Abstract
Papaya is economically important cultivated fruit crop grown in all tropical countries, having enormous nutritional values. Papaya Ring Spot Virus imposes a significant crop loss in terms of quality and quantity. To encounter the virus vector (Aphis gossypii), indiscriminate use of chemical pesticides creates severe environmental hazards whereas biological control is a perfect alternative to this problem. The objectives of our present study were isolation and characterization of indigenous fungi and their comparative analysis of entomopathogenic fungi against papaya aphid and finding its pathogenicity. Fungal isolates collected from natural sources were characterized and identified by the cultural and morphological study. Potential EPF genera were molecularly identified by PCR (ITS1-5.8S-ITS2) method. Entomopathogenic fungi were screened against A. gossypii for their pathogenecity by incised leaf disc method. LD50 (median lethal dose) and LT50 (median lethal time) were analyzed by regression analysis. Phylogenetic relationship among EPF was evaluated by MEGA software. Out of forty isolated entomopathogenic fungi, three (Beauveria bassiana deb4, Penicillium verrucosum Nlg1, and Fusarium equiseti khr4) were highly effective entomopathogen. The LD50 value of B. bassiana, P. verrucosum and F. equiseti were 1.4 × 104, 9.8 × 104, 1.0 × 106 spores ml−1, and LT50 values were 32.14, 37.5, 32.14 h respectively. Their phylogenetic analysis indicates related closeness on the basis of their conserved internal transcribed spacer region. In conclusion, the indigenous isolated strain of B. bassiana (deb4) has shown highest biocontrol potentiality amongst three indigenous entomopatogenic fungi under lab condition against A. gossypii and can be applied in agrifields.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Papaya (Carica papaya Linn.) is one of the most cultivated fruit crops having abundant prophylactic, nutritional and therapeutic values, in almost all tropical countries. Despite taking several precautions, Papaya Ring Spot Virus (PRSV) causes huge destruction and crop loss (10–100%) every year (Tennant et al. 2007). Aphids; the insect vectors transmit this virus in a non-persistent manner (Chakrabarti and Raychaudhuri 1978). Several chemical pesticides are being used to control this disease, but they cause environmental pollution and health hazards (Kalleshwaraswamy et al. 2012). In the present scenario, biological control strategies are essential for increasing human awareness against the hazardous effects of chemical control of pest and disease (Fang et al. 2009; Garkoti et al. 2014). Insect management with the help of entomopathogenic fungi (EPF) is the modern approach as an alternative to chemical control (Han et al. 2014a, b; Verma et al. 2007). In the biological control approach, EPFs are applied in a lesser amount in pre-harvesting season, (in case of papaya, it is pre-monsoon season) expecting flourishment of growth whenever their natural enemy of pest appears in the field. This mycopesticide/mycoinsecticide is supposed to prevent the aphid pest to grow, multiply (ovipary or vivipary) (Ogawa and Miura 2014) and thereby preventing the capacity of carrying and transferring PRSV virus by means of primary (direct transmission by feeding) and secondary transfer method (flying to another healthy plant) (Ng and Perry 2004). Use of indigenous fungal isolate is very helpful against various pests, as EPFs are predominant in that particular geographical niche. Long-term control is the best strategy to prevent further pest attack. The pest population should be controlled below the economic threshold level, by using EPF as a bio-control tool.
The objectives of our present study are to isolate and characterize indigenous entomopathogenic fungi, to find out entomopathogenic potentiality, to compare antagonistic potentiality among isolates and to find out the mechanism of action against A. gossypii under lab condition.
Materials and methods
Collection of samples
Soil samples were collected by discarding the topsoil up to one inch in a randomized manner from different rhizospheric soils of agrifields of North 24 Parganas district during the pre-monsoon time mixed them well and then brought to the laboratory in a biodegradable polythene bag and shade dried, grinned and sieved (2 mm pore size) (Sharma et al. 2012). Insect cadavers collecting from different zones of this district were used for isolation of entomopathogenic fungi.
