, consequently, contributing to the improvement of the population’s quality of life [44].
S. Enteritidis is a serovar of great public health concern in several countries around the world. Since the 1980s, food surveillance agencies have reported several outbreaks by this serovar, most of them caused by products of poultry origin, such as chicken meat and eggs [45,46,47]. Phenotypic serotyping, phage typing, and pulsed field gel electrophoresis have been used for a long time to investigate foodborne outbreaks by S. Enteritidis. Now SNP-based WGS analyses are contributing to improve the tracking of foodborne outbreaks by this pathogen in different countries, due to the possibility of sharing information and online databases [41, 48].
Here we described a large WGS study of S. Enteritidis associated to foodborne outbreaks in Southern Brazil in the last 18 years. Noteworthy, all genomes were highly similar to previously sequenced isolates of the global epidemic lineage that has been circulating since the 1980s in Brazil and other countries in the world. All S. Enteritidis isolates here sequenced seem to have the same common ancestor that started to spread around 1983 in Brazil and more specifically around 1986 in Southern Brazil. A previous study has already shown the worldwide dissemination of this lineage by foods in the 1980s and 1990s [20].
Chicken, eggs, and other avian-source foods have been associated as the major vehicles for increasing infections by the global epidemic lineage of S. Enteritidis in the western world [6, 49]. Undoubtedly, the poultry production chain seems to have a pivotal role in the introduction and dissemination of this lineage in Brazil. Results presented here demonstrated a high increase of S. Enteritidis population exactly in this same period (1980s to 1990s), clearly reinforcing the wide spreading of this serovar by this production chain. International poultry and foods trade probably further contributed to several introductions of this lineage in the country in this period. Other data and epidemiological studies also proposed that the entry of S. Enteritidis in Brazil occurred in the late 1980s with a wide spread soon after in the whole country as well as in some specific regions [49,50,51].
Additionally, all Brazilian S. Enteritidis isolates clearly clustered into one monophyletic cluster. All recent isolates belonged to a more specific cluster together with the isolate SE86 (SE86-RS-99), recently sequenced [52]. Previous studies have already shown that this strain was the first best characterized of this lineage highly spread in Southern Brazil. It was also demonstrated that SE86 has a great capacity for adaptation and survival compared to strains from other Salmonella serovars, especially regarding its resistance to acid, heat, sanitizers, and antimicrobials [53,54,55].
Although S. Enteritidis remains a significant problem in poultry, the prevalence of this serovar has declined in poultry flocks since the 2000s in the USA. The implementation of guidelines to control S. Enteritidis increased flock immunity from bird exposure or vaccination likely contributed to this decline [2]. In our study, there was a reduction in the population of S. Enteritidis after 2007 in Southern Brazil that probably was a consequence of the implementation of the National Poultry Health Plan (PNSA, Programa Nacional de Sanidade Avícola) and the adoption of sanitary measures instituted in instructions from the Brazilian government [56]. Noteworthy, the introduction of vaccination against S. Enteritidis in breeding and laying hens together with the frequent control of poultry-producing flocks seems to have also contributed to the reduction of the occurrence of foodborne outbreaks by this serovar in Southern Brazil in recent years, as clearly observed in the skyline plot (Fig. 6). Further, a food regulation was launched in Rio Grande do Sul State prohibiting the preparation of food meals containing raw or undercooked eggs in 2009. The same regulation established the necessity of a specific training for food handlers, focusing on good hygienic practices and how to prevent foodborne diseases, mainly salmonellosis [57]. These preventive and control measures in Southern Brazil were necessary (and now seem to have been very successful) to reduce the occurrence of foodborne outbreaks caused by S. Enteritidis. Data from the UK’s Health Protection Agency (HPA) also demonstrated a decline in the number of foodborne outbreaks caused by S. Enteritidis between 1997 and 2008 [58]. All this data provides additional evidences that the introduction and adherence to control the spreading of this serovar minimized foodborne infection risk in Southern Brazil too.
