Abstract
Sandflies (Diptera: Psychodidae) are medically important insects prevalent in tropical and temperate regions of the world. About 30 species of these flies have been recorded from the Western Ghats region of Kerala. While carrying out epidemiological investigations on Leishmaniasis in this region, we found two species of field-collected sandflies viz., Phlebotomus argentipes and Sergentomyia monticola infected with Tylenchid nematode parasites. The infection rates were 0.89% and 0.9%, respectively. The parasite density of nematode juvenile stages was more than 1000 in all the specimens. Both males and females were found infected. We performed molecular characterization of these nematode parasites using multiple genetic markers, mitochondrial COI, and two different regions of 18S rDNA. The genetic analysis revealed that the nematode belonged to the genus Howardula of family Allantonematidae, one infesting P. argentipes (major vector species of Leishmania) and the other, S. monticola. This is the first report of natural infection with the entomo-parasitic nematode species, belonging to Allantonematidae among these sandfly species from India. Genetically related and unclassified species of nematodes belonging to this family had been reported elsewhere from termite species. As larval stages of sandflies develop in the organic matter of termite mounds, this finding may have significant implications on their bionomics and control.
Graphical Abstract

Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Phlebotomine sand flies are a group of insects that are of significant public health importance (Lane 1993). There are around 700 species of phlebotomine sand flies of which about 70 of them are considered important vectors that transmit diseases (Adler and Theodor 1957). Sand flies are able to transmit several viral, bacterial, and protozoal disease-causing organisms to man and other animals. Leishmaniasis is the most known disease transmitted by sandflies that can infect the human population, dogs, cats, horses, and other livestock (Schantz 2004; Travi et al. 2002). The leishmaniasis is parasitic diseases with a wide range of clinical symptoms: cutaneous, mucocutaneous, and visceral. Leishmaniasis currently affects about 350 million populations in 88 countries globally (World Health Organization 2021).
Sandflies are found mainly in tropical and subtropical Countries. However, some species are reported from temperate regions of the world also. In India distribution of sand flies has been reported in several areas of the northern region, the eastern coastal region, and the western coastal region (Srinivasan et al. 2015; Srinivasan and Subramanian 2015). Since the last decade, sporadic cases of cutaneous leishmaniasis (CL) and visceral leishmaniasis (VL) cases have been reported from the tribal belts of the Western Ghats region (Kumar et al. 2015; Saini et al. 2020). Around 19 species of sand flies were recorded from the Western Ghats region so far (Srinivasan et al. 2015; Srinivasan and Subramanian 2015). The study also reported that P. argentipes, the major vector of Leishmaniasis is the predominant species in this area (Srinivasan et al. 2016). The distribution and abundance of sand flies vary depending upon environmental factors such as rainfall, temperature humidity, and altitudes (Srinivasan et al. 2013).
The disease transmission potential of sand flies varies among different species. They can transmit bacterial diseases like bartonellosis in humans and dogs (Lannino et al. 2018). In addition, several viral diseases like Sandfly Fever Virus, Toscana virus, Rift Valley Fever Virus, Chagres virus, and Punta Toro virus were reported to be transmitted by sand flies (Azar and Nel 2003; World Health Organization 2021). Infections in sand flies with certain nematodes were also reported from different parts of the world (McConnell and Correa 1964; Killick-Kendrick et al. 1989; Warburg 1991; Poinar et al. 1993; Alves et al. 2020; Brilhante et al. 2020). The Nematodes (Phylum: Nematoda) are small slender worms that are about 5 to 100 µm thick, and 0.1 to 2.5 mm long (Weischer and Brown 2000). They are the most diverse pseudocoelomates which occur as parasites in animals and plants or as free-living forms in soil, freshwater, and marine environments. More than 28,000 species of nematodes have been reported globally (Hugot et al. 2001), of which over 16,000 are parasitic.
