Abstract
Mycoplasma hyopneumoniae is the etiologic agent of porcine enzootic pneumonia, responsible for major production losses worldwide. The bacteria have a limited metabolism and need to obtain molecules from the growth environment, which causes multiple difficulties for in vitro culture. These limitations have a negative influence on the ability to carry out research for the development of the rational use of antimicrobials and vaccines. The objective of this investigation was to evaluate the genetic profile and in vitro susceptibility of field isolates of M. hyopneumoniae to different antimicrobials. All 16 isolates obtained from the samples presented 100% of identity in the partial sequence of 16S rRNA gene when compared to M. hyopneumoniae. A dendrogram was created using the PCR results of the genes related to pathogenicity, and the isolates were distributed into four clusters, suggesting genetic variability among four different isolates circulating on the same farm. The minimum inhibitory concentration of the isolates was higher for the antimicrobials tylosin (< 0.001–16 mg/L) and spiramycin (< 0.001–16 mg/L) than for enrofloxacin (< 0.001–0.125 mg/L) and tiamulin (< 0.001–0.125 mg/L). Our results demonstrate the genetic variability among M. hyopneumoniae isolates from pigs of the same farm, with differences in their susceptibility to antimicrobial agents.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Brazil, as a substantial producer of pork meat, is the fourth largest producer and exporter of this product in the world [1] and has highly skilled pig breeding and varied production systems. The Brazilian pork industry consists of small independent producers, regional companies, or vertically integrated production complexes that cater to internal and external markets [2].
Respiratory diseases are the most important health problems associated with pig production. Enzootic pneumonia is a highly contagious chronic respiratory disease of pigs, caused by Mycoplasma hyopneumoniae. This disease is characterized by dry cough, delayed growth, poor feed conversion, and a predisposition to other pulmonary infections [3, 4]. The clinical manifestations associated with the agent have been frequent and common in the main producing regions of the country, which lead to a decrease in productivity and significant economic losses [5].
M. hyopneumoniae has few biosynthetic pathways. The lack of biosynthetic pathways implies that the bacteria need to obtain amino acids, purines, pyrimidines, and components of the cell membrane from the growth environment. These needs show a complex nutritional requirement and dependence on the host. These characteristics explain the great difficulty of in vitro M. hyopneumoniae culture [6, 7]. Due to the fastidious nature of M. hyopneumoniae, only a few laboratories are capable of performing isolation and susceptibility tests on antimicrobial molecules [7]. Even international reference guides, such as the Clinical and Laboratory Standards Institute (CLSI), which sets standards for antimicrobial susceptibility testing, do not provide information on Mycoplasma species of veterinary importance, making it difficult to evaluate results and leading to a lack of consensus among researchers.
Therefore, to be able to evaluate the antimicrobial susceptibility of field isolates of M. hyopneumoniae, the ability to monitor antimicrobial resistance and implement more efficient medication programs in Brazil is essential. This work is the first study carried out with M. hyopneumoniae Brazilian field isolates and aims to understand the genetic profile and evaluate in vitro susceptibility to the main classes of antimicrobials.
Materials and methods
Samples and M. hyopneumoniae isolation
Lung samples from pigs with lesions suggestive of enzootic pneumonia were collected in November 2017, from a slaughterhouse located at Vale do Piranga in the region of Zona da Mata, Minas Gerais State, Brazil, which is an important region of swine production. The samples were from the same batch and farm.
The affected lung fragments were sectioned into fragments of ± 10 cm2, macerated, and added to a tube containing liquid medium; the medium used was Friis 1x and 2.8 [8]. After membrane filtration (porosity: 0.45 μm) and serial dilutions of the contents, the tubes were kept in an incubator at 37 °C and observed daily for 30 days. Color changes of the liquid medium evidenced bacterial growth. The samples were submitted to DNA extraction and PCR for confirmation, and the positive samples were maintained at− 80 °C.