Isolation and purification of fungi
Soil samples were then used for isolating fungi using serial dilution technique (Dhingra and Sinclair 1985) and by an insect bait method (Zimmermann 1986). Cadavers were surface sterilized with 75% ethanol for 30 s and then subjected to washing with sterilized water, followed by blotting the extra water in blotting paper, cut into small pieces, placed in Petri dishes containing Potato Dextrose Agar (PDA HiMedia-GM096) medium (pH 6.8). Plates were supplemented with 0.01% Rose Bengal and Streptomycin to prevent the growth of unwanted fastidious fungi and bacteria respectively. Purification of fungal isolates was done by transferring a hyphal tip from freshly growing single colony to a new PDA containing plate and incubated at 28 ± 2 °C. Purified fungal cultures were re-transferred in PDA slant and preserved at 4 °C for the further experimental purpose. Plates were kept in BOD incubator at 28 ± 2 °C for significant growth coverage.
Spore suspension preparation
All fungal genera isolated were applied for primary screening. Fungi were grown in Erlenmeyer flask containing wheat grain medium and kept in the incubator at 28 ± 2 °C for total medium coverage and sporulation. After relevant days of incubation 40 ml of sterilized distilled water was poured and vortex vigorously to detach the spores from hyphae/conidia. Water suspension containing spores were collected in a separate sterilized capped centrifuge tube (15 ml). Higher spore concentration was obtained by short spinning (1000 g for 3 min) in the centrifuge. Spore concentration was measured by haemocytometer and adjusted to 1 × 104, 105, 106, 107 and 108 spores ml−1.
Screening of fungal isolates against papaya aphid
Incised Leaf Disk Method (ILDM) was adopted here (Ghosh et al. 2014). In brief, sterilized glass Petri dishes (9 cm diameter) were taken and sterilized Whatman filter paper (no. 1) soaked with sterile distilled water was placed. Fresh healthy papaya leaves were cut into pieces and surface sterilized with 3% Sodium hypochlorite (NaOCl) solution followed by twice of washing with sterilized distilled water. Leaves were placed on the filter paper (dorsal part down) and served as feed for test insect. Aphis gossypii Glover (3–4th instar), collected from fully grown infected papaya plant leaf (North 24 Parganas, West Bengal), were surface sterilized with 3% Sodium hypochlorite (NaOCl) solution followed by twice washing with sterilized distilled water and ten aphids were placed into each Petri dish containing papaya leaves and sterilized Whatman filter paper (no. 1).
One milliliter spore suspension (1 × 107 spore’s ml−1 concentration) of each tested fungal isolates was spread uniformly on each Petri dish by a small glass chromatographic sprayer. The control sets were sprayed with sterile distilled water. The experiment was conducted at room temperature.
Aphid mortality was recorded by counting dead insect daily until total fatality. Dead aphids were further taken out to investigate fungal growth under the microscope. Only aphids which exhibited fungal sporulation were considered to have died from the fungus treatment. Entomopathogenecity of EPFs was graded on the basis of the mortality of the aphids. Best isolates were selected for further screening.
After primary screening an I–IV grade was constructed like Grade I = 70–100% mortality; Grade II = 40–50%; Grade III = 10–30% and Grade IV = non-effective.
Identification of selected entomopathogen by the cultural and morphological method
The fungal isolates were identified by cultural characteristics and microscopic morphological characters (Domsch et al. 1980; Nagamani et al. 2006; Humber 2005). For the microscopic study, fungal mycelium and spores were stained with a Lactophenol-Cotton-Blue solution (HiMedia-S016) on a glass slide and observed under bright field compound microscope (40X) (Olympus-CX31).
Determination of LD50 and LT50
EPF treatment was done to test the mortality rate of aphid. The three top graded isolates which showed best entomopathogenic activity were cultured and their spore suspensions were prepared (1 × 104, 105, 106, 107, 108 spores ml−1). Previous Incised Leaf Disc technique was followed here in triplicate along with control set and means value was taken. Comparative regression analysis was carried out using SPSS (v. 21). Different treatments of normally distributed data were compared by using one-way ANOVA. In addition, which isolates were shown the highest mortality in which time duration, was also evaluated.