In India, there are only a few reports of nematode infestation in sand flies. Srinivasan et al. (1992) reported the presence of nematode in the natural population of Phlebotomus papatasi in Pondicherry for the first time in India. The study reported that the nematode belonged to the superfamily Tylenchoidea. Dinesh et al. (2013) reported the presence of a nematode in P. argentipes in Bihar State, and the species of nematode involved in these infections were not characterised further.
In the present study, we report the presence of insect parasitic nematodes of the genus Howardula belonging to the Family Allantonematidae (Order: Tylenchida) among the natural populations of different species of sand flies from the Western Ghats region of Kerala State for the first time. P. argentipes, the major vector of Leishmaniasis were also found infected with these entomo-pathogenic nematodes.
Material and methods
Study area: Routine surveillance of sand flies are being carried out in the foothills of the Western Ghats region since 2018. The study villages were selected in different districts of Kerala with the objective of the epidemiological investigations of Leishmaniasis in these emerging foci of transmission.
Collection of sand flies: Indoor resting collections were carried out in human dwellings during the morning hours using mechanical aspirators. Light traps were fixed in the cattle sheds from 6.00 pm to 6.00 am for collecting the sand flies. Collected sand flies were transferred to the ICMR-VCRC field laboratory at Kottayam. The collected sand flies were sorted Genus wise as well as sex-wise and identified up to species level using identification keys (Lewis 1978). Individual sand flies were kept at 70% alcohol (50 µl) for further processing.
Dissection of sand flies: The dissections were carried out under the binocular microscope. Before the dissection of female specimens their abdominal conditions were classified into un-fed (UF), full-fed (FF), semi-gravid (SG), and Gravid (G), following Sella’s method, described for mosquitoes (WHO 1975). While dissecting, we found that the gut of some sand flies was infected with nematode parasite species. All those sand flies infected with nematodes were processed separately. For morphological identification of the species, the head and last segment of the individual sand flies were separated and mounted in Hoyer’s medium. The legs of the sand flies were also kept separately in 70% alcohol for species confirmation by DNA barcode analysis (Kumar et al. 2012).
Identification of nematode: The nematodes isolated from the haemocoel of the sand flies were kept in 70% alcohol for molecular analysis. A portion along with the nematodes was mounted in a glass slide and kept for future reference. The nematodes were observed under a 40 × objective of the binocular microscope and the morphological characteristics were recorded.
Molecular characterization of nematodes and sand flies: DNA of the nematodes and sandflies were extracted using Gen Elute mammalian DNA mini prep kit (manufactured by Sigma Aldrich). The extracted nematode DNA was then amplified using specific primers (Holterman et al. 2006). Three different sets of primers were used; one which amplified the cytochrome C oxidase subunit 1 (COI) gene for identifying the family of nematode; COI F (5’-CCTACTATGATTGGTGGTTTTGGTAATTG-3’) & COI R (5’-GTAGCAGCAGTAAAATAAGCACG-3’) (Kanzaki and Futai 2002) and other two primer pairs targeting both fragments of small subunit ribosomal DNA (SSU rDNA) gene was used for confirmation of nematode species; 1096 F (5’-GGTAATTCTGGAGCTAATAC-3’) & 1912 R 5’-TTTACGGTCAGAACTAGGG-3’ (Holterman et al. 2006) and 1813 F (5’-CTGCGTGAGAGGTGAAAT-3’) & 2646 R (5’-GCTACCTTGTTACGACTTTT-3’) (Holterman et al. 2006). For mitochondrial COI gene amplification, the PCR cycling conditions were as follows: an initial denaturation step of 5 min at 95 °C followed by 38 cycles each of 95 °C for 50 s, 51 °C for 1 min, and 72 °C for 2 min, and a final extension step at 72 °C for 7 min.