Furthermore, the comparison of the complete genomes data of the S. Enteritidis isolates demonstrated a strong epidemiological relationship among the isolates of the outbreaks that occurred in different places and periods in Southern Brazil. In the analysis of the hqSNP differences, there was not any genetic difference (0 SNPs) among many isolates from the same outbreak site. In addition, similar result was obtained for isolates from different cities, indicating strict epidemiological relationships. Unfortunately, there was a lack of data that could contribute to a better investigation of these outbreaks as previously published [40]. However, all these data generated here will help in future studies in Brazil to trace back and forward S. Enteritidis foodborne outbreaks. A recent study in Poland identified eggs as the vehicle of infection and the source of the outbreak. This study has also demonstrated that in the implementation of control measures, there was a change in population dynamics and the number of human salmonellosis case reports decreased, indicating a practical application in foodborne disease control [19]. This approach can be applied in tracking foodborne outbreaks to diagnose the source of the contamination. This would be very useful for real-time tracking of foodborne outbreaks.
Finally, the present study demonstrated the occurrence of some genes for antimicrobial resistance (mainly for aminoglycosides and tetracyclines) in the S. Enteritidis isolates from Southern Brazil. Previous reports have already demonstrated by phenotypic methods an intermediate resistance in foodborne outbreaks S. Enteritidis isolates, which may corroborate the detection of antimicrobial resistance genes in isolates from Southern Brazil [21, 59]. It is important to continue monitoring the use of antibiotics in animal production, mainly in commercial poultry flocks in Southern Brazil, an emerging hotspot for antimicrobial resistance [60].
In conclusion, the study shows that the global epidemic lineage continued to circulate in Brazil in the last two decades, causing community outbreaks due to the consumption of food contaminated with S. Enteritidis.
References
Jajere SM (2019) A review of Salmonella enterica with particular focus on the pathogenicity and virulence factors, host specificity and antimicrobial resistance including multidrug resistance. Vet World 12:504–521. https://doi.org/10.14202/vetworld.2019.504-521
Foley SL, Nayak R, Hanning IB, Johnson TJ, Han J, Ricke SC (2011) Population dynamics of Salmonella enterica serotypes in commercial egg and poultry production. Appl Environ Microbiol 77:4273–4279. https://doi.org/10.1128/AEM.00598-11
Antunes P, Mourão J, Campos J, Peixe L (2016) Salmonellosis: the role of poultry meat. Clin Microbiol Infect 22:110–121. https://doi.org/10.1016/j.cmi.2015.12.004
World Organisation for Animal Health (2019) Prevention, detection and control of Salmonella in poultry. https://www.oie.int/index.php?id=169&L=0&htmfile=chapitre_prevent_salmonella.htm. Accessed 07 Feb 2020
Rampling A, Upson R, Peters E, Anderson JR, Ward LR, Rowe B (1989) Salmonella enteritidis phage type 4 infection of broiler chickens: a hazard to public health. Lancet 334:436–438. https://doi.org/10.1016/S0140-6736(89)90604-1
Rodrigue DC, Tauxe RV, Rowe B (1990) International increase in Salmonella Enteritidis: a new pandemic? Epidemiol Infect 105:21–27. https://doi.org/10.1017/s0950268800047609
Giessen AW, Dufrenne JB, Ritmeester WS, Berkers PATA, Leeuwen WJ, Notermans SHW (1992) The identification of Salmonella Enteritidis-infected poultry flocks associated with an outbreak of human salmonellosis. Epidemiol Infect 109:405–411. https://doi.org/10.1017/s0950268800050391