Linear regression is a way to model the relationship between two variables. Finding a linear regression equation is to determine if there is a relationship between the two variables. Regression analysis was done by Probit method to determine the mortality, based on the formula of straight line equation [Y = aX + b]. Here X is the dependent variable (that’s the mortality rate that plotted on the X-axis), Y is the independent variable (the spore concentration, plotted on the Y-axis), “a” is the slope of the line and “b” is the X-intercept (Abbott 1925, Gomez and Gomez 1984). Median Lethal Dose (LD50) and Median Lethal Time (LT50) (Han et al. 2014a, b) were estimated aftermath by using SPSS (v.21) and Microsoft Office Excel 2013. On the other hand, LT50 was determined using LD50 value as standard dosage. The regression equation was derived using Microsoft Office Excel 2007.
Molecular characterisation by (ITS1-5.8S-ITS2) PCR based technique
Genomic DNA of these three fungi was extracted by a modified CTAB method (Chutima et al. 2011), purified and visualized under gel electrophoresis (1% agarose). These internal transcribed spacers (ITS1-5.8S-ITS2) regions were amplified by PCR technology using DNA amplification reagent kit (GeNei) with the help of fungus-specific forward primer ITS-1(5′-CTTGGTCATTTAGAGGAAGTAA-3′) (Gardes and Bruns 1993) and the reverse primer ITS-2(5′-TCCTCCGCTTATTGATATGC-3′) (White et al., 1990). The amplified PCR product was purified by DNA purification kit (HiMedia), and run on 1.2% agarose gel Sequencing of PCR products was carried out in Sci Genome Labs Pvt. Ltd., Kerala, India. These sequences were analyzed by homology searching (BLAST) against the Gene Bank nucleotide database (http://blast.ncbi.nlm.nih.gov) and the most similar sequences were selected to compare with reference sequences from the online bioinformatics tools of Gene Bank from NCBI. The sequences obtained were submitted to NCBI Gene Bank to obtain the Accession numbers.
Establishment of a phylogenetic relationship
Sequence homology searched by putting nucleotide sequences (considered as FASTA format) in n-BLAST within NCBI online search tool for each isolate. Fungal species were determined by the highest similarity of ITS sequences to known strains. For each fungal isolate, highest similar strain (with accession number) was used as a reference sequence to our subject strain to determine evolutionary likeliness (Lv et al. 2011; Kumar et al. 2016) the maximum alike sequence has been taken as a reference organism and their accession number was noted (Nei and Kumar 2000).
Sequence alignment was done with the help of MEGA 7 software by Muscle alignment method. Maximum parsimony phylogenetic tree of these genera was constructed using the UPGMA method with the help of MEGA 7 software (Tamura et al. 2013). Tajima’s relative rate test (Tajima 1993) was done to establish the divergence of sequence distribution among these isolates.
Interaction of entomopathogenecity
To find out the interaction of entomopathogenecity, Incised Leaf Disk Method (ILDM) as described earlier was performed and critically examined under light compound microscopy (Olympus-CX31) for host-parasite interaction.
Results and discussion
Total 40 isolates have been isolated from the rhizospheric soil (15 cm below soil surface, near the root zone) and insect cadaver. Out of 40 isolates, we have found 12 genera as per our phenotypical identification. Out of them, Trichoderma has 11 isolates, Aspergillus 9 isolates and Penicillium 8 isolates (Table 1). Out of 40 isolates, 20 isolates have shown entomopathogenic property against papaya aphid (3–4th instar) under laboratory condition. Out of them, three fungal isolates (ded4, Nlg1 and khr4) showed potent entomopathogenic activity (Table 1).
The detail cultural and morphological characteristics of the three profound fungal isolates of entomopathogens against aphid were presented in Table 2 and Fig. 1 and they were phenotypically identified as Beauveria sp (deb4), Penicillium sp. (Nlg1) and Fusarium sp. (khr4). These three isolates were identified molecularly.