PCR and DNA sequencing
The samples were submitted to PCR to confirm the presence of M. hyopneumoniae. An additional PCR was used for the detection of M. hyorhinis and M. flocculare, which present faster growth in Friis medium and could inhibit the growth of M. hyopneumoniae [9]. DNA extraction from the samples was performed with phenol-chloroform [10], and PCR was performed using primers to the 16S ribosomal RNA gene of M. hyopneumoniae that amplify a partial sequence between regions V2 and V4 [11]. For M. hyorhinis PCR, we used the protocol described by Stakenborg et al. [12], and the protocol we used for M. flocculare PCR was based on Stemke [13].
All reactions were performed in a thermocycler and composed of 12.5 μL of Go Taq Green Master Mix 2x PCR kit (Promega Corporation). PCR products from each sample were purified using 0.8% agarose gel with the “GFX™ PCR DNA and Gel Band Purification” kit and sent for sequencing at Myleus Biotechnology. The samples were sequenced by capillary electrophoresis in the ABI3730 apparatus using POP7 and BigDye v3.1 polymer. The generated sequences were aligned and edited using CLC Main Workbench 6.7.1 (CLC Bio), then compared to sequences already deposited in GenBank using BLAST software.
Mycoplasma hyopneumoniae genetic profile
The genetic profile of the isolated samples was applied to PCR, directed to the M. hyopneumoniae genes involved with mechanisms of virulence or immune response. The primers used for PCR assays are described in Table A.1 [14, 15].
The results obtained in the PCR were analyzed by BioNumerics (version 6.6.11-Applied Maths NV, Sint-Martens-Latem, Belgium), with the following similarity parameters: Dice correlation, with a cutoff value of 100% similarity and applying the unweighted paired group method with the arithmetic mean (UPGMA).
Antimicrobial susceptibility testing
The antimicrobial susceptibility test was performed using the microbroth dilution method to determine the minimum inhibitory concentration (MIC) [16]. Antimicrobials belonging to the three most commonly used classes to control infection in vivo, and that were available for delivery in the Brazilian market, were selected. Therefore, the following antimicrobials were chosen: tylosin and spiramycin (macrolides), tiamulin (pleuromutilins), and enrofloxacin (fluoroquinolones), all of which originated from Sigma Aldrich, Brazil.
The isolates of M. hyopneumoniae had a count between 103 and 105 CCU/mL, assessed using the microbroth dilution method [17]. Stock solutions were prepared in sterile distilled water and stored at− 20 °C. Sixteen dilutions of the antibiotics were performed, ranging from 0.001 to 64 mg/L. The microbroth dilution test was performed using a 96-well plate. Previously, 100 μL(103–105 CCU/mL) of the culture was added to the plate and incubated for at least 1 h. All plates had a positive control (culture without antibiotic) and negative control (modified Friis medium). After 1 h of incubation, 100 μL of the antibiotic at each concentration (0.001–64 mg/L) was added to the wells. The plates were then sealed and incubated at 37 °C for 21 days. The plates were monitored daily for any color change in the wells. The MIC was established as the lowest antibiotic concentration at which there was no color change in the medium, characterized by no bacterial growth.
Results
Isolation and Mycoplasma hyopneumoniae culture
Sixteen isolates of M. hyopneumoniae were obtained. After three days, acidification of the culture was observed in some samples, characterizing bacterial grow thin of the Friis-modified medium. In the solid culture medium, diverse types of colonies and growth times were observed under a stereoscopic microscope. The morphology of the M. hyopneumoniae colonies varied from circular to oval and irregular. Moreover, it was small to large, smooth to rough, pale to stained, and single or in combination (Fig. 1).
Identification and partial sequencing
PCR, which amplified a 649-base pair fragment of 16S rRNA gene, was performed to identify the isolates. All samples were positive for M. hyopneumoniae, and only one sample was positive for M. hyopneumoniae and M. flocculare. All isolates for M. hyopneumoniae were purified in solid medium. After the second inoculation in a solid medium, there was no more contamination for M. flocculare.
The results of the M. hyopneumoniae16S partial gene alignment showed that the samples had 100% identity, compared to other isolates deposited into GenBank.
