Abstract
Citrus tristeza virus (CTV) is the causal agent of the most important viral disease of citrus. Symptoms that may affect the productive potential of citrus plants are observed in Uruguayan orchards even though resistant rootstocks are used. CTV is fully eliminated in propagative materials by the National Sanitation and Certification Program, but since the virus and its vector are widespread in the country, the risk of infection in the field persists. In this situation, using mild CTV strains in a cross-protection program would be a useful alternative to attempt to increase yield and quality of the local citrus industry. To this aim, this study assessed the biological and molecular characteristics of 32 local CTV isolates. Bioassays were conducted in a greenhouse with controlled conditions. Each isolate was graft-inoculated on Mexican lime, sweet orange, sour orange and Duncan grapefruit indicator plants. Symptoms and their intensity were evaluated. Molecular characterization was carried out by RT-PCR amplification, using primers for the p25, p20 and p23 genes. PCR products were sequenced, nucleotide sequences were aligned with international reference strains and phylogenetic trees were constructed. Results of the biological and molecular analysis showed the prevalence of severe CTV isolates with a high genetic variability. Two out of 32 characterized isolates were selected as mild CTV isolates to be tested as candidates for future cross-protection experiments. The survey showed a complex scenario for the management of CTV in Uruguay.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
In Uruguay, citrus is the main fruits crop and its production is mainly exported to fresh fruit markets, thus the achievement of high yields with outstanding fruit quality are key steps to maintain a competitive industry. However, the presence of several diseases in the country and their effect, represent a main limitation to achieve this task.
Citrus tristeza virus (CTV), a member of genus Closterovirus (family Closteroviridae), is the causal agent of tristeza, the most important viral disease of citrus (Bar-Joseph and Lee 1989; Roistacher et al. 2010). The disease is widely distributed in most citrus growing areas of the world and is endemic in Uruguay. CTV is a phloem limited virus and it is transmitted by aphids and grafting (Roistacher and Bar-Joseph 1987a, b). The virus has showed a high genetic and biological diversity (Moreno et al. 2008). Its genome consists of a 19.3 kb, single-stranded, positive-sense RNA. This RNA is encapsidated by two capsid proteins (CP); a major CP of 25 kDa covering 97% of the length of the virion and the minor CP of 27 kDa that covers the 5′ extreme (Moreno et al. 2008). Seven CTV genotypes have been described (Harper 2013; Dawson et al. 2015) and often field isolate contain a mixture of them (Albiach-Marti et al. 2000).
The expression and intensity of symptoms depends on the isolate, the scion-rootstock combination, the climate and the vector population (Rocha-Peña et al. 1995). The range of symptoms can vary from asymptomatic plants to different degrees of seedling yellows, stunting, stem pitting and decline (Moreno et al. 2008). The main symptoms that have been reported are quick decline of trees grafted onto sour orange (Citrus aurantium) and stem pitting (Rocha-Peña et al. 1995; Roistacher et al. 2010). Decline has been restrained using resistant rootstocks as Poncirus trifoliata and its hybrids (Moreno et al. 2008), while stem pitting can be controlled by cross-protection (Da Graça and Van Vuuren 2010; Folimonova 2013). Cross-protection uses mild virus isolates to protect plants against the damage caused by the infection with a severe strain(s) of the same virus (Gonsalves and Garnsey 1989).
CTV was first reported in Uruguay in 1955 (Koch de Brotos and Boasso) causing the death of thousands of plants and thus, forcing the use of decline-resistant scion-rootstocks combinations. Nowadays CTV is widespread as well as Toxoptera citricida (Hemiptera: Aphididae), its most efficient known vector (Bentancour et al. 2009). Since more than 90% of citrus crop are grafted onto P. trifoliata, typical decline symptoms are not observed, however, stem pitting in grapefruits and sweet oranges as well as small-sized fruit, have been reported (Müller and Campiglia 1981; Francis et al. 1997). These symptoms could be attributed to the infection with severe CTV isolates, which could be limiting the potential productivity of the plants (Da Graça and van Vuuren 2010; Roistacher et al. 2010). Generally, in regions with a similar situation, tolerant rootstocks and the presence of T. citricida, the prevalence of severe CTV isolates has been reported (Roistacher 1988; Broadbent et al. 1991; Koizumi 1991). In fact, VT genotype, which have been frequently associated with severe SP symptoms (Biswas et al. 2012) was widespread detected in Uruguay (Benítez-Galeano et al. 2015). Moreover, the CTV strain that overcome the resistance of P. trifoliata previously reported in New Zealand (Dawson and Money 2000; Harper et al. 2010) and recently in California (Yokomi et al. 2017), also was detected in Uruguay (Hernández-Rodríguez et al. 2017; Benítez-Galeano et al. 2018).