For confirmation of nematode species, SSU rDNA was amplified. For this, the PCR reactions adopted was an initial denaturation step of 5 min at 94 °C, followed by 5 cycles each of 94 °C for 30 s, 45 °C for 30 s and 72 °C for 1 min and 35 cycles each of 94 °C for 30 s, 54 °C for 30 s and 72 °C for 1 min. This was followed by a final extension step at 72 °C for 7 min. The PCR products were purified and custom sequenced after lyophilisation. The amplified gene sequences were further analyzed and compared with the NCBI database results. The partial gene sequences of COI and SSU rDNA were deposited at GenBank.
Molecular phylogenetic analysis was carried out in MEGA 7.0 software. The phylogenetic tree was inferred by using the Maximum Likelihood method based on the Tamura-Nei model, along with similar sequences obtained from GenBank. In the COI gene, the analysis involved 14 nucleotide sequences which included 8 sequences for this study. In this analysis, 591 base pair consensus sequences were used. In the case of SSU rDNA 1096 F/ 1912R gene, out of 8 nucleotide sequences analysed and three were from this study. There was a total of 793 consensus sequences. For the 1813F / 2646R SSU rDNA sequences, 10 nucleotide sequences were considered of which three were from this study, and 793 consensus regions were considered.
Results
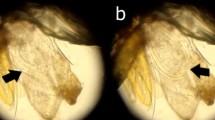
A total of 1193 sand fly species belonging to two genera; Phlebotomus and Sergentomyia were collected during the study period. The collection included three species of Phlebotomus and 11 species of Sergentomyia. Out of these, 9 (0.75%) were found infected with nematodes (Fig. 1A, B, and C). The species of sand flies infected were P. argentipes (n = 8) and S. monticola (n = 1). Though male and female sandflies were found infected, the latter were found to be more infected (78%). Two male sandflies which were found to be infected with nematode were P. argentipes and S. monticola species. All the infected females were P. argentipes (n = 7). We found nematode infection in 0.89% of the collected P. argentipes and 0.9% of S. monticola. Out of 7 females infected, two were unfed, one was fully engorged with blood and four were gravid (Table 1).
The length of the nematode ranged from 127 to 230 µm an average width of 0.75 to 1.25 µm. The anterior portion of nematode being sheath and tail portion end pointed tip. The visible morphological characters like stylet at the opening of the dorsal oesophageal duct and pharyngeal glands suggested that the nematode belonged to the order Tylenchidae (Fig. 1D).
Genetic analysis of COI and two different regions of SSU rDNA
The GenBank accession numbers generated in the study are ON063477-ON063484 (COI), ON65561-ON65566 (SSU rDNA). The phylogenetic analysis of the COI and SSU rDNA 1096/1912 gene showed that the species belongs to the order Tylenchida and family Allantonematidae (Fig. 2A, B). The 1813/2646 gene sequences of SSU rDNA showed that the species belong to the genus Howardula (NCBI Accession NO. AF519232) with a query coverage of 84% and genetic similarity of 98.31% (Fig. 2C). A unique insertion of 115 base pair corresponding to the 2284–2400 bp positions in the SSU rDNA region was recorded among our isolates, which was not noted in any of the GenBank submissions of these three genera. However, 292 bp (corresponding to 1991–2212 bp positions) of the sequences of our isolates exactly matched with GenBank accession no. AB663183 nucleotide sequences of Allantonematode species (reported from termites) with query coverage of 100% and 100% identity and therefore we infer that the nematode isolated belongs to Howardula sp. of the family Allantonematidae.
Discussion
In this study, we have observed that two sand fly species P. argentipes and S. monticola collected from the Western Ghats region of Kerala naturally infected with nematodes. Molecular analysis of the nematode confirmed that the entomoparasite belonged to genus Howardula of the family Allantonematidae and order Tylenchida.