Genetic profile of Mycoplasma hyopneumoniae isolates
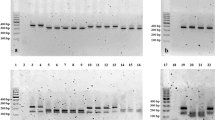
The presence or absence of genes involved with pathogenicity was evaluated. The genes mhp0461, mhp0272, mhp0487, mhp0107, mhp0660, mhp372, mhp199, mhp0443, p42, p97, and p46 were present in 100% of the isolates. To NrdF, p95, mnuA, and mhp418 genes, the positivity rate was 93%, and for p37, it was 81.25%. Figure 2 shows that the isolate named “O” did not have the mhp418, p37, mnuA, and NrdF genes. There was an absence of the gene that encodes P37 protein in isolates “N” and “P”. Only isolate “I” was negative for the gene encoding the P95 lipoprotein. In the evaluation of the gene distribution among the isolates (Fig. 2a), the isolates were classified into four different genogroups (Fig. 2b). The isolate “O” had the greatest genetic distance, compared to the other isolates, and it was negative for three of the 16 genes tested.
Minimal inhibitory concentration
The values for the antimicrobial susceptibility of M. hyopneumoniae are presented as MICrange, MIC50, and MIC90. Table 1 presents the comparative results between this study and other research. For tylosin and spiramycin, a similar range of MIC was found, with values < 0.001 to 16 mg/L. The enrofloxacin and tiamulin MIC values ranged from < 0.001 to 0.125 mg/L. Regarding the MIC50/MIC90 ratios, tylosin showed values of 0.125/16 mg/L, whereas spiramycin had values of 0.5/16 mg/L. Enrofloxacin and tiamulin had a proportion of 0.003/0.125 mg/L and < 0.001/0.125 mg/L, respectively.
The analyses of the MIC range values (Table 1) showed a wide range of distribution for all tested antimicrobials, mainly for tylosin and spiramycin; antimicrobial susceptibility values for 90% of the isolates were the highest values for these two antimicrobials. For enrofloxacin, a low MIC50value was obtained. The lowest inhibitory concentration for 50 and 90% of the isolates was observed for tiamulin.
Discussion
This work characterized 16 isolates of M. hyopneumoniae from lung samples with lesions suggestive of enzootic pneumonia from a single pig farm. The medium for culture, developed by Cook et al. [8], proved to be efficient for the growth of M. hyopneumoniae. M. hyopneumoniae isolates were genetically characterized, and the antimicrobial resistance was evaluated.
The partial sequence of 16S ribosomal RNA gene analyses showed 100% identity among the J, 232, 7448, 7442, and 168 strains. After alignment and comparison with other isolates deposited into GenBank, it could be observed that there was no variation. Therefore, the sequences are highly conserved for the 16S gene in M. hyopneumoniae isolates. Ribosomal RNA (rRNA) genes have been used worldwide to establish phylogeny because they are present in all cells and maintain high identity in conserved regions [18].
Figure 2 shows the isolates that were grouped into four clusters based on the presence or absence of genes encoding proteins involved with the pathogenicity of the bacterium. Two isolates did not have the gene that encodes P37 protein. One isolate did not have the gene p95 lipoprotein-related protein. The isolate called “O” did not have the mhp418, p37, mnuA, and NrdF genes. Results indicate the presence of at least four different M. hyopneumoniae isolates within the same farm. Other studies have also shown that different strains of M. hyopneumoniae can circulate on the same farm [19, 20].
P37 protein is characterized as an ABC transporter or ATP-binding cassette, one of the major carrier families. It is located on the plasma membrane and binds and hydrolyzes adenosine triphosphate (ATP), providing energy for nutrient absorption. ABC transporters have already demonstrated immunogenic properties stimulating specific immune responses, and they may be related to pathogenesis in some species of mycoplasma [21]. It is believed that most lipoproteins, as well as P95, are exposed to the extracellular environment and can perform various functions, such as adhesion, signal transduction, and nutrient transport [22].
The mnuA nuclease is considered a surface virulence factor. Virginio et al. [23] demonstrated that mnuA induced a strong humoral immune response when IgG was tested in the murine model; it was shown to be a strong immunogenic antigen as it significantly stimulated high levels of interferon-gamma and increased the amount of interleukin-10. When evaluating the antigenicity of proteins secreted by M. hyopneumoniae, mhp418 was recognized in serum from swine that were naturally infected by the agent [24]. The obtained results, with the presence of isolates from the same batch of animals belonging to four different genetic groups, demonstrated the possible genetic diversity of M. hyopneumoniae strains within the same batch, farm, city, and region, as has been also observed in other genetic diversity studies [20, 25].
