Abstract
The black rockfish Sebastes schlegelii, a valuable commercial fish, is one of the most important fish species for marine fishery stock enhancement in China. In this study, genetic diversity of the hatchery-released stock and two restored-natural stocks (SSNAC1 and SSNAC2) of S. schlegelii from Rongcheng Bay, Yellow Sea, China was analyzed using 15 microsatellite DNA loci, and the genetic differentiation between natural and hatchery-released stocks was also investigated. The mean effective number of alleles (Ae) in the hatchery-released stock was 3.6, the observed heterozygosity (Ho) was 0.506, and the expected heterozygosity (He) was 0.606, while the Ae, Ho and He values in the SSNAC1 and SSNAC2 stocks were 2.9 and 3.9, 0.381 and 0.577, and 0.569 and 0.692, respectively. The FST values between the hatchery-released and natural stock and the natural stocks before and after restocking were 0.055 and 0.013, respectively. The results showed that there was low differentiation between the natural and hatchery-released stocks even though the hatchery stock displayed slightly higher genetic diversity and heterozygosity than the natural stock. Furthermore, there was no differentiation on genetic diversity indices between the S. schlegelii natural stock before release and the mixed stocks after release in the short term. However, larger differentiation was found between the natural stock and restored-natural stock SSNAC2. Consequently, more detailed surveys are required to elucidate the long-term genetic effects of the stock enhancement. The present study would be helpful for the future stock enhancement strategy of black rockfish.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Demand for seafood will continue to rise until 2050 as a result of population growth. Nevertheless, global fish stocks have steadily declined since the 1980s through factors such as over-fishing, habitat degradation, and global warming (Botsford et al. 1997; Nellemann 2009). Nowadays, the stocks of many fish species have become depleted. Data show that 35.4% of fish stocks were estimated to be overfished (FAO 2022). Fisheries will not easily improve even if we reduce fishing and instate better management. The release of hatchery-reared juveniles into the wild has been used as a tool to recover marine fish stocks along with the augmentation of an existing fishery to enable larger catches to be taken (Blankenship and Leber 1995; Welcomme and Bartley 1998; Leber 2004; Bell et al. 2005, 2008). These hatchery-reared juveniles were released into the wild for three distinct purposes: restocking, stock enhancement, and sea ranching (Bell et al. 2008). Then, more and more species and mass of marine organisms have been released into the wild in recent years. More than 190 billion aquatic animals were released nationwide between 2016 and 2020 in China (Lin et al. 2023). However, the pros and cons of this approach have been widely debated (Blaxter 2000; Hedgecock and Coykendall 2007; Lorenzen et al. 2012; Camp et al. 2017).
There are growing concerns about the genetic disturbance to natural populations with stock enhancement activities (Kitada 2018). Most of the individuals used for stock enhancement came from the cultured stock. As the broodstock individuals used to produce hatchery-released juveniles are usually from limited numbers, genetic diversity often decreases, resulting in observed genetic differences. The farmed stock often differs genetically from its natural counterpart, showing reduced genetic diversity (Champagnon et al. 2012; Lorenzen et al. 2012). Application of hatchery individuals to reverse the decline of marine fisheries is likely to reduce the genetic diversity of the mixed stock after release, and may even erode the long-term genetic diversity of the wild stock through mating with wild fish (Hedgecock and Coykendall 2007). Therefore, stock enhancement activities can reduce the overall amount of genetic variation in the stocked species, leading to a loss of adaptive potential (Christie et al. 2012). Negative genetic impacts of stock enhancement have been documented in some fish species (Laikre et al. 2010; Hagen et al. 2021). The long-term extensive hatchery release program in red sea bream (Pagrus major) has affected the genetic diversity of wild populations, even if the genetic effects were low and appeared to remain within Kagoshima Bay (Hamasaki et al. 2010). Romo et al. (2006) found that there were significant pairwise differences between the wild captive broodstock and the tentative recaptured groups in spotted halibut (Verasper variegatus) based on the FST estimator. Although the genetic risks associated with these releases are well known (Bell et al. 2005; Lorenzen et al. 2012), they have rarely been monitored (Laikre et al. 2010). Thus, the consequences of genetic interactions between hatchery-reared and wild individuals remain poorly understood and are of great concern in conservation genetics. For each released fish species, the genetic variability of the released stock and the genetic divergence of the restored stock after release should be monitored to elucidate the genetic effects of such activities on wild stocks and ensure a responsible stock enhancement program (Romo et al. 2006; Ward 2006; Lorenzen et al. 2012).