Tylenchid nematodes are widely prevalent in plants and animals mainly maintain a parasitic relationship. The presence of tylenchid nematodes in sandflies were first reported in P. papatasi from Puducherry (Srinivasan et al. 1992). Later in 2013, Dinesh et al. also reported nematode infestation in P. argentipes in Bihar State (Dinesh et al. 2013). In the present study, we have carried out the molecular characterisation of the nematode up to the family level. Phylogenetic analysis of the nematode sequences showed it belonged to a new taxonomic clade (family: Allantonematidae). As far as our knowledge, this is the first-ever report of species characterisation of nematode infection in sand flies. Different studies have reported spirurid, filarid, tylenchid, and tetradonematid nematodes from the body cavities of phlebotomine sandflies (Killick-Kendrick et al. 1989; Poinar et al. 1993).
Allantonematidae species are mainly insect parasites found in beetles, flies, butterflies etc. Tylenchid nematodes occur widely in all habitats like soil, water, etc. These are the most important group of plant nematodes that attack all the plant parts (Siddiqui 2000). Tylenchida leaves their host body as juveniles or eggs to develop in the outside environment. The fertilized females seek out and penetrate the body of the host larva or pupa to establish themselves in the haemocoel (Siddiqui 2000).
In our study, we observed that both male and female sand flies were infected with nematodes. The nematodes were seen in the hemocoel of sandflies. In the Western Ghats area, sand flies are prevalent in different indoor and outdoor habitats. The female sandfly lays its eggs in tree barks, burrows, termite mounds, and cracks of houses (Warburg et al. 1991). The larvae of sandflies feed on moist organic matter. The prevailing climatic conditions in the study area are also favorable for the development of many entomopathogens (Warburg et al. 1991). Genetically related Allantonematide has been recorded from termites elsewhere (Kanzaki and Futai 2002). As the immature stages of sandflies are associated with termite mounds they would have been infested with nematodes during this stage of their life cycle.
There are only a few reports of tylenchid nematode infestations in sandfly worldwide (McConnell and Correa 1964; Warburg 1991; Srinivasan et al. 1992; Dinesh et al. 2013; Qing and Bert 2019). Though entomoparasitic nematodes of the order Tylenchida are known to control populations of various insect hosts (Bedding et al. 1974, 1993; Siddiqi 1986, 2000; Hurley et al. 2007), there are also reports that tylenchids are unsuitable for biological control due to their heteroxenous life cycle (Warburg 1991). Several studies reported the possibility of tylenchid nematode infestation in sand flies, this is the first record of sand flies harboring Allantonematide in India. Further studies are required to explore whether the entomoparasitic infestation by nematodes could be implemented as a biological control agent or not in the study area.
Availability of data and material
All the data submitted in this manuscript are original and available at the ICMR Vector Control Research Centre field station data bank.
References
Adler S, Theodor O (1957) Transmission of disease agents by phlebotomine sand flies. Annu Rev Entomol 2:203–226
Alves JRC, Nascimento do Carmo C, Menezes RC, Vilela ML, Santos-Mallet JRD, (2020) First report of nematodes in Lutzomyia edwardsi (Diptera: Psychodidae, Phlebotominae) in cave, in the municipality of Sumidouro. State of Rio of Janeiro, Brazil. https://doi.org/10.1101/2020.09.20.305151
Azar D, Nel A (2003) Fossil psychodoid flies and their relation to parasitic diseases. Mem Inst Oswaldo Cruz Rio De Janeiro 98(1):35–37