Even though in Uruguay the commercial citrus plants are produced under a Sanitation and Certification Program, the presence of CTV and its vector in the field would lead to the infection of them in the short time. In this situation, the only means to protect commercial citrus plants against stem pitting CTV isolates is the cross-protection (Moreno et al. 2008; Da Graça and Van Vuuren 2010; Folimonova 2013). In South Africa, Australia, Brazil and Peru, cross-protection programs have succeeded (Costa et al. 2010; Da Graça and Van Vuuren 2010), increasing yield and fruit size of important varieties such as Pera sweet orange (Citrus sinensis) in San Pablo, Brazil and Marsh grapefruit (Citrus paradisi) in South Africa. However, in other citrus regions or with other varieties it has shown limited results (Müller et al. 1988; Broadbent et al. 1991).
In many cases mild CTV isolates have been difficult to achieve, while in others mild CTV isolates provided only short-term protection (Folimonova 2013). In the past, most of candidate CTV isolates for cross-protection were selected empirically. Nowadays, more information is available to implement it, being required that the isolate to be controlled, and the protective isolate, belong to the same strain of the virus (Folimonova 2013). In our conditions, a cross-protection program would be an interesting alternative to complement the Certification Program, in order to decrease the harmful effects of CTV and thus to improve the yield and quality of the citrus crop. To this aim, we studied the aggressiveness of the local CTV isolates and the genotypes prevalence by evaluating their biological and genetic characteristics.
Materials and methods
Plant material and biological indexing
A total of 32 CTV isolates were collected from the main citrus growing of the country. Sixteen of them were collected between 1993 and 1998 and kept in an insect-proof glasshouse grafted on rough lemon (C. jambhirí) and other 16 isolates were collected during 2012–2014 (Table 1). Samples consisted of twigs from a) trees with poor vegetative development, showing stem pitting and small fruit size (as potential carriers of severe CTV strains) and b) healthy looking trees, vigorous and with high yield and fruit quality (as possible hosts of mild CTV strains). All isolates were kept, by grafted pieces of bark, on rough lemon seedlings.
Biological indexing was carried out in an insect-proof and temperature-controlled glasshouse (ranging 18–26 °C) according to Garnsey et al. (1987). The biological characteristics of each isolate were assessed on Mexican lime (ML) [Citrus aurantifolia (Christ.) Swing.], sour orange (SO), grapefruit cv. ‘Duncan’ (DG) and sweet orange cv. ‘Madame Vinous’ (SW) indicator plants, each of which produces a specific set of symptoms. The combination sweet orange/sour orange was not included. Each isolate was inoculated by grafting two pieces of bark in three seedlings of each indicator species. Four negative controls (CTV free-seedlings, growth under controlled conditions) and four positive controls were included per indicator species. The positive controls used (UY-10 and UY-35) are local isolates used as positive controls for CTV by the Citrus Sanitation and Certification Program. CTV infection was confirmed by DAS-ELISA using the Magic-DAS ELISA kit from Plant Print Diagnostics S.L. (Valencia, Spain) following the manufacturer’s instructions. Two months after inoculation, plants were pruned, conducted to a single stem. Foliar evaluated symptoms were: vein clearing (VC), leaf cupping (LC) and vein corking (Vck) in Mexican lime (LM); seedling yellows in SO and DG, which were registered periodically after growth flushes. Stem pitting (SP) was evaluated at the end of tests (10–12 months post-inoculation) by peeling stems above inoculation point in LM, DG and SW. Stunting was assessed visually in SO and DG. Intensity of each symptom was rated according to the following scale: 0 = no symptoms, 1 = mild symptoms, 2 = moderate symptoms, 3 = severe symptoms. Total evaluation period was of one year.