Molecular characterization
Figure 2a showed the DNA bands under UV-transilluminator. DNA band of each EPFs were near 3 kb comparing with DNA ladder. The PCR product showed band near 650 bp under UV-transilluminator (Fig. 2b).
Visualization of DNA bands by agarose gel electrophoresis. a Band of interest under UV-transilluminator. From the left 3 kb Ladder (Lane 1) and genomic DNA of Beauveria sp. (Lane 2), Penicillium sp. (Lane 3), Fusarium sp. (Lane 4). b Banding pattern of PCR product from ITS regions. From the left 1 kb ladder (Lane 1) and PCR product from Beauveria sp. (Lane 2), Penicillium sp. (Lane 3), Fusarium sp. (Lane 4)
PCR amplified products were sent for sequencing in Eurofins Genomics India Private Limited, Bengaluru, India. Obtained FASTA sequences were searched for homology by BLAST in NCBI nucleotide database. B. bassiana isolate deb4 (567 nucleotides) showed the highest similarity with B. bassiana (Accn. No. AJ560690.1) and identified as same species. Similarly, P. verrucosum Nlg1 isolate (607 nucleotides) and F. equiseti khr4 isolate (548 nucleotides) showed the highest similarity with P. verruculosum (Accn. No. HM469420.1) and F. equiseti (Accn. No. KY523100.1) respectively. These sequences were submitted to NCBI to get the accession number and P. verrucosum has got the Accession number KY966028.1 so far.
Construction of phylogenetic tree and determination of the evolutionary relationship
B. bassiana (deb4) query sequence was compared with the most related isolate having Accession number AJ560690.1. P. verrucosum (Nlg1) has reference isolate having accession number HM469420.1. F. equiseti (khr4) was compared with isolate accession number KY523100.1.
The evolutionary history was inferred using the Neighbor-Joining method (Saitou and Nei 1987). The optimal tree with the sum of branch length = 1.16130728 is created. The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to conclude the phylogenetic tree. The evolutionary distances were computed using the Maximum Composite Likelihood method (Tamura et al. 2004) and are in the units of the number of base substitutions per site. The analysis involved 6 nucleotide sequences. All positions comprising gaps and missing data were rejected. There were a total of 446 positions in the final dataset. Evolutionary analyses were conducted in MEGA6 (Tamura et al. 2013) (Fig. 3). Result specifies that, regardless of their genus distinction, the conserved ITS1-5.8s-ITS2 region of these three fungi have significant sequence similarity.
The equality of evolutionary rate between sequences A (B. bassiana deb4) and (P. verrucosum Nlg1), with sequence C (F. equiseti khr4) used as an outgroup in Tajima’s relative rate test (Table 3) (Tajima 1993). The χ2 test statistic was 92.47 (P = 0.00 with 1 degree of freedom). The P value less than 0.05 used to reject the null hypothesis of equal rates between lineages. A total of 480 positions calculated in the final dataset. These analyses were conducted in MEGA7 (Kumar et al. 2016).
Bio-assay and mortality of aphid by EPF
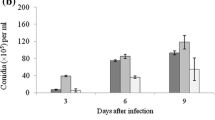
Data obtained from Fig. 4 showed the different concentrations of spore suspension (1 × 104 to 1 × 108) of three EPFs, have different effects on aphids. Ten aphids were tasted under lab condition for each isolate and their respective spore concentration. Figure 4 clearly showed that B. bassiana had the highest effectiveness followed by P. verrucosum and F. equiseti. Some of the isolates showed high fatality against papaya aphid, others showed minimal to null effect (Martinuz et al. 2012; Mishra et al. 2013).
Regression analysis and evaluation of LD50
The results presented in Table 4 clearly explained that B. bassiana was diagnosed with lowest LD50 value (1.4 × 104 spores ml−1) followed by P. verrucosum (9.8 × 104 spores ml−1) and F. equiseti (1.0 × 106ml−1). Similarly, LT50 values were also evaluated (Table 5) and the lowest LT50 (32.14 h at 1 × 104 spores ml−1) indicated B. bassiana as best EPF followed by P. verrucosum (37.5 h at 1 × 105ml−1) and F. equiseti (32.14 h at 1 × 106 spores ml−1).