Bedding RA (1974) Five new species of Deladenus (Neotylenchidae), entomophagous–mycetophagous nematodes parasitic in siricid woodwasps. Nematologica 20:204–225
Bedding RA, Akhurst RJ, Kaya HK (1993) Nematodes and the biological control of insect pests. CSIRO Press, Melbourne, Australia, 178 pp
Brilhante AF, Lima de Albuquerque A, de Brito Cézar, Rocha A, Ayres CFJ, Henrique Paiva MHS, de Ávila MM, de Oliveira Cardoso C, Mauricio AL, Galati EAB (2020) First report of an Onchocercidae worm infecting Psychodopygus carrerai carrerai sandfy, a putative vector of Leishmania braziliensis in the Amazon. Sci Rep 10(1):15246–15255
Dinesh DS, Kumar V, Das P (2013) Infestation of Nematodes in Phlebotomus argentipes Annandale and Brunetti (Diptera: Psycodidae), Bihar, India. Global J Med Res Dis 13(6):7–9
Holterman M, van der Wurff A, van den Elsen S, van Megen H, Bongers T, Holovachov O, Bakker J, Helder J (2006) Phylum-wide analysis of SSU rDNA reveals deep phylogenetic relationships among nematodes and accelerated evolution toward crown clades. Mol Biol Evol 23(9):1792–1800
Hugot JP, Baujard P, Morand S (2001) Biodiversity in helminths and nematodes as a field of study: an overview. Nematology 3(3):199–208
Hurley BP, Slippers B, Wingfield MJ (2007) A comparison of control results for the alien invasive woodwasp, Sirex noctilio, in the southern hemisphere. Agric for Entomol 9:159–171
Kanzaki N, Futai K (2002) A PCR primer set for determination of phylogenetic relationships of Bursaphelenchus species within Xylophilus group. Nematology 4(1):35–44
Killick-Kendrick R, Killick-Kendrick M, Quala NA, Nawi RW, Ashford RW, Tang Y (1989) Preliminary observations of a tetradonematid nematode of phlebotomine sandflies of Afghanistan. Ann Parasitol Hum Comp 64:332–339
Kumar NP, Srinivasan R, Anish TS, Nandakumar G, Jambulingam P (2015) Cutaneous leishmaniasis caused by Leishmania donovani in the tribal population of the Agasthyamala Biosphere Reserve forest, Western Ghats, Kerala, India. J Med Microbiol 64:157–163
Kumar NP, Srinivasan R, Jambulingam P (2012) DNA barcoding for identification of sand flies (Diptera: Psychodidae) in India. Mol Ecol Resour 12(3):414–420
Lane RP (1993) Sand flies (Phlebotominae). In: Lane RP, Crosskey RW (eds) Medical insects and arachnids. Chapman and Hall, London, pp 78–119
Lannino F, Salucci S, Di Provvido A, Paolini A, Ruggieri E (2018) Bartonella infections in humans dogs and cats. Vet Ital 54(1):63–72
Lewis DJ (1978) The phlebotomine sandflies (Diptera: psychodidae) of the oriental region. Bull Br Mus (Natural History), London 1–343
McConnell E, Correa M (1964) Trypanosomes and other microorganisms from Panamanian Phlebotomus sand flies. J Parasitol 50:523–528
Poinar GO, Ferro C, Morales A, Tesh RB (1993) Anandarema phlebotophagan.gen; n.sp. (Allantonematoda: Tylenchida), a new nematode parasite of phlebotomine sand flies (Psychodidae: Diptera) with notes on experimental infections of these insects with parasitic rhabditoids Fundam. Appl Nematol 16:11–16
Qing S, Bert W (2019) Family Tylenchidae (Nematoda): an overview and perspectives. Org Divers Evol 19:391–408
Saini P, Kumar N, Ajithlal PM, Joji A, Rajesh KR, Reena KJ, Kumar A (2020) Visceral leishmaniasis caused by Leishmania donovani Zymodeme MON-37, Western Ghats, India. Emerg Infect Dis 26(8):1956–1958
Schantz PM (2004) Visceral leishmaniasis in dogs. Spinone Club of America. http://www.spinone.com/Health/Leishmaniasis/cdc_Schantz.htm
Siddiqi MR (1986) Tylenchida parasites of plants and insects, vol 645. Commonwealth Agricultural Bureaux, Farnham Royal, Slough, UK
Siddiqi MR (2000) Tylenchida: parasites of plants and insects, 2nd edn. CABI, Wallingford, UK