Poppe C (1994) Salmonella Enteritidis in Canada. Int J Food Microbiol 21:1–5. https://doi.org/10.1016/0168-1605(94)90193-7
Sakai T, Chalermchaikit T (1996) The major sources of Salmonella enteritidis in Thailand. Int J Food Microbiol 31:173–180. https://doi.org/10.1016/0168-1605(96)00979-8
Scuderi G, Fantasia M, Filetici E, Anastasio MP (1996) Foodborne outbreaks caused by Salmonella in Italy, 1991–4. Epidemiol Infect 116:257–265. https://doi.org/10.1017/s0950268800052559
Herikstad H, Motarjemi Y, Tauxe RV (2002) Salmonella surveillance: a global survey of public health serotyping. Epidemiol Infect 129:1–8. https://doi.org/10.1017/s0950268802006842
Oliveira SD, Santos LR, Schuch DM, Silva AB, Salle CT, Canal CW (2002) Detection and identification of salmonellas from poultry-related samples by PCR. Vet Microbiol 87:25–35. https://doi.org/10.1016/s0378-1135(02)00028-7
Humphrey TJ, Baskerville A, Maver S, Rove B, Hopper S (1989) Salmonella Enteritidis phage type 4 from the contents of intact eggs: a study involving naturally infected hens. Epidemiol Infect 103:415–423. https://doi.org/10.1017/s0950268800030818
Hickman-Brenner FW, Stubbs AD, Farmer JJ 3rd (1991) Phage typing of Salmonella enteritidis in the United States. J Clin Microbiol 29:2817–2823. https://doi.org/10.1128/JCM.29.12.2817-2823.1991
Irino K, Fernandes SA, Tavechio AT, Neves BC, Dias AMG (1996) Progression of Salmonella Enteritidis phage type 4 strains in São Paulo State, Brazil. Rev Inst Med trop Sao Paulo 38:193–196. https://doi.org/10.1590/S0036-46651996000300005
Nunes IA, Helmuth R, Schroeter A, Mead GC, Santos MA, Solari CA, Silva OR, Ferreira AJ (2003) Phage typing of Salmonella enteritidis from different sources in Brazil. J Food Prot 66:324–327. https://doi.org/10.4315/0362-028x-66.2.324
Cavalcanti Júnior RC (2013) Instrução Normativa nº 10 de 11 de abril de 2013. Diário Oficial da União. http://www.avimig.com.br/galeria_imagens/LEGISLACAO_04112016_094044.pdf. Accessed 14 Feb 2021
Rangel LEP (2017) Instrução Normativa nº 8 de 17 de fevereiro de 2017. Diário Oficial da União. https://www.in.gov.br/materia/-/asset_publisher/Kujrw0TZC2Mb/content/id/20472445/do1-2017-03-03-instrucao-normativa-n-8-de-17-de-fevereiro-de-2017-20472317. Accessed 14 Feb 2021
Pijnacker R, Dallman TJ, Tijsma ASL, Hawkins G, Larkin L, Kotila SM, Amore G, Amato E, Suzuki PM, Denayer S, Klamer S, Pászti J, McCormick J, Hartman H, Hughes GJ, Brandal LCT, Brown D, Mossong J, Jernberg C, Müller L, Palm D, Severi E, Gołębiowska J, Hunjak B, Owczarek S, Le Hello S, Garvey P, Mooijman K, Friesema IHM, van der Weijden C, van der Voort M, Rizzi V, Franz E (2019) An international outbreak of Salmonella enterica serotype Enteritidis linked to eggs from Poland: a microbiological and epidemiological study. Lancet Infect Dis 19:778–786. https://doi.org/10.1016/S1473-3099(19)30047-7
Feasey NA, Hadfield J, Keddy KH, Dallman TJ, Jacobs J, Deng X, Wigley P, Barquist L, Langridge GC, Feltwell T, Harris SR, Mather AE, Fookes M, Aslett M, Msefula C, Kariuki S, Maclennan CA, Onsare RS, Weill FX, Le Hello S, Smith AM, McClelland M, Desai P, Parry CM, Cheesbrough J, French N, Campos J, Chabalgoity JA, Betancor L, Hopkins KL, Nair S, Humphrey TJ, Lunguya O, Cogan TA, Tapia MD, Sow SO, Tennant SM, Bornstein K, Levine MM, Lacharme-Lora L, Everett DB, Kingsley RA, Parkhill J, Heyderman RS, Dougan G, Gordon MA, Thomson NR (2016) Distinct Salmonella Enteritidis lineages associated with enterocolitis in high-income settings and invasive disease in low-income settings. Nat Genet 48:1211–1217. https://doi.org/10.1038/ng.3644