The black rockfish (Sebastes schlegelii) is an important commercial fish inhabiting the coasts of Asia and is considered a promising target fish species for stock enhancement. In Japan (Yoshida et al. 2005) and Korea (An et al. 2009), stock enhancement programs for this species started in the 1980s and mid-1990s, respectively. In China, hatchery-reared juveniles have been released into coastal waters every year since 1995 to recover the wild resources (Wang et al. 2020). According to incomplete statistics, a total of 3.86 million seeds were released in the coastal region of Shandong Province, China in 2007 and 2008, 3.85 million seeds in 2010, and approximately 1 million juveniles in 2012 (Lü et al. 2014). And from 2015 to 2019, over 30 million S. schlegelii were released annually in Shandong (Liu et al. 2022). Although there are a few papers reported on the population genetic diversity and differentiation of this species (Yoshida et al. 2005; An et al. 2009; Zhang et al. 2016), the genetic variability of released rockfish and recipient stocks has only been examined by Wang et al. (2020) and Liu et al. (2022). In 2009, more than 100 thousand hatchery-reared juvenile black rockfish were released into the sea of Rongcheng Bay, Shandong Province, China, as part of the National Special Research Fund for Non-Profit Marine Sector project. The present study examines the genetic variability of the hatchery-released stock in Rongcheng Bay as well as the divergence between the natural and the mixed restored-natural stocks. The aim of the study is to assess the potential genetic impact of hatchery-reared fish on the wild population. This work will provide basic data to ensure a healthy and sustainable restoration of S. schlegelii in Rongcheng Bay. The genetic data of local stocks will establish a baseline that could provide valuable suggestions for future fish stock enhancement strategy.
Materials and Methods
Sample Collection and DNA Extraction
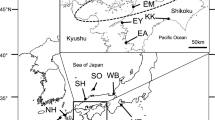
Mature female spawning fish were collected from the natural coastal habitat of Yantai City, Shandong, China, adjacent to Rongcheng Bay, and then were transferred to an aquaculture farm (Taihua Marine Technology Co., Ltd, Yantai, Shandong, China). After the female fish spawned in May 2009, the hatchery-reared individuals were reared until release. The hatchery-reared S. schlegelii individuals were released into Rongcheng Bay in Oct. 2009. Before stock enhancement began, 60 individual specimens were collected from the hatchery stock for release (SSR). These individuals were stored at -20℃ after anesthesia with tricaine methanesulfonate (MS222). Approximately 1 and 1.5 years after the release, 2 restored-natural stock samples (SSNAC1 and SSNAC2) were collected using motorized bottom trawls and cage nets from same location in Sept. and Nov. 2010 (n = 60) and Feb. and May 2011 (n = 60), respectively. The sampling locations (122º39′11″-122º46′16″E, 37º21′32″-37º13′41″N) are shown in Fig. 1. The 52 natural individuals, captured in Rongcheng Bay in May and Aug. 2009, as reported in our previous study (Wang et al. 2012), refer to the natural population (SSNB).
Muscle tissue of the dorsal region of the fish was excised from each specimen. Total genomic DNA was extracted from the tissue samples using rapid salt-extraction of genomic DNA according to Aljanabi and Martinez (1997). The extracted DNA was checked using 1% agarose gel electrophoresis and then stored at -20 °C for PCR amplification. The study was approved by the Ethics Committee of the Institute of Oceanology, Chinese Academy of Sciences.
Microsatellite Analysis
A total of 15 previously published microsatellite loci of S. schlegelii: KSs2A, KSs3, KSs6, KSs7, KSs11B, KSs12B, KSs17, KSs18A, KSs20, KSs26 (An et al. 2009) and Ssc12, Ssc23, Ssc51, Ssc69, Ssc71 (Yoshida et al. 2005) were selected for analysis. The primers were synthesized by Sangon Biological Engineering Technology & Services CO. Ltd (Shanghai, China).