RNA extraction, cDNA synthesis, and PCR amplification
Plant tissue was homogenized with liquid nitrogen and total RNA was extracted using RNeasy Plant Mini kit (QIAGEN, Hilden, Germany), following manufacturer’s instructions. The cDNA was synthesized using 2 μg of RNA, 0.5 μl of random primers (10 μM) and sterile water to complete the final volume (20 μl); the mixture was incubated for 5 min at 95 °C and for 4 min on ice. Then, 3.75 μl of the mixture: 2 μl of RT (10X) buffer, 1 μl of dNTP (10 mM), 0.25 μl of Ribolock RNAase Inhibitor (Thermo Fisher Scientific) and 0.5 μl of M-MuLV Reverse Transcriptase (200 U / μl) were added to each tube. For the amplification of the three studied regions, 2 μl of cDNA was PCR-amplified in a final reaction volume of 25 μl, with a reaction mix containing: 2.5 μl of 10X Taq buffer with (NH4)2SO4 (750 mM Tris-HCl (pH 8.8 at 25 °C), 1 μl of each primer (10 uM), 2 μl MgCl2 (25 mM), 0.7 μl dNTP (10 mM each), 18.4 μl water and 0.4 μl Taq Polymerase (5 U / μl) (Thermo Scientific Inc., Hanover, MD, USA). The p20, p25 and p23 genes were selected to determine phylogenetic relationships among Uruguayan isolates and isolates from the South American region (Iglesias et al. 2008), following the procedure described by Benítez-Galeano et al. 2015, using the primers p20F/p20R (561 bp), p25F/p25R (677 bp) and PM50/PM51 (697 bp), respectively (Sambade et al. 2003; Benítez-Galeano et al. 2015). Thermal cycling conditions for p20 and p25 genes were: an initial denaturation phase at 95 °C for 4 min; 35 cycles of 30 s at 95 °C, 45 s at 50 °C, 60 s at 72 °C, with a final extension of 2 min at 72 °C. For the p23 gene, cycling conditions were the same as described above but with an annealing temperature of 54 °C. Amplification products were visualized by GoodView™ (SBS Genetech Co., Beijing, China) staining in 2% agarose gels, after electrophoresis. The amplified fragments were purified using the “AxyPrep DNA Gel Extraction” kit (Axygen, Corning, NY, USA), according to manufacturer’s recommendations.
Nucleotide sequences and phylogenetic analysis
Amplicon-sequencing was performed in both directions (5′-3 ‘and 3’-5′), using for each gene the specific primers used for the amplification. Sequences were assembled and edited with SeqMan program (Lasergene, DNASTAR) and compared with the reference sequences of the six CTV genotypes: VT, T30, T3, RB, T36, T68, described by Harper (2013), and HA16–5 for the group (NC) proposed by Benítez-Galeano et al. in 2015 (T36: AY340974, U16304, VT: U16304, EU937519, AB046398, T3: KC525952, EU857538, RB: FJ525431, FJ525432, FJ525433, FJ525434, FJ525435, T68: JQ965169, EU076703, FJ525436, T30: AF260651, Y18420, and NC: GQ454870), retrieved from the GenBank database (http://www.ncbi.nml.nih.gov/genbank). Alignments were obtained with Clustal W algorithm in MEGA 6.0 (Tamura et al. 2013). The model of nucleotide substitution that best fit the dataset (HKY) was selected using the JModelTest program (Posada 2008) according to the Akaike Information Criterion (Akaike 1974). Maximum Likelihood (ML) phylogenetic trees were reconstructed with PhyML program (https://www.hiv.lanl.gov/content/sequence/PHYML/interface.html) (Guindon et al. 2010). The branches support was estimated with the approximate likelihood-ratio test (aLRT) (Anisimova and Gascuel 2006).