Oliveira FA, Brandelli A, Tondo EC (2006) Antimicrobial resistance in Salmonella enteritidis from foods involved in human salmonellosis outbreaks in southern Brazil. New Microbiol 29:49–54
Campioni F, MorattoBergamini AM, Falcão JP (2012) Genetic diversity, virulence genes and antimicrobial resistance of Salmonella Enteritidis isolated from food and humans over a 24-year period in Brazil. Food Microbiol 32:254–264. https://doi.org/10.1016/j.fm.2012.06.008
European Food Safety Authority, European Centre for Disease Prevention and Control (2014) European Union Summary Report on antimicrobial resistance in zoonotic and indicator bacteria from humans, animals and food 2012 published. EFSA J 19:20748. https://doi.org/10.2903/j.efsa.2014.3590
Food and Drug Administration (2018) Whole Genome Sequencing (WGS) Program. Available at: https://www.fda.gov/food/science-research-food/whole-genome-sequencing-wgs-program. Accessed 02 Feb 2020
Zankari E, Hasman H, Cosentino S, Vestergaard M, Rasmussen S, Lund O, Aarestrup FM, Larsen MV (2012) Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother 67:2640–2644. https://doi.org/10.1093/jac/dks261
Toro M, Retamal P, Ayers S, Barreto M, Allard M, Brown EW, Gonzalez-Escalona N, Dudley EG (2016) Whole-genome sequencing analysis of Salmonella enterica serovar Enteritidis isolates in Chile provides insights into possible transmission between gulls, poultry, and humans. Appl Environ Microbiol 82:6223–6232. https://doi.org/10.1128/aem.01760-16
Campioni F, Cao G, Kastanis G, Janies DA, Bergamini AMM, Rodrigues DDP, Stones R, Brown E, Allard MW, Falcão JP (2018) Changing of the genomic pattern of Salmonella Enteritidis strains isolated in Brazil over a 48 year-period revealed by whole genome SNP analyses. Sci Rep 8:1–7. https://doi.org/10.1038/s41598-018-28844-6
Souza MN, Lehmann FKM, De Carli S, Kipper D, Fonseca ASK, Ikuta N, Lunge VR (2019) Molecular detection of Salmonella serovars Enteritidis, Heidelberg and Typhimurium directly from pre-enriched poultry samples. Br Poult Sci 60:388–394. https://doi.org/10.1080/00071668.2019.1614525
Bolger AM, Lohse M, Usadel B (2014) Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120. https://doi.org/10.1093/bioinformatics/btu170
Andrews S (2014) FastQC. Babraham Bioinformatics. Available at: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/. Accessed 02 Feb 2020
Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA (2012) SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. https://doi.org/10.1089/cmb.2012.0021
Bushnell B (2015) BBMap. Source Forge. Available at: https://sourceforge.net/projects/bbmap/. Accessed 02 Feb 2020
Yoshida CE, Kruczkiewicz P, Laing CR, Lingohr EJ, Gannon VP, Nash JH, Taboada EM (2016) The Salmonella In silico typing resource (SISTR): an open web-accessible tool for rapidly typing and subtyping draft Salmonella genome assemblies. PLoS ONE 11:e0147101. https://doi.org/10.1371/journal.pone.0147101
Gardner SN, Slezak T, Hall BG (2015) kSNP3.0: SNP detection and phylogenetic analysis of genomes without genome alignment or reference genome. Bioinformatics 31:2877–2878. https://doi.org/10.1093/bioinformatics/btv271
Davis S, Pettengill JB, Luo Y, Payne J, Shpuntoff A, Rand H (2015) CFSAN SNP Pipeline: an automated method for constructing SNP matrices from next-generation sequence data. Peer J Comput Sci 1:e20. https://doi.org/10.7717/peerj-cs.20