PCR were carried out in 25 µL volume comprising 1x PCR buffer [10 mmol/L Tris-HCl (pH 9.0) and 50 mmol/L KCl], 100 ng template DNA, 1.5 mmol/L MgCl2, 100 µmol/L dNTPs, 10 pmol/L of each forward and reverse primers, and 1 U of Taq DNA polymerase (Promega, USA). The thermal cycling conditions for each locus were as follows: 7 min at 94℃, followed by 35 cycles of 94 °C for 30 s, annealing for 45 s at the specific annealing temperature of the primers and extension at 72 °C for 45 s, with a final extension temperature of 72 °C for 10 min. Details of the primer sequences and specific annealing temperatures are available in An et al. (2009) and Yoshida et al. (2005). The PCR products were added to an equal volume of loading buffer. Three microliters of each PCR product mixture were electrophoresed on a 12% nondenaturing polyacrylamide gel for 3 h at 300 V with a DYCZ-24B vertical electrophoresis tank and a DYY-6B constant voltage electrophoresis instrument (Liuyi Instrument Factory, Beijing, China). The DNA fragments were visualized using the modified silver staining method (Xu et al. 2002). Alleles were designated according to the PCR product size relative to a molecular size marker (M1041 50-500 bp, Dongsheng Biotech, China) in combination with Quantity One software (Bio-Rad). Minimum 2%, blindly scored, and sample repeats were performed within and across plates to reduce genotyping errors. Reruns were conducted for approximately 30% of all individuals to ensure reproducibility of allele scoring, and these replicates yielded identical results for each trial.
Microsatellite DNA data from fifty-two S. schlegelii individuals as SSNB were taken from our previous work (Wang et al. 2012).
Data Analysis
Measurements of genetic diversity within stock: standard genetic diversity parameters, including number of alleles (A), effective alleles (Ae), observed heterozygosity (Ho), and expected heterozygosity (He) were calculated using GenAlEx 6.5 (Peakall and Smouse 2012). Allelic richness (Ar) was calculated by using the rarefaction method based on a minimum sample size of 52 individuals with FSTAT v.2.9.3.2 (Goudet 2001). Deviations from linkage disequilibrium and Hardy-Weinberg equilibrium (HWE) for each microsatellite locus were tested by using the GENEPOP v.4.0 package (Saitoh and Rousset 2008), and significance was adjusted by applying the sequential Bonferroni correction. The polymorphic information content (PIC) was calculated according to Botstein et al. (1980) using the program Cervus v.3.0.6 (Kalinowski et al. 2007). ADZE v.1.0 (Szpiech et al. 2008) was used to determine the number of private alleles with standardized sample sizes. Micro-Checker software (Van Oosterhout et al. 2004) was used to search for null alleles at loci.
Measurements of genetic differentiation among stocks: estimation of Nei’s unbiased genetic distance (Nei 1978), genetic identity, and the Principal component analysis (PCA) of S. schlegelii samples were performed by using GenAlEx 6.5. Population structure was analyzed using the model-based Bayesian clustering method in STRUCTURE v.2.3.4 (Pritchard et al. 2000; Falush et al. 2003), which assigned individuals to K populations. STRUCTURE was run for K = 1–4 with a burn-in length of 100,000 and a run of 500,000 steps. All runs were repeated in triplicate for each K and the results were consistent across runs. The most probable K value was inferred by calculating ΔK (Evanno et al. 2005). To assess the extent of differentiation within and among stocks, pairwise FST was determined as an estimator of inbreeding using FSTAT v.2.9.3.2 (Goudet 2001) with the analysis of variance method of Weir and Cockerham (1984). The Markov chain method (10,000 dememorization steps, 100 batches, 5000 iterations) was utilized to obtain unbiased estimates of the exact P-value. SMOGD v. 1.2.5 (Crawford 2010) was used to calculate the GST (recommended by Jost 2008) in order to validate the FST results (bootstrap replicates = 1000).
Estimations of effective population size of S. schlegelii stocks were performed in NeEstimator v. 1.3 (Do et al. 2014) by using the linkage disequilibrium and heterozygote excess methods. Bottleneck 1.2 software (Cornuet and Luikart 1996) was used to detect the presence of bottlenecks using the infinite allele model (IAM), stepwise-mutation model (SMM), and two-phased model of mutation (TPM) (Luikart and Cornuet 1998). The significant heterozygosity excess was tested using the Wilcoxon signed-rank test. Estimation of full- and half-sib family pairs in 3 S. schlegelii stocks was performed in kingroup v.2 (Konovalov et al. 2004). Here, 2 restored-natural stocks from Rongcheng Bay (SSNAC1 and SSNAC2) were combined into one stock (SSNAC) to be compared with the SSR and SSNB stocks.