Results
Biological indexing
The results of the biological assays are summarized in Table 2. All the isolates produced at least one of the expected symptoms on indicator plants (Fig. 1), but with different intensity. Foliar symptoms appeared two months post-inoculation, reaching the highest intensity four months later. The pattern of symptoms in ML allowed a clear differentiation between isolates. While some of them only showed vein clearing, others also produced different degrees of leaf cupping, vein corking, stunting and stem pitting. In SO 72% of the isolates developed SY reaction, ranging from moderate to intense. This symptom was also observed in DG, but with a mildest reaction. Finally, SP, the most important symptom to determine the aggressiveness of the isolates, was developed by most of isolates in ML, DG and SW; being moderate to intense in SW for 47% of the evaluated isolates.
According to symptom pattern isolates grouped into four remarkable different groups (Table 2). Only two isolates showed mild symptoms in ML and SO, so they could be considered as mild isolates (group 1). A second group (12.5% of isolates) showed mild symptoms in ML and mild or moderate SY and SP in SO and DG, respectively. The third group (34.5% of the isolates) showed moderate to intense symptoms in ML, some SY reactions in SO, moderate SP in DG and mild SP in SW. Group four (47% of the isolates) developed moderate to intense symptoms in all the indicators plants.
Phylogenetic comparisons
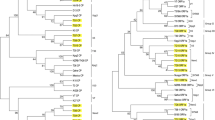
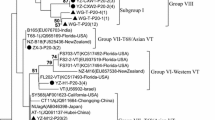
The phylogenetic comparisons were based on the sequences of the genes p20, p23 and p25 of the major viral components in CTV populations of the collected isolates. To build p25 gene phylogenetic tree, 25 CTV isolate sequences of 572 nt in length were generated (GenBank accession Nos. MH321296 to MH321320) and phylogenetically compared with reference sequences. A tree with six defined clades was obtained (Fig. 2a). The sequences UY-17 and UY-18 grouped in the clade VT with the NZ-M16 sequence (VT-T3 recombinant); sample UY-26 grouped within the RB genotype, and the rest 72% of the Uruguayan sequences obtained were included in the New Clade (NC). Samples UY-20, UY-33, UY-8 and UY-11 were not included into any of the established genotypes.
Phylogenetic tree for CTV-p25 (a), p20 (b) and p23 (c) genes. Colored and highlighted branches represent CTV genotypes: T3 (yellow), T30 (green), T36 (violet), T68 (orange), VT (pink), RB (blue), NC (turquoise). Principal node aLRT values are indicated. Uruguayan samples are identified with the initials UY- number and were deposited in the Genbank with accession numbers: p20 from MH321249 to MH321271, p23 from MH321272 to MH321295, p25 from MH321296 to MH321320
Regarding to the p20 gene, were obtained 23 CTV isolate sequences of 411 nt in length (MH321249 to MH321271) and the corresponding phylogenetic tree showed seven clades (Fig. 2b). Samples UY-17 and UY-18 grouped with the VT clade; sequences UY-22 and UY-28 belong to genotype T3; 26% of the samples were included within the NC cluster; and the rest 56% of the sequences obtained were not assigned to any genotype.
In the case of the p23 gene, the phylogenetic tree was constructed using 24 Uruguayan CTV sequences of 528 nt in length (MH321272 to MH321295). Six clusters were observed (Fig. 2c). In this analysis the T68 genotype was not observed as a monophyletic lineage. The 50% of Uruguayan sequences were grouped into the VT/T68 clade; 33% of the samples were associated with reference sequence of the T3 genotype; and the sample UY-14 is within the RB clade. The sequences UY-5, 11 and 34 could not been assigned to any genotype.
Most of the isolates harbored a mixture of the CTV genotypes in their genetic composition, meaning that the sequences of the three analyzed genes belonging to a specific isolate were assigned to different genotypes (Table 2). UY-17 and UY-18 were the exception since their p25, p20 and p23 sequences were assigned to the VT genotype.
Discussion
Since the first report of CTV in Uruguay in 1955 (Koch de Brotos and Boasso), this is the first study that characterizes the aggressiveness and genetic variability of the prevailing isolates in the country. In addition to the high percentage of plants infected with CTV and the presence of T. citricida, this work confirmed the prevalence of severe isolates as well as the high genetic variability of the virus, indicating a complex scenario for the management of the disease.