Trifinopoulos J, Nguyen LT, von Haeseler A, Minh BQ (2016) W-IQ-TREE: a fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res 44:232–235. https://doi.org/10.1093/nar/gkw256
Rambaut A, Lam TT, Max Carvalho L, Pybus OG (2016) Exploring the temporal structure of heterochronous sequences using TempEst (formerly Path-O-Gen). Virus Evol 2: vew007. https://doi.org/10.1093/ve/vew007
Drummond AJ, Suchard MA, Xie D, Rambaut A (2012) Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol Biol Evol 29:1969–1973. https://doi.org/10.1093/molbev/mss075
D’Alessandro B, Pérez Escanda V, Balestrazzi L, Iriarte A, Pickard D, Yim L, Chabalgoity JA, Betancor L (2018) A novel prophage identified in strains from Salmonella enterica serovar Enteritidis is a phylogenetic signature of the lineage ST-1974. Microb Genom 4:e000161. https://doi.org/10.1099/mgen.0.000161
Pightling AW, Pettengill JB, Luo Y, Baugher JD, Rand H, Strain E (2018) Interpreting whole-genome sequence analyses of foodborne bacteria for regulatory applications and outbreak investigations. Front Microbiol 9:1482. https://doi.org/10.3389/fmicb.2018.01482
Taylor AJ, Lappi V, Wolfgang WJ, Lapierre P, Palumbo MJ, Medus C, Boxrud D (2015) Characterization of foodborne outbreaks of Salmonella enterica serovar Enteritidis with whole-genome sequencing single nucleotide polymorphism-based analysis for surveillance and outbreak detection. J Clin Microbiol 53:3334–3340. https://doi.org/10.1128/JCM.01280-15
World Health Organization (2008) Foodborne disease outbreaks: guidelines for investigation and control. Geneva: WHO Library
Klein LR, Bisognin RP, Figueiredo DMS (2017) Estudo do perfil epidemiológico dos surtos de doenças de transmissão hídrica e alimentar no Rio Grande do Sul: uma revisão dos registros no Estado. Hygeia 13:48–64. https://doi.org/10.14393/Hygeia132504
Brasil (2010) Manual integrado de vigilância, prevenção e controle de doenças transmitidas por alimentos. Brasília: Editora do Ministério da Saúde
Galanis E, Lo Fo Wong DM, Patrick ME, Binsztein N, Cieslik A, Chalermchikit T, Aidara-Kane A, Ellis A, Angulo FJ, Wegener HC, World Health Organization Global Salm-Surv (2006) Web-based surveillance and global Salmonella distribution, 2000–2002. Emerg Infect Dis 12:381–388. https://doi.org/10.3201/eid1205.050854
Jackson BR, Griffin PM, Cole D, Walsh KA, Chai SJ (2013) Outbreak-associated Salmonella enterica serotypes and food commodities. Emerg Infect Dis 19:1239–1244. https://doi.org/10.3201/eid1908.121511
Centers for Disease Control and Prevention (2017) Surveillance for Foodborne Disease Outbreaks, United States, 2017: Annual Report. Centers for Disease Control and Prevention. https://www.cdc.gov/fdoss/annual-reports/2017-report-highlights.html. Accessed 13 Feb 2021
Inns T, Ashton PM, Herrera-Leon S, Lighthill J, Foulkes S, Jombart T, Rehman Y, Fox A, Dallman T, de Pinna E, Browning L, Coia JE, Edeghere O, Vivancos R (2017) Prospective use of whole genome sequencing (WGS) detected a multi-country outbreak of Salmonella Enteritidis. Epidemiol Infect 145:289–298. https://doi.org/10.1017/S0950268816001941
Tavechio AT, Fernandes AS, Neves BC, Dias AMG, Irino K (1996) Changing patterns of Salmonella serovars: increase of Salmonella Enteritidis in São Paulo, Brazil. Rev Inst Med trop S Paulo 38:315–322. https://doi.org/10.1590/s0036-46651996000500001
Silva EN, Duarte A (2002) Salmonella Enteritidis em Aves: Retrospectiva no Brasil. Rev Bras Cienc Avic 4:85–100. https://doi.org/10.1590/S1516-635X2002000200001