Results
Population Genetic Variation
The measures of genetic diversity calculated from the observed allele distribution for each stock are presented in Table 1. In the studied stocks, the average number of alleles per stock per locus varied from 4.7 to 6.0. The average effective allelic numbers (Ae) and allelic richness values (Ar) ranged from 2.9 to 3.9 and 4.7 to 5.9, respectively. The overall observed heterozygosity (Ho) was lower than expected (He), indicating heterozygote deficiency. Null alleles were detected at loci Ssc12, Ssc51, Ssc69, KSs11, KSs12, KSs17, and KSs18. The values of the expected heterozygosity ranged from 0.569 to 0.692. And the lowest average expected heterozygosity was in the SSNAC1 stock (0.569), while the highest was in the SSNAC2 stock (0.692). The mean observed heterozygosity values showed similar results. The lowest value was in the SSNAC1 stock (0.381), whereas the highest was in the SSNAC2 stock (0.577). Most microsatellite markers in this study were unconfirmed to HWE. In which, the heterozygote of Ssc71 was excess and the heterozygotes of the others were deficient. Overall, the hatchery SSR stock presented a slightly higher genetic diversity than the natural SSNB stock. And the mixed stock SSNAC1 exhibited almost similar genetic diversity to the SSNB stock before release.
The number of private alleles per stock when the sample size is standardized was different. The value in the SSR stock was 0.5, whereas the value of the SSNB stock was considerably lower at 0.07. Among the stocks examined, the SSRAC2 stock achieved the highest value of 0.8, whereas the SSRAC1 stock exhibited the lowest value of 0.01 (Fig. 2). The analyzed stocks contained a total of 23 private alleles, of which one was exclusively present in the SSNB stock, nine exclusively in the SSR stock, and thirteen exclusively in the SSNAC2 stock.
Population Structure and Gene Flow among Stocks
The results of the exact testing of the genetic differentiation between population pairs revealed that there was a clear genetic differentiation in these studied stocks (Table 2). The genetic differentiation between the SSNB and SSR was relatively low (FST = 0.055), and the lowest genetic divergence was observed between SSNAC1 and SSNB (FST = 0.013). However, the FST between SSNAC2 and SSNB (FST = 0.152) was much larger than that between SSNAC1 and SSNB. The results also showed that the largest differentiation was observed between the two restored-natural stocks SSNAC1 and SSNAC2 (FST = 0.167). The GST values, genetic similarity, and genetic distance values presented similar results (Table 2; Fig. 3).
Furthermore, the scatter plots of PCA (Fig. 4) clearly revealed three main clusters (i.e. SSNB-SSNAC1, SSR, and SSNAC2). Similar to the results of the FST analysis, the PCA results showed that the SSNAC1 stock was not genetically distinct from the SSNB stock, and the two restored-natural stocks were genetically distinct from each other. According to ΔK, the optimal value for K was determined as K = 2 (Fig. 5). This result was consistent across all replicate runs of the analysis. STRUCTURE estimated that the samples from the SSNB, SSR, and SSNAC1 stocks derived on about 97%, 96%, and 95% of their ancestry from the group one, respectively, while the SSNAC2 samples derived only 98% of their ancestry from the group two. Analysis of molecular variance (AMOVA) showed that most of the genetic variance occurred among individuals within stocks, providing 88.98% variation, while only 11.02% of the variance occurred between stocks.
The estimation of the effective population size (Ne) using the linkage disequilibrium method indicated that the Ne value was the highest in the SSNB, while of the values were lower in the SSNAC1 and SSNAC2. However, the Ne value in the SSR could not be calculated (Table 3). The estimates for all stocks were infinite by using heterozygote excess method.
The IAM, TPM, and SMM models were applied to test for population bottlenecks. The results are shown in Table 4. The sign, standardized difference, and Wilcoxon tests all showed significant heterozygosity excess in all stocks under the IAM model (P < 0.05). The Wilcoxon test, which is considered more reliable than the sign test and the standardized difference test, showed significant results for population bottleneck under the IAM and TPM models in all stocks. But under the SMM model, only the SSNAC2 stock showed a significant result. Furthermore, a mode shift was detected in the frequency distribution of alleles, resulting in a normal L-shaped curve.
The results of estimating full-sib family pairs in the S. schlegelii stocks showed that the ratio of full-sib and half-sib pairs was the lowest between SSR and SSNAC, while it was the highest between SSNB and SSNAC. However, all the ratios were relatively low. There were 509 full-sib family pairs between the SSNAC and SSNB stocks, accounting for 8.2% of the total. Between the SSNAC and SSR stocks, there were 188 full-sib family pairs, representing 2.6% of the total (Tables 5 and 6).