The bioassays showed the induction of a wide range of symptoms in the different indicators plants used, a typical behavior of CTV infections causing diverse and complex syndromes that can differ markedly among isolates (Garnsey et al. 2005). Most of the characterized isolates produced symptoms such as SY, ST and SP in SO, SW and DG, which have been described associated with the infection by severe CTV isolates (Rocha-Peña et al. 1995; Roistacher et al. 2010). The production of intense SP indicates the presence of isolates capable of causing damage in sweet orange independently of the rootstock used (Rocha-Peña et al. 1995; Broadbent et al. 1996) in Uruguay. In addition, many of these isolates (with SP) caused severe ST, strong foliar chlorosis, growth restrictions and vein corking in various indicators plants, so it is feasible to assume that severe CTV isolates could be limiting the quality and production (Rocha-Peña et al. 1995; Roistacher et al. 2010) of the local citrus industry.
Despite of these results, there was no relationship between the CTV symptoms induced by each isolate on the indicator hosts, and those observed in the original field tree. Probably, this differential expression of symptoms in the indexing was influenced by optimal environmental conditions to which the isolate was exposed and by the highest susceptibility of the indicators (Garnsey et al. 1987) used (e.g. MV) against the commercial field sweet orange varieties (e.g. Valencia and Navel). Nevertheless, most isolates collected from trees without vigor, or with SP and small fruit, showed moderate or severe SP in DG and / or SW and SY in SO, confirming their infection with severe strains of this virus. The results demonstrated the predominance of severe isolates of CTV in the Northern and Southern citrus regions of Uruguay, collected either in 1998 or 2014. In this situation, probably the presence of T. citricida contributed to the prevalence and distribution of the most severe variants of the virus (Rocha-Peña et al. 1995; Broadbent et al. 1996; Gottwald et al. 1996). However, we also found some mild isolates (UY-12 and UY-13), with desirable characteristics such as mild VC, LC or SY and without SP in SW, to be used in cross protection.
The study of genetic diversity of the three spanned regions allowed us to confirm the highly heterogeneous populations in Uruguayan CTV isolates, as previously described by Benítez-Galeano et al. (2015). Five (T3, T68, VT, RB and NC) out the seven CTV described genotypes (Harper 2013; Dawson et al. 2015) were detected and indeed, 30 out the 32 characterized isolates were composed by more than one genotype. This genetic diversity could be explained by the constant flow of citrus propagation material (i.e. plants, bud woods) in Uruguay for several decades before the implementation of the National Sanitation and Certification Program. The detection of mixtures of the T3, T68, VT, RB and NC genotypes in the samples collected in 1998 from 15 to 20-year-old trees, suggested that these CTV strains are present in mixed infections in the Uruguayan citrus orchards at least since 40 years ago.
The genetic variability of each isolate (considering the three analyzed genes) is generally related to the presence of a complex population in the sampled tree composed by several CTV strains (Hilf et al. 1999; Iglesias et al. 2008), a foreseeable phenomenon since plants are continuously exposed to re-infections by the vector in the fields. Besides, the co-existence of multiple CTV genotypes is a direct cause of the genetic variability, since recombination among viral genomes has been described as one of the main mechanisms for the quasispecies generation forming the isolates (Rubio et al. 2001; Weng et al. 2007; Harper 2013). Moreover, for genotypes found in this study (VT, T3, T68, RB and NC), recombination events have been reported (Harper 2013; Benítez-Galeano et al. 2018). In fact, our data showed that samples included within the VT clade in the three phylogenetic trees, always clustered closer to the sequence NZ-M16, which indeed is a recombinant among VT and T3 genotypes. In addition, several sequences could not be included into any of the established genotypes.