Antimicrobial resistance tests were carried out under routine pig management. The use of antibiotics, despite all its peculiarities and resistance reports, continues to be of great importance since vaccination alone is not able to inhibit the colonization of M. hyopneumoniae in the respiratory tract. Therefore, the treatment of infected herds with antibiotics is frequently used as an alternative or in a complementary manner to vaccines, to improve clinical signs and pig performance [26, 27]. Regarding the evaluation of antimicrobial susceptibility, although the M. hyopneumoniae isolates used in this study came from the same farm and may not represent the population of the bacteria in Brazil, our study performed, for the first time, the evaluation of antimicrobial susceptibility using field samples.
We compared the minimum inhibitory concentrations of antimicrobials that were obtained in this study with several authors (Table 1) because the reference guide (CLSI) does not list cutoff values for M. hyopneumoniae. Tavío et al. [27] found a MICrange for tylosin and spiramycin in isolates from Spain that was consistent with the values of < 0.001 to 16 mg/L, different from that observed by Klein et al. [16], who obtained much smaller values. For enrofloxacin and tiamulin, Klein et al. [16] found MIC50 values of 0.0031 mg/L and 0.0016 mg/L, respectively, with concentrations similar to those found in our work.
The rate of change for the MIC obtained by the cited authors [16, 27, 28] was generally 0.008–25 mg/L for enrofloxacin and 0.002–0.78 mg/L for tiamulin. The present study demonstrated rates below < 0.001–0.125 mg/L for both antimicrobials and the lowest inhibitory concentration for 90% of the isolates for enrofloxacin (Table 1). The MIC90 values for tiamulin were similar to those obtained in other studies [27,28,29].
A recent study showed the susceptibility of field isolates of M. hyopneumoniae from Central Europe, and, in comparison with the results obtained from our work, the MIC range for tylosin was even higher, with a maximum limit of 64 μg/mL, than that obtained in this study, which was 16 mg/L [26]. A large divergence, in comparison with these results, was found in a 1993 study in which 25 isolates were evaluated for susceptibility to tylosin: 100% of the isolates had MIC values ≤ 0.125, with the exception of strain 232, with a MIC of 0.25 μg/mL [30].
Analyzing these results and those obtained in our study, there is a decrease in the susceptibility of M. hyopneumoniae over time, with the need to increase concentrations to inhibit the growth of M. hyopneumoniae, demonstrating resistance; this occurrence is more pronounced for the macrolides (tylosin and spiramycin), followed by the fluoroquinolones (enrofloxacin). MICs did not exceed 0.3 μg/mL for pleuromutilins (tiamulin), which have been shown to have a better effect in vitro, with low inhibitory concentrations, corroborating with the results obtained by other authors [7, 16, 27, 28].
Sensitive, resistant, or intermediate designations are common to almost all laboratory-testing methods and are distinguishable using breakpoints of antibiotic concentrations. However, these factors have not been determined for mycoplasma infections, owing to the variety of hosts that can cause disease and because there is little available pharmacological data [17].
The highest level of inhibitory concentrations of the major veterinary Mycoplasma species was observed for macrolides. Although resistant strains have been described for fluoroquinolones, most strains remain susceptible to this family of antibiotics. Pleuromutilins are the most effective antibiotics in vitro [7]. Fluoroquinolones are antimicrobial agents that are potentially active against M. hyopneumoniae through the inhibition of the enzyme DNA gyrase and topoisomerase IV [26]. In the present study, a low MIC50 value was recorded for enrofloxacin, similar to other results [16, 27, 28]; however, this was different from the level found by Thongkamkoon et al. [29], who obtained MIC50 values above 1.56 μg/mL. According to the results obtained by Felde et al. [26], and in agreement with the results of this work, pleuromutilins remain the most effective antimicrobial against M. hyopneumoniae because they have a low inhibitory concentration in vitro.