A group of scientists (Kim et al. 2013) recorded that B. bassiana isolate Bb08 showed LT50 in 3.0 ± 0.3 days against green peach aphid. In our laboratory, previously isolated B. bassiana (BB1) strain showed ED50 value at 106 spores ml at 3.2 days after inoculation (Ghosh et al. 2014). Supporting that information, in our present study, B. bassiana (deb4) isolate showed the lowest LD50 value (1.4 × 104 spores ml−1) on the 2nd day after inoculation. P. verruculosum can produce cellulase enzyme that in turn could be used as a potential biocontrol agent against aphids (Morozova et al. 2010). Some scientist (Nicoletti and De Stefano 2000) reported P. verruculosumto have potential killing effect against aphids. Some other Penicillium. spp. are also capable of killing several pests (Shah and Iqbal 2017; Patil and Jadhav 2015). In our present study, P. verrucosum showed significant LD50 value i.e., 9.8 × 104 spores ml−1 on the 2nd day after inoculation. Fusarium semitectum was also used against Aphis gossypii as a biocontrol agent (Jayasimha et al. 2013). Our F. equiseti isolate has also successfully controlled the A. gossypii insect having the LD50 value of 1.0 × 106 spores ml−1 on 2nd day after inoculation (Table 4).
Host-parasitic interaction
These fungi harbour entomopathogenecity through adhesion, germination, ramification and penetration as shown in Fig. 5. For the mechanical study, EPFs are well known for their capacity to produce several extracellular enzymes and biologically active molecules (Freimoser et al. 2003, Ganassi et al. 2001). Fungal spores first adhere to the cuticle of Aphis gossypii and then penetrate the cuticle layer by secreting chitinase, hydrolase etc. (Quesada-Moraga et al. 2006). Thereafter entering the host, they ramify and secrete secondary metabolites (Ortiz-Urquiza and Keyhani 2013; Han et al. 2014a). Surface interaction and subsequent penetration, germination of these fungi on aphid cuticle have also established by the microscopical study as revealed in an earlier study (Ortiz-Urquiza and Keyhani 2013; Ghosh et al. 2014). Our mechanical studies by microscopical observations are assertive as par the other workers. In brief, these three EPF isolates also exhibited attachment and penetration of aphid cuticle, observed under a compound microscope. The details mechanical studies on respect to fungus insect interaction were done by few workers and a correlation between toxin secretion and reduction of PPO, which is responsible for insect immunity, has been recorded by some authors (Binggeli et al. 2014; Karthikeyan and Selvanarayanan 2011; Podder and Ghosh 2019).
Conclusions
Among 40 isolates of different fungi, 3 isolates (B. bassiana deb4, P. verrucosum Nlg1 and F. equiseti khr4) were best effective against papaya aphid. B. bassiana (deb4) isolate was most pathogenic against A. gossypii on the 2nd day after inoculation and this may be tested under field condition for biological control of this pest. The mechanistic study showed that these fungal EPFs produced spores and these spores germinated on aphid surface and by penetration directly to kill the host. Moreover, as per phylogenetic tree analysis, it showed that these three EPFs had a close ancestral relationship, whereas Tajima’s relative rate test of sequences showed significant diverseness among them.