In our survey, most of the isolates were assigned to T3, T68 and VT genotypes (p20 and p23) and induced severe symptoms in the biological assays. Although the presence of a CTV genotype has not been associated with the intensity in symptoms induced (Harper 2013), isolates of these strains are frequently related with the expression of severe symptoms such as stem pitting (Sambade et al. 2003; Biswas et al. 2012). In addition, the T30 genotype was not detected among the characterized isolates, strain that have been associated with mild symptoms (Sambade et al. 2003). One of the frequent genotypes was NC, and due to its recent finding (Harper 2013; Benítez-Galeano et al. 2015, 2017) a described symptomatology is not available yet; noteworthy is that our study reveals the presence of this genotype in 20-year old samples. Several regional isolates grouped also in NC clade for the p25 gene (Iglesias et al. 2008; Benítez-Galeano et al. 2015) and were distant from other reference genotypes, suggesting a phylogenetic analogy of the virus within the region. Finally, in low frequency, sequences of RB genotype were found in isolates collected in 1997 and 2014. The 1997’s samples were kept in a greenhouse without being exposed to aphid re-inoculations producing genetic exchange, therefore we can conclude that the RB genotype has been circulating in the country for a long time before its first detection (Hernández-Rodríguez et al. 2017). This genotype is the only that breaks the natural resistance of P. trifoliata (Dawson and Money 2000; Harper et al. 2010), the main citrus rootstock in Uruguay.
Conclusions
Our results revealed the high variability of the CTV isolates in Uruguay. Regardless of the geographical growing region or citrus species, this work evidenced the prevalence of severe CTV isolates; the presence, in low frequency, of the RB genotype capable to overcome P. trifoliata resistance our main rootstock; and we also identified mild CTV strains. Taking together our findings, we can conclude that conditions exist to implement a cross-protection program, preventing healthy plants of being re-infested with severe CTV isolates. Therefore, some of the mild CTV isolates identified in this work will be challenged with severe strains in future works.
References
Akaike H (1974) A new look at the statistical model identification. IEEE Trans Autom Control 19:716–723
Albiach-Marti M, Mawassi M, Gowda S, Satyanarayana T, Hilf M, Shanker S, Almira E, Vives M, Lopez C, Guerri J, Flores R, Moreno P, Garnsey S, Dawson W (2000) Sequences of Citrus tristeza virus separated in time and space are essentially identical. J Virol 74:6856–6865
Anisimova M, Gascuel O (2006) Approximate likelihood-ratio test for branches: a fast, accurate, and powerful alternative. Syst Biol 55:539–552
Bar-Joseph M, Lee R (1989) The continuous challenge of Citrus tristeza virus control. Annu Rev Phytopathol 27:291–316
Benítez-Galeano M, Rubio L, Bertalmío A, Maeso D, Rivas F, Colina R (2015) Phylogenetic studies of the three RNA silencing suppressor genes of south American CTV isolates reveal the circulation of a novel genetic lineage. Viruses 7:4152–4168
Benítez-Galeano MJ, Castells M, Colina R (2017) The evolutionary history and spatiotemporal dynamics of the NC lineage of Citrus tristeza virus. Viruses 9:272. https://doi.org/10.3390/v9100272
Benítez-Galeano M, Vallet T, Carrau L, Hernández-Rodriguez L, Bertalmío A, Rivas F, Rubio L, Maeso D, Vignuzzi M, Moratorio G, Colina R (2018) Complete genome sequence of a novel recombinant Citrus tristeza virus: a resistance-breaking isolate from Uruguay. Genome Announc 6:e00442–e00418. https://doi.org/10.1128/genomeA.00442-18
Bentancour C, Scatoni I, Morelli E (2009) Insectos del Uruguay. Universidad de la República, Facultad de Agronomía, Facultad de Ciencias, 658p
Biswas K, Tarafdar A, Sharma S (2012) Complete genome sequence of mandarine decline Citrus tristeza virus of the northeastern Himalaya hill region if India: comparative analyses determine recombinant. Arch Virol 157:579–583