Discussion
Nowadays, the growing demand for seafood can no longer be met by natural captures alone. Consequently, many stock enhancement programs have been developed to augment the biomass of target species. It is well recognized that enhancement programs should be coupled with habitat restoration and should attempt to ensure the greatest possible genetic diversity (McCay and Rowe 2003). The maintenance of genetic diversity affects the ability of a population to survive (Schindler et al. 2010). Loss of genetic variation affects adaptive potential, i.e. the ability of wild populations to respond to ecological challenges. Ideally, these enhancement programs should be achieved with little or no negative impact on the local gene pool. Several strategies have been proposed to minimize genetic alterations and reduce the loss of genetic diversity. To prevent rapid genetic shifts in the hatchery population, it is recommended to use native individuals that are genetically compatible with the individuals at the release site as broodstock (Grant et al. 2017). Small broodstock sizes can rapidly alter the genetic composition of the hatchery population. Therefore, using a large number of broodstock to generate hatchery populations is an effective approach for preserving genetic diversity (Kitada et al. 2009). In addition, brood stock must be derived from naturally produced individuals rather than hatchery-reared individuals to avoid the accumulation of domestication effects over generations (Willoughby and Christie 2019). Regrettably, most hatchery restocking results in a decline in combined wild and hatchery populations (Grant et al. 2017). The negative effects on the genetic diversity of recipient populations caused by large-scale releases have been documented in many plant and animal species (Laikre et al. 2010), such as Acacia saligna lindleyi (Millar and Byrne 2007), eucalypt (Barbour et al. 2008), red drum (Sciaenops ocellatus) (Gold et al. 2008), and steelhead trout (Oncorhynchus mykiss) (Araki et al. 2007). However, despite the genetic risks of stock enhancement programs have long been recognized, they have rarely been monitored appropriately and effectively (Laikre et al. 2010). Such is the case with the stock enhancement program of S. schlegelii. The program has been conducted for more than 30 years, but there is a lack of studies evaluating its genetic impact.
In this study, significant deviations from HWE were detected in all the S. schlegelii stocks analyzed, even after sequential Bonferroni correction. These departures may be mainly caused by heterozygote deficiencies. Thirteen significant HWE deviation loci show heterozygote deficiencies, while only 1 shows heterozygote excess. In wild stocks, heterozygote deficiencies could have several causes, such as the decreases of wild population size, inbreeding, or possibly assortative mating due to repeated interbreeding between the natural population and mass-released hatchery strains (Marchant et al. 2009; An et al. 2014). As for hatchery stocks, non-random mating, the limited number of founders, inbreeding, or artificial selection forces during cultivation might result in heterozygote deficiency (Kohlmann et al. 2005; An et al. 2014). Null alleles are also likely to be an important explanation for the deviation observed in marine fish (Dick et al. 2014) and could potentially be responsible for heterozygote deficiency. In our study, the presence of null alleles was also observed at some loci. All the above mechanisms may have contributed to the heterozygote deficiencies observed in the S. schlegelii stocks.
Genetic evaluations have been recommended for both natural stocks and hatchery-released stocks before and after release to assess the genetic impact of the subsequent stock enhancement. This is necessary to describe the genetic structure of the natural stock to determine the most suitable broodstock to provide hatchery-reared juveniles for stock enhancement. In general, the number of broodstock used to produce hatchery-released juveniles is limited due to the constraints imposed by hatchery techniques. Maintenance of broodstock in hatcheries leading to hatchery progeny usually yields low genetic diversity compared to the natural population (Ryman and Laikre 1991; Alarcón et al. 2004). However, unlike other farmed fish species, no deep depression was observed in the hatchery stock of S. schlegelii in this study. The hatchery-released stock (SSR, A = 6.0, Ae = 3.6, Ar = 5.9, PIC = 0.565, Ho = 0.506, He = 0.606) displayed slightly higher genetic diversity and heterozygosity compared to the natural stock (SSNB, A = 5.2, Ae = 3.3, Ar = 5.2, PIC = 0.552, Ho = 0.414, He = 0.592). And the genetic differentiation between the natural and hatchery-released stocks is relatively low (FST = 0.055). One possible reason for the high genetic diversity and limited genetic differentiation between the natural and SSR stocks was the relatively short domestication history of S. schlegelii in China. Another reason was that the broodstock amount in the artificial selective breeding of the black rockfish was large due to its viviparous reproduction. Overall, the genetic diversity indices of the S. schlegelii stocks in this study did not differ significantly (average allele A varied from 4.7 to 6.0, mean average He varied from 0.569 to 0.692) and are lower than those reported for the Korean coastal stocks (average allele A = 20.1; average He = 0.81) (An et al. 2012). The variations are also lower than other Sebastes species and other marine fish species (average allele A = 19.96 ± 6.6, average He = 0.77 ± 0.19, 12 species) (DeWoody and Avise 2000). This may be due to the limited sample size (n = 60 in each stock) and nondenaturing PAGE method, which reduced the possibility of detecting more novel and rare alleles.