It has already been observed that strains of the same herd can show different DNA sequences. Based on genotyping analyses, Felde et al. [26] showed different susceptibility profiles and genetic alterations correlated with this antimicrobial susceptibility. The results of this study were consistent with those of Felde et al. [26]. The isolates that presented different genetic profiles showed differences in their susceptibility to the antimicrobials tylosin, spiramycin, enrofloxacin, and tiamulin.
MIC values are not necessarily correlated with antimicrobial efficacy in vivo, and the interpretation of MIC distributions is difficult because Mycoplasma species with veterinary relevance do not have official clinical breakpoints. In addition, strains with different susceptibilities to antibiotics may coexist within a herd [26]. However, the obtained results can help veterinarians make decisions about which antimicrobial agent to choose, thus performing a more targeted treatment. It is important to note that, unlike most studies that are carried out with classical strains that were isolated decades ago, this study evaluated susceptibility in recently obtained isolates from the lungs with lesions of enzootic pneumonia.
Based on the results of this study, high minimum inhibitory concentrations were found for some isolates in relation to the antimicrobials tylosin and spiramycin. Authorities from various countries, such as the World Health Organization (WHO) and the Food and Agriculture Organization of the United Nations (FAO), have been working to reduce the use of these molecules, mainly their use as additives to improve performance. In this sense, the Ministry of Agriculture, Livestock, and Supply (MAPA) from Brazil has also taken some measures to reduce the use of antimicrobials, such as IN 14/2016 and Ordinance 171/2018, which created more rigid criteria for the handling and use of veterinary drugs in production animal feed, and Order 171/2018, which outlines the intention to ban the use of antimicrobials as additives for the purpose of performance enhancement.
Due to different antimicrobial handling and use practices, resistance frequencies can vary considerably from one country to another but also within the same country with isolates of different origins and farms from the same country’s region. About the protocol of antimicrobial treatment performed on the farm, we did not had access to this information because the samples were not collected from the farm but from the slaughterhouse certified by the Brazilian official inspection service (MAPA). However, if we had access to the antimicrobial protocol used by farm, it could improve the results’ discussion herein presented.
Conclusion
The Mycoplasma hyopneumoniae field strains isolated in our study showed variations in the NrdF, p95, mnuA, mhp418, p42, and p37 genes, which could be possibly involved in the pathogenesis of the bacteria, giving rise to at least four different genetic groups. The MIC determination assay showed lower inhibitory concentrations for enrofloxacin and tiamulin than for spiramycin and tylosin. There was a wide range of variation among isolates, showing the importance of constant monitoring to avoid resistance to the antimicrobials used in the field.
References
ABPA AB de PA. Relatório Anual 2018.; 2018.
Guimarães D, Amaral G, Maia G, Lemos M, Ito M, Custodio S (2017) Suinocultura: estrutura da cadeia produtiva, panorama do setor no brasil e no mundo e o apoio do BNDES. Agroindústria/ BNDES Setorial 45:85–136
Hillen S, von Berg S, Köhler K, Reinacher M, Willems H, Reiner G (2014) Occurrence and severity of lung lesions in slaughter pigs vaccinated against Mycoplasma hyopneumoniae with different strategies. Prev Vet Med 113(4):580–588. https://doi.org/10.1016/j.prevetmed.2013.12.012