Broadbent P, Bevington KB, Coote BG (1991) Control of stem pitting of grapefruit in Australia by mild strain cross protection. In: Brlansky R, Lee R, Timmer L (eds) Proceedings of 11th conference of the International Organization of Citrus Virologists. IOCV, Riverside CA, pp 64–70
Broadbent P, Brlansky R, Indsto J (1996) Biological characterization of Australian isolates of Citrus tristeza virus and separation of subisolates by single aphid transmissions. Plant Dis 80:329–333
Costa A, Nunes W, Corazza M, Zanutto C, Müller G (2010) Biological and molecular characterization of isolates of Citrus tristeza virus with potential for use in preimmunization programs. Summa Phytopathol 36:81–82
Da Graça J, van Vuuren S (2010) Managing Citrus tristeza virus losses using cross protection. In: Citrus tristeza virus complex and Tristeza diseases. APS Press, St. Paul MN, pp 247–260
Dawson T, Money P (2000) Evidence for trifoliate resistence breaking isolates of Citrus tristeza virus in New Zealand. In: da Graca J, Lee R, Yokomi R (eds) Proceedings of the 14th conference of the International Organization of Citrus Virologists. IOCV, Riverside, CA, pp 69–76
Dawson WO, Bar-Joseph M, Garnsey SM, Moreno P (2015) Citrus tristeza virus: making an ally from an enemy. Annu Rev Phytopathol 53:37–55
de Koch Brotos L, Boasso C (1955) Lista de las enfermedades de los vegetales en el Uruguay. Ministerio de Ganadería y Agricultura. Dirección de Agronomía. Laboratorio de Fisiología y Patología Vegetal. Publicación N° 106, 65p
Folimonova S (2013) Developing an understanding of cross-protection by Citrus tristeza virus. Front Microbiol 4(75):1–9
Francis M, Peyrou M, Perea B, Borde J, Fosali Y (1997) Caracterización de aislamientos del virus de la Tristeza de los cítricos (CTV) que causan pérdidas en pomelo injertado sobre Trifolia en Uruguay. In: IX Congreso Latinoamericano de Fitopatología.10/97. Montevideo, Uruguay
Garnsey S, Gumpf D, Roistacher C, Civerolo E, Lee R, Yokomi R, Bar-Joseph M (1987) Toward standarized evaluation of the biological properties of Citrus tristeza virus. Phytophylactica 19:151–157
Garnsey M, Civerolo E, Gumpf D, Paul C, Hilf M, Lee R, Brlansky R, Yokomi R, Hartung J (2005) Biological characterization of an international collection of Citrus tristeza virus (CTV) isolates. In: Hilf M, Duran-Vila N, Rocha-Peña M (eds) Proceedings of the 16th conference of International Organization of Citrus Virologists. IOCV, Riverside, CA, pp 75–93
Gonsalves D, Garnsey S (1989) Cross-protection techniques for control of plant virus diseases in the tropics. Plant Dis 73:592–597
Gottwald T, Garnsey S, Cambra M, Moreno P, Irey M, Borbón J (1996) Differential effects of Toxoptera citricida vs. Aphis gossypii on temporal increase and spatial patterns of spread of citrus tristeza. In: Moreno P, da Graca J, Timmer L (eds) Proceedings of the 13th Conference of International Organization of Citrus Virologists. IOCV, Riverside, pp 120–129
Guindon S, Dufayard J, Lefort V, Anisimova M, Hordijk W, Gascuel O (2010) New algorithms and methods to estimate maximum-likelihood phylogenies: 3.0. Syst Biol 59:307–321
Harper S (2013) Citrus tristeza virus: evolution of complex and varied genotypic groups. Front Microbiol 4(93):1–18
Harper S, Dawson T, Pearson M (2010) Isolates of Citrus tristeza virus that overcome Poncirus trifoliata resistance comprise a novel strain. Arch Virol 155:471–480
Hernández-Rodríguez L, Bertalmío A, Arruabarrena A, Rubio L, Rivas F, Benítez-Galeano MJ, Colina R, Maeso D (2017) First report of the Citrus tristeza virus trifoliate resistance-breaking (RB) genotype in 'Newhall' sweet Orange in South America. Plant Dis 101:1063
Hilf M, Karasev A, Albiach-Marti M, Dawson W, Garnsey S (1999) Two paths of sequence divergence in the Citrus tristeza virus complex. Phytopathology 89:336–342
Iglesias NG, Gago-Zachert SP, Robledo G, Costa N, Plata MI, Vera O, Grau O, Semorile LC (2008) Population structure of Citrus tristeza virus from field Argentinean isolates. Virus Genes 36:199–207