Srinivasan R, Jambulingam P, Kumar NP, Selvakumar M, Edwin B, Dilip Kumar T (2015) Temporal distribution and behaviour of sand flies (Diptera: Psychodidae) in a cutaneous leishmaniasis focus of the Kani Tribe settlements in the Western Ghats, India. Acta Trop 148:147–155
Srinivasan R, Jambulingam P, Vanamail P (2013) Sand fly (Diptera: Psychodidae) abundance in relation to environmental factors in parts of the coastal plains of southern India. J Med Entomol 50(4):758–763
Srinivasan R, Kumar NP, Jambulingam P (2016) Detection of natural infection of Leishmania donovani (Kinetoplastida: Trypanosomatidae) in Phlebotomus argentipes (Diptera: Psychodidae) from a forest ecosystem in the Western Ghats, India, endemic for cutaneous leishmaniasis. Acta Trop 156:95–99
Srinivasan R, Panicker KN, Dhanda V (1992) Occurrence of entomophilic nematode infestation among phlebotomid sandfly, Phlebotomies papatasi- a preliminary report. J Commun Dis 24(1):8–11
Srinivasan R, Subramanian S (2015) Sand flies diversity in association with human activities in the Kani tribe settlements of the Western Ghats, Thiruvananthapuram, Kerala, India. Mem Inst Oswaldo Cruz 110(4):174–180
Travi BL, Ferro C, Cadena H, Montoya-Lerma J, Adler GH (2002) Canine visceral leishmaniasis: dog infectivity to sand flies from non-endemic areas. Res Vet Sci 72(1):83–86
Warburg A (1991) Entomopathogens of phebotomine sand flies: laboratory experiments and natural infections. J Invert Pathol 58(2):189–202
Warburg A, Ostrovska K, Lawyer PG (1991) Pathogens of Phlebotomine sandflies: a review. Parasitologica 33:519–526
Weischer B, Brown DJ (2000) An introduction to nematodes: general nematology. A students textbook. Pensoft publishers 187 pp
World Health Organization (1975) Manual on practical entomology in malaria, part II, vol 87. WHO, Geneva, Switzerland, 947 pp
World Health Organization (2021) Leishmaniasis. Vector control. Available at www.who.int › leishmaniasis › vector-controls
Acknowledgements
We are grateful to the technical staff of ICMR Vector Control Research Centre Field Station at Kottayam for their technical help and to the Directorate of Health Services, Government of Kerala, India, for facilitating the survey.
Funding
This study was supported by the ICMR-Vector Control Research Centre intramural funding (Project grant no.: IM-1905).
Author information
Authors and Affiliations
Contributions
All Authors contributed to the study conception and design, conceived the ideas and designed the methodology. Prasanta Saini, P.M.Ajithlal, and Jessu Mathew performed the experiments. Prasanta Saini, Sonia T and N. Pradeep Kumar analysed the data and drafted the manuscript. Prasanta Saini, N. Pradeep Kumar, and Ashwani Kumar supervised the practical and theoretical works. All authors contributed to the preparation of the final manuscript and gave approval for publication.
Corresponding author
Ethics declarations
Ethics approval
The study was approved by ICMR-Vector Control Research Centre, Puducherry scientific advisory committee and no such ethics were involved in this study.
Consent to participate
Not applicable.
Consent for publication
The manuscript was approved by the research integrity unit of ICMR-Vector Control Research Centre, Puducherry. Publication related content will be provided to the journal upon acceptance of the manuscript.
Conflict of interest
The authors hereby declared that no conflict of interest is involved in this study.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Saini, P., PM, A., Mathew, J. et al. Genetic characterization of a new entomo-pathogenic nematode (Tylenchida: Allantonematidae) parasite in wild-caught sandflies (Diptera: Psychodidae) from Western Ghats, India. Int J Trop Insect Sci 43, 2145–2150 (2023). https://doi.org/10.1007/s42690-023-01124-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s42690-023-01124-1