References
Abbott WS (1925) A method of computing the effectiveness of an insecticide. J Econ Entomol 18:265–267
Binggeli O, Neyen C, Poidevin M, Lemaitre B (2014) Prophenoloxidase activation is required for survival to microbial infections in Drosophila. PLoS Pathog 10:e1004067
Chakrabarti S, Raychaudhuri D (1978) New and little known aphids (Homoptera: Aphididae) from Kumaon Himalaya, India. Entomon 3(1):95–103
Chutima R, Dell B, Vessabutr S, Bussaban B, Lumyong S (2011) Endophytic fungi from Pecteilis susannae (L.) Rafin (Orchidaceae), a threatened terrestrial orchid in Thailand. Mycorrhiza 21:221–229
Dhingra OD, Sinclair JB (1985) Culture media and their formulas. Basic plant pathology methods. CRC, Boca Raton
Domsch KH, Gams W, Anderson TH (1980) Compendium of soil fungi, vol 1. Academic, London
Fang W, Pava-Ripoll M, Wang S, Leger RS (2009) Protein kinase A regulates production of virulence determinants by the entomopathogenic fungus, Metarhizium anisopliae. Fung Genet Biol 46:277–285
Freimoser FM, Screen S, Bagga S, Hu G, St. Leger RJ (2003) Expressed sequence tag (EST) analysis of two subspecies of Metarhizium anisopliae reveals a plethora of secreted proteins with potential activity in insect hosts. Microbiology 149:239–247
Ganassi S, Moretti A, Stornelli C, Fratello B, Pagliai AB, Logrieco A, Sabatini MA (2001) Effect of Fusarium, Paecilomyces and Trichoderma formulations against aphid Schizaphis graminum. Mycopathologia 151:131–138
Gardes M, Bruns TD (1993) ITS primers with enhanced specificity for basidiomycetes-application to the identification of mycorrhizae and rusts. Mol Ecol 2:113–118
Garkoti A, Kumar V, Tripathi H (2014) Control of wilt disease of lentil through bio control agents and organic amendments in Tarai region of Uttarakhand, India. J Environ Biol 35:1067–1070
Ghosh SK, Chakraborty N, Biswas PP (2014) In vitro biological control of aphid of Papaya by Beauveria bassiana. III Int Symp Papaya 1022:113–117
Gomez KA, Gomez AA (1984) Statistical procedures for agricultural research. Wiley, Oxford
Han JH, Jin BR, Kim JJ, Lee SY (2014a) Virulence of entomopathogenic fungi Metarhizium anisopliae and Paecilomyces fumosoroseus for the microbial control of Spodoptera exigua. Mycobiology 42:385–390
Han Z, Wang Q, Fu J, Chen H, Zhao Y, Zhou B, Gong Z, Wei S, Li J, Liu H, Zhang X, Liu C, Yu H (2014b) Multiple bio-analytical methods to reveal possible molecular mechanisms of developmental toxicity in zebrafish embryos/larvae exposed to tris (2-butoxyethyl) phosphate. Aquat Toxicol 150:175–181
Humber RA (2005) Fungal identification USDA-ARS plant protection research 103 Unit US plant. Soil & Nutrition Laboratory, Ithaca
Jayasimha G, Rachana R, Rajkumar V, Manjunatha M (2013) Evaluation of fungal pathogen, Fusarium semitectum Berk and Ravenel against okra aphid, Aphis gossypii Glover under laboratory and green house conditions. Pest Manag Hort Ecosyst 18:139–142
Kalleshwaraswamy CM, Krishnakumar NK, Chandrashekara KN, Vani A (2012) Efficacy of insecticides and oils on feeding behaviour of Aphis gossypii Glover and transmission of Papaya ringspot virus (PRSV). K J A S 25(1):63–67
Karthikeyan A, Selvanarayanan V (2011) In vitro efficacy of Beauveria bassiana (Bals.) Vuill. and Verticillium lecanii (Zimm.) viegas against selected insect pests of cotton. Recent Res Sci Technol 3(2):142–143
Kim JJ, Jeong G, Han JH, Lee S (2013) Biological control of aphid using fungal culture and culture filtrates of Beauveria bassiana. Mycobiology 41:221–224
Kumar S, Stecher G, Tamura K (2016) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874
Lv C, Huang B, Qiao M, Wei J, Ding B (2011) Entomopathogenic fungi on Hemiberlesia pitysophila. PLoS One 6:e23649