Koizumi M (1991) Citrus tristeza virus field isolates from declined of dwarfed citrus trees in Japan. In: Brlansky R, Lee R, Timmer L (eds) Proceedings of the 11th conference of the International Organization of Citrus Virologists. IOCV, Riverside, pp 25–30
Moreno P, Ambros S, Albiach-Marti M, Guerri J, Peña L (2008) Plant diseases that changed the world Citrus tristeza virus: a pathogen that changed the course of the citrus industry. Mol Plant Pathol 9:251–268
Müller IA and Campiglia HG (1981) Nivel de "stem pitting" (SP) en naranja 'Valencia' (Citrus sinensis (L) Obs.) y tangor 'Ellendale' (C. reticulata blanco x C. sinensis (L) Obs.) injertados en diferentes portainjertos. In Müller IA, ed. Investigaciones agronómicas Año 2 (1), pp. 79–82. Uruguay
Müller G, Costa A, Castro J, Guirado N (1988) Results from preimmunization tests to control the Capao Bonito strain of tristeza. In: Timmer L, Garnsey S, Navarro L (eds) Proceeding of the 10th conference of the International Organization of Citrus Virologists. IOCV, Riverside, CA, pp 82–85
Posada D (2008) jModelTest: phylogenetic model averaging. Mol Biol Evol 25:1253–1256
Rocha-Peña M, Lee R, Lastra R, Niblett C, Ochoa-Corona F, Garnsey S, Yokomi R (1995) Citrus tristeza virus and its aphid vector Toxoptera citricida: threats to citrus production in the Caribbean and central and North America. Plant Dis 79:437–443
Roistacher C (1988) Observations on the decline of sweet orange trees in coastal Peru caused by stem-pitting tristeza. Plant protection bulletin India 36:19–26
Roistacher C, Bar-Joseph M (1987a) Transmission of Citrus tristeza virus (CTV) by Aphis gossypii and by graft inoculation to and from Passiflora spp. Phytophylactica 19:179–182
Roistacher C, Bar-Joseph M (1987b) Aphid transmission of tristeza virus: a review. Phytophylactica 19:163–167
Roistacher C, da Graça J, Müller G (2010) Cross protection against Citrus Tristeza Virus a review. In: Brlansky R, Lee R, Timmer L (eds) Proceedings of the 17th conference of the International Organization of Citrus Virologists. IOCV, Riverside, CA, pp 1–27
Rubio L, Ayllon M, Kong P, Fernandez A, Polek ML, Guerri J, Moreno P, Falk B (2001) Genetic variation of Citrus tristeza virus isolates from California and Spain: evidence for mixed infections and recombination. J Virol 75:8054–8062
Sambade A, Lopez C, Rubio L, Flores R, Guerri J, Moreno P (2003) Polymorphism of a specific region in gene p23 of Citrus tristeza virus allows discrimination between mild and severe isolates. Arch Virol 148:2325–2340
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729
Weng Z, Barthelson R, Gowda S, Hilf ME, Dawson WO, Galbraith DW et al (2007) Persistent infection and promiscuous recombination of multiple genotypes of an RNA virus within a single host generate extensive diversity. PLoS One 2(9):e917
Yokomi R, Selvaraj V, Maheshwari Y, Saponari M, Giampetruzzi A, Chiumenti M, Hajeri S (2017) Identification and characterization of Citrus tristeza virus isolates breaking resistance in trifoliate orange in California. Phytopathology 107:901–908
Acknowledgements
This research was funded by Instituto Nacional de Investigación Agropecuaria, Uruguay (project CT-06), through the National Program of Citrus, in collaboration with the Laboratorio de Virología Molecular, Centro Universitario Regional Litoral Norte de la Universidad de la República.
Funding
This study was funded by Instituto Nacional de Investigación Agropecuaria, Uruguay (project CT-06).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflicts of interest.
Ethical approval
The authors declare no animals were used in this research.
Rights and permissions
About this article
Cite this article
Rubio, L., Bertalmío, A., Hernández-Rodríguez, L. et al. Biological and molecular characterization of Uruguayan citrus tristeza virus field isolates. J Plant Pathol 101, 97–105 (2019). https://doi.org/10.1007/s42161-018-0149-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s42161-018-0149-0