Kuhnert P, Overesch G (2014) Molecular epidemiology of Mycoplasma hyopneumoniae from outbreaks of enzootic pneumonia in domestic pig and the role of wild boar. Vet Microbiol 174(1-2):261–266. https://doi.org/10.1016/j.vetmic.2014.08.022
Yamaguti M (2009) Isolamento de micoplasma de suínos com problemas respiratórios e tipificação dos isolados pela PFGE e seqüenciamento do gene 16S rRNA. Tese, Univ Fed São Paulo, São Paulo- SP, Bras
Simionatto S, Marchioro SB, Maes D, Dellagostin OA (2013) Mycoplasma hyopneumoniae: from disease to vaccine development. Vet Microbiol 165(3-4):234–242. https://doi.org/10.1016/j.vetmic.2013.04.019
Gautier-Bouchardon AV (2018) Antimicrobial Resistance in Mycoplasma spp. Microbiol Spectr 6:1–21. https://doi.org/10.1128/microbiolspec.ARBA-0030-2018
Cook BS, Beddow JG, Manso-Silván L, Maglennon GA, Rycroft AN (2016) Selective medium for culture of Mycoplasma hyopneumoniae. Vet Microbiol 195:158–164. https://doi.org/10.1016/j.vetmic.2016.09.022
Kobisch M, Friis NF (1996) Swine mycoplasmoses. Rev Sci Tech 15(4):1569–1605. https://doi.org/10.1021/pr201115v
Sambrook J, Russel DW (2001) Molecular Cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, New York
Mattsson JG, Bergström K, Wallgren P, Johansson KE, Wallgren PER (1995) Detection of Mycoplasma hyopneumoniae in nose swabs from pigs by in vitro amplification of the 16S rRNA gene;33(4)
Stakenborg T, Vicca J, Butaye P, Imberechts H, Peeters J, de Kruif A, Haesebrouck F, Maes D (2006) A multiplex PCR to identify porcine mycoplasmas present in broth cultures. Vet Res Commun 30(3):239–247. https://doi.org/10.1007/s11259-006-3226-3
Stemke GW (1997) Gene amplification (PCR) to detect and differentiate mycoplasmas in porcine mycoplasmal pneumonia. Lett Appl Microbiol 25(5):327–330. https://doi.org/10.1046/j.1472-765X.1997.00243.x
Assao VS (2017) Detecção e variabilidade genética de mycoplasma hyopneumoniae em amostras de pulmão do estado de Minas Gerais, Brasil. Diss mestrado, Univ Fed Viçosa, Viçosa- MG, Bras. http://www.albayan.ae.
Moreira TS (2016) Perfil genético e epidemiologia molecular de Mycoplasma hyopneumoniae no estado de Minas Gerais, Brasil. Diss mestrado, Univ Fed Viçosa, Viçosa- MG, Bras
Klein U, de Jong A, Moyaert H et al (2017) Antimicrobial susceptibility monitoring of Mycoplasma hyopneumoniae and Mycoplasma bovis isolated in Europe. Vet Microbiol 204(February):188–193. https://doi.org/10.1016/j.vetmic.2017.04.012
Hannan PCT (2000) Guidelines and recommendations for antimicrobial minimum inhibitory concentration (MIC) testing against veterinary mycoplasma species. Vet Res 31(4):373–395. https://doi.org/10.1051/vetres:2000100
Lim K, Furuta Y, Kobayashi I (2012) Large variations in bacterial ribosomal RNA genes. Mol Biol Evol 29(10):2937–2948. https://doi.org/10.1093/molbev/mss101
Dubosson CR, Conzelmann C, Miserez R, Boerlin P, Frey J, Zimmermann W, Häni H, Kuhnert P (2004) Development of two real-time PCR assays for the detection of Mycoplasma hyopneumoniae in clinical samples. Vet Microbiol 102(1-2):55–65. https://doi.org/10.1016/j.vetmic.2004.05.007
Stakenborg T, Vicca J, Butaye P, Maes D, Peeters J, de Kruif A, Haesebrouck F (2005) The diversity of Mycoplasma hyopneumoniae within and between herds using pulsed-field gel electrophoresis. Vet Microbiol 109(1-2):29–36. https://doi.org/10.1016/j.vetmic.2005.05.005
Nicolás MF, Barcellos FG, Hess PN, Hungria M (2007) ABC transporters in Mycoplasma hyopneumoniae and Mycoplasma synoviae: insights into evolution and pathogenicity. Genet Mol Biol 30(SUPPL. 1):202–211. https://doi.org/10.1002/hrm.20267
Reolon LA, Martello CL, Schrank IS, Ferreira HB (2014) Survey of surface proteins from the pathogenic mycoplasma hyopneumoniae strain 7448 using a biotin cell surface labeling approach. PLoS One;9(11). doi:https://doi.org/10.1371/journal.pone.0112596.