Martinuz A, Schouten A, Menjivar R, Sikora R (2012) Effectiveness of systemic resistance toward Aphis gossypii (Hom., Aphididae) as induced by combined applications of the endophytes Fusarium oxysporum Fo162 and Rhizobium etli G12. Bio Control 62:206–212
Mishra DS, Kumar A, Prajapati CR, Singh A, Sharma S (2013) Identification of compatible bacterial and fungal isolate and their effectiveness against plant disease. J Environ Biol 34:183
Morozova VV, Gusakov AV, Andrianov RM, Pravilnikov AG, Osipov DO, Sinitsyn AP (2010) Cellulases of Penicillium verruculosum. Biotechnol J 5:871–880
Nagamani A, Kunwar IK, Manoharachary C (2006) Hand book of soil fungi. IK International Pvt. Ltd., New Delhi, p 477
Nei M, Kumar S (2000) Molecular evolution and phylogenetics. Oxford University Press, Oxford
Ng JC, Perry KL (2004) Transmission of plant viruses by aphid vectors. Mol Plant Pathol 5:505–511
Nicoletti R, De Stefano S (2000) Peptidi ciclici di origine fungina. IL Tabacco 8:33–59
Ogawa K, Miura T (2014) Aphid polyphenisms: trans-generational developmental regulation through viviparity. Front Physiol 5:1
Ortiz-Urquiza A, Keyhani NO (2013) Action on the surface: entomopathogenic fungi versus the insect cuticle. Insects 4:357–374
Patil NS, Jadhav JP (2015) Significance of Penicillium ochrochloron chitinase as a biocontrol agent against pest Helicoverpa armigera. Chemosphere 128:231–235
Podder D, Ghosh SK (2019) A new application of Trichoderma asperellum as an anopheline larvicide for eco friendly management in medical science. Sci Rep 9:1108. https://doi.org/10.1038/s41598-018-37108-2
Quesada-Moraga E, Carrasco-Díaz JA, Santiago-Álvarez C (2006) Insecticidal and antifeedant activities of proteins secreted by entomopathogenic fungi against Spodoptera littoralis (Lep., Noctuidae). J Appl Entomol 130:442–452
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Shah HU, Iqbal Z (2017) Biological screening of crude extract of Penicillium sp. EU0013. J Anim Plant Sci 27(4):1209–1216
Sharma P, Saini MK, Deep S, Kumar V (2012) Biological control of groundnut root rot in farmer’s field. J Agric Sci 4(8):48
Tajima F (1993) Simple methods for testing the molecular evolutionary clock hypothesis. Genetics 135:599–607
Tamura K, Nei M, Kumar S (2004) Prospects for inferring very large phylogenies by using the neighbor-joining method. Proc Natl Acad Sci 101:11030–11035
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729
Tennant PF, Fermin GA, Roye ME (2007) Viruses infecting papaya (Carica papaya L.): etiology, pathogenesis and molecular biology. Plant Viruses. 1:178–188
Verma M, Brar SK, Tyagi RD, Surampalli RY, Valero JR (2007) Antagonistic fungi, Trichoderma spp.: panoply of biological control. Biochem Eng J 37:1–20
White TJ, Bruns T, Lee S, Taylor J (1990) Amplification and direct sequencing of fungal ribosomal RNA genesfor phylogenetics. PCR Protoc 18:315–322
Zimmermann G (1986) The ‘Galleria bait method for detection of entomopathogenic fungi in soil. J Appl Entomol 102:213–215
Acknowledgements
Authors are thankful to the Principal of Ramakrishna Mission Vivekananda Centenary College (Autonomous), Rahara, Kolkata, for providing the lab facility. The financial support was received from “Department of Science and Technology & Biotechnology (DSTBT)”, Government of West Bengal, India, as major research project (Sanction Number: 820 (Sanc.)/ST/P/S&T/1G-2/2014 dated 5/1/16); authors are also thankful to them.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Mukherjee, A., Debnath, P., Ghosh, S.K. et al. Biological control of papaya aphid (Aphis gossypii Glover) using entomopathogenic fungi. Vegetos 33, 1–10 (2020). https://doi.org/10.1007/s42535-019-00072-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s42535-019-00072-x