Borges KA, Furian TQ, de Souza SN, Tondo EC, Streck AF, Salle CT, de Souza Moraes HL, do Nascimento VP (2017) Spread of a major clone of Salmonella enterica serotype Enteritidis in poultry and in salmonellosis outbreaks in Southern Brazil. J Food Prot 80:158–163. https://doi.org/10.4315/0362-028X.JFP-16-299
Ritter AC, Tondo EC, Siqueira FM, Soggiu A, Varela APM, Mayer FQ, Brandelli A (2019) Genome analysis reveals insights into high-resistance and virulence of Salmonella Enteritidis involved in foodborne outbreaks. Int J Food Microbiol 306:108269. https://doi.org/10.1016/j.ijfoodmicro.2019.108269
Geimba MP, Tondo EC, Brandelli A (2005) Antimicrobial resistance in Salmonella Enteritidis isolated from foods involved in human foodborne outbreaks that occurred in the south of Brasil, 1999–2000. J Food Safety 25:173–182. https://doi.org/10.1111/j.1745-4565.2005.00573.x
Ritter AC, Bacciu D, Santi L, Da Silva WOB, Vainstein MH, Rubino S, Tondo EC (2012) Investigation of rpoS and dps genes in sodium hypochlorite resistance of Salmonella Enteritidis SE86 isolated from foodborne illness outbreaks in southern Brazil. J Food Prot 75:437–442. https://doi.org/10.4315/0362-028X.JFP-11-286
Ritter AC, Bacciu D, Santi L, Rubino S, Uzzau S, Tondo EC (2014) Expression of ompR gene in the acid adaptation and thermal resistance of Salmonella Enteritidis SE86. J Infect Dev Ctries 8:474–479. https://doi.org/10.3855/jidc.3584
Guazelli S (1994) Portaria nº 193 de 19 de setembro de 1994. Ministério da Agricultura, Pecuária e Abastecimento. https://www.emdagro.se.gov.br/wp-content/uploads/2020/06/PORTARIA-193-DE-19-DE-SETEMBRO-DE-1994.pdf. Accessed 13 Feb 2021
Bergmann A (2009) Portaria nº 78 de 30 de janeiro de 2009. Diário Oficial. https://www.cevs.rs.gov.br/upload/arquivos/201612/26090340-portaria-ses-rs-nu-78-de-2009.pdf. Accessed 13 Feb 2021
Gormley FJ, Little CL, Rawal N, Gillespie IA, Lebaigue S, Adak GK (2011) A 17-year review of foodborne outbreaks: describing the continuing decline in England and Wales (1992–2008). Epidemiol Infect 139:688–699. https://doi.org/10.1017/S0950268810001858
Vaz CS, Streck AF, Michael GB, Marks FS, Rodrigues DP, Dos Reis EM, Cardoso MR, Canal CW (2010) Antimicrobial resistance and subtyping of Salmonella enterica subspecies enterica serovar Enteritidis isolated from human outbreaks and poultry in southern Brazil. Poult Sci 89:1530–1536. https://doi.org/10.3382/ps.2009-00453
Van Boeckel TP, Pires J, Silvester R, Zhao C, Song J, Criscuolo NG, Gilbert M, Bonhoeffer S, Laxminarayan R (2019) Global trends in antimicrobial resistance in animals in low- and middle-income countries. Science 365:eaaw1944. https://doi.org/10.1126/science.aaw1944
Acknowledgements
We are grateful to Central Laboratory of Rio Grande do Sul (LACEN/RS. Laboratório Central do Estado do Rio Grande do Sul) for the assignment of isolates and to FDA GenomeTrakr network for support through the collaborative research agreement U18 FD006229 and the Wadsworth Center Advanced Genomic Technologies Cluster for sequencing 52 isolates.
Funding
The authors would like to thank Simbios Biotecnologia for the financial support. This study was financed in part by the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior—Brasil (CAPES)—Finance Code 001.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Responsible Editor: Cristiano Gallina Moreira
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Mascitti, A.K., Kipper, D., dos Reis, R.O. et al. Retrospective whole-genome comparison of Salmonella enterica serovar Enteritidis from foodborne outbreaks in Southern Brazil. Braz J Microbiol 52, 1523–1533 (2021). https://doi.org/10.1007/s42770-021-00508-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s42770-021-00508-0