Virginio VG, Gonchoroski T, Paes JA, Schuck DC, Zaha A, Ferreira HB (2014) Immune responses elicited by Mycoplasma hyopneumoniae recombinant antigens and DNA constructs with potential for use in vaccination against porcine enzootic pneumonia. Vaccine. 32(44):5832–5838. https://doi.org/10.1016/j.vaccine.2014.08.008
Galli V, Simionatto S, Marchioro SB, Klabunde GHF, Conceição FR, Dellagostin OA (2013) Recombinant secreted antigens from Mycoplasma hyopneumoniae delivered as a cocktail vaccine enhance the immune response of mice. Clin Vaccine Immunol 20(9):1370–1376. https://doi.org/10.1128/CVI.00140-13
Charlebois A, Marois-Créhan C, Hélie P, Gagnon CA, Gottschalk M, Archambault M (2014) Genetic diversity of mycoplasma hyopneumoniae isolates of abattoir pigs. Vet Microbiol 168(2-4):348–356. https://doi.org/10.1016/j.vetmic.2013.11.006
Felde O, Kreizinger Z, Sulyok KM, et al. (2018) Antibiotic susceptibility testing of Mycoplasma hyopneumoniae field isolates from Central Europe for fifteen antibiotics by microbroth dilution method. PLoS One.:1-13. doi:https://doi.org/10.1371/journal.pone.0209030.
Tavío MM, Poveda C, Assunção P, Ramírez AS, Poveda JB (2014) In vitro activity of tylvalosin against Spanish field strains of Mycoplasma hyopneumoniae. Vet Rec 175(21):539. https://doi.org/10.1136/vr.102458
Vicca J, Stakenborg T, Maes D, Butaye P, Peeters J, de Kruif A, Haesebrouck F (2004) In vitro susceptibilities of Mycoplasma hyopneumoniae field isolates. Antimicrob Agents Chemother 48(11):4470–4472. https://doi.org/10.1128/AAC.48.11.4470-4472.2004
Thongkamkoon P, Narongsak W, Kobayashi H, Pathanasophon P, Kishima M, Yamamoto K (2013) In vitro susceptibility of Mycoplasma hyopneumoniae field isolates and occurrence of fluoroquinolone, macrolides and lincomycin resistance. J Vet Med Sci 75(8):1067–1070. https://doi.org/10.1292/jvms.12-0520
Tanner AC, Erickson BZ, Ross RF (1993) Adaptation of the Sensititre® broth microdilution technique to antimicrobial susceptibility testing of Mycoplasma hyopneumoniae. Vet Microbiol 36(3-4):301–306. https://doi.org/10.1016/0378-1135(93)90096-P
Acknowledgments
We would like to thank EMBRAPA, who gently ceded the Mycoplasma hyopneumoniae strain J and Dr. Matheus Gandra Campos for assistance with the statistical analyses. We thank Thiago Augusto Teles de Souza and Wendel Mayer Rezende for the technical support in the analyses.
Availability of data and materials
There is supplementary material.
Funding
We thank the Brazilian Government Agencies. This research was funded by the Coordination for the Improvement of Higher Education Personnel – CAPES (Ph.D. fellowship), the National Council for Scientific and Technological Development (Grant: CNPq304727/2016-4) and the Foundation for Research Support of the State of Minas Gerais (Grants: FAPEMIG-PPM-006618-17 and FAPEMIG-APQ-01327-14).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare that they have no competing interests.
Ethical approval
Project approved by CEUA-UFV under process n°78/2016.
Statement of informed consent
All authors read and agreed to the final manuscript.
Additional information
Responsible Editor: Miliane Moreira Soares de Souza.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
ESM 1
(DOCX 14 kb)
Rights and permissions
About this article
Cite this article
Gonzaga, N.F., de Souza, L.F.L., Santos, M.R. et al. Antimicrobial susceptibility and genetic profile of Mycoplasma hyopneumoniae isolates from Brazil. Braz J Microbiol 51, 377–384 (2020). https://doi.org/10.1007/s42770-019-00185-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s42770-019-00185-0