After release, the contribution of the released individuals to the gene pool of the mixed stock and the changes in genetic structure should also be assessed. In this study, the genetic effects of stock enhancement for the natural stock were examined. The results showed that two mixed stocks after the release of S. schlegelii (SSNAC1 and SSNAC2) exhibited similar average genetic diversity, although slightly lower in SSNAC1 and slightly higher in SSNAC2, compared to the natural stock before the release (SSNB) (Table 1). No significant difference in average PIC (SSNB = 0.553, SSNAC1 = 0.528) or average He (SSNB = 0.592, SSNAC1 = 0.569) was observed when comparing the diversity level of the natural stock with that of the restored-natural stock SSNAC1. Both the exact tests of genetic differentiation (Table 2) and FST (0.013) indicate that the SSNB and SSNAC1 stocks are genetically closely related. AMOVA also indicated that most of the variation in microsatellite alleles of these two stocks is explained within stocks and not between stocks. Moreover, the results of phylogenetic tree clustering and PCA analyses showed weak genetic subdivision of the SSNB and SSNAC1 stocks. All the above results demonstrated that genetic variation in the natural stock did not decrease after the stock enhancement and that stock enhancement did not have a negative genetic effect on the natural population of S. schlegelii in Rongcheng Bay in a short time. This could be due to the relatively high genetic diversity of the hatchery-released stock. To reduce the difference between the released offspring and the wild populations, it is optimal to select the broodstock for the hatchery-released stock from the native population at the release site. However, it is not feasible (Ward 2006) and animals are usually transported from other areas for release. Previous studies have shown that there is no genetic differentiation among populations in wild stocks of S. schlegelii (Wang et al. 2012; Zhang et al. 2016; Gao et al. 2018), which could minimize the genetic effect of using non-native individuals as broodstock. It is worth noting that the mean FST values between the SSNAC2 samples and the other two stock samples are much larger compared with SSNAC1, and the largest differentiation was observed between the two restored-natural stocks. The results showed that there was no obvious genetic impact on the wild stock 1 year after stock enhancement, but genetic divergence was observed 1.5 years after stock enhancement. This suggests that the hatchery-released individuals may have a lasting impact on the genetic structure of the mixed stock as time goes on. In this study, differentiation was found between the hatchery stock (SSR) and the wild stock (SSNB) (FST =0.055), suggesting that the hatchery-reared released population may influence the original population structure of the wild population. Differentiation between the hatchery-released and wild black rockfish stocks before and after enhancement were also reported in Lidao Bay (Wang et al. 2020) and Liaoning Peninsula (Liu et al. 2022), but their FST values were all low. Another possible reason for SSNAC2 differentiation is the behavior of S. schlegelii. Although it is a demersal rockfish that does not migrate over long distance, S. schlegelii can disperse with the complex oceanic current systems during its early developmental stage (Gao et al. 2018). Moreover, S. schlegelii moves seasonally over short distances. Therefore, the aggregation of rockfish populations may not be completely uniform in different years. Some new alleles could be introduced into the restored-natural stock. In addition, the Ne values of the SSNAC stocks after release in this study were much smaller than those of the wild stock in this study. In general, populations with smaller Ne values tend to be more inbred and improverished in terms of local genetic variability due to strong genetic drift, limiting evolutionary potential (Waples et al. 2016). Further study is required to assess the influence of the long-term effects of the stock enhancement.
This study examined the genetic variability of the hatchery-released and natural S. schlegelii stocks after artificial release in Rongcheng Bay, Yellow Sea, China. Our results showed that there was no marked decrease of genetic diversity indices in the natural stock of S. schlegelii in Rongcheng Bay after stock enhancement. More detailed surveys for elucidating the long term genetic effects of S. schlegelii stock enhancement are still required. Nevertheless, the evaluation of the genetic variability of S. schlegelii in this study may provide valuable insights into the strategy for wild stock management and stock enhancement in Rongcheng Bay.
References
Alarcón JA, Magoulas A, Georgakopoulos T, Zouros E, Alvarez MC (2004) Genetic comparison of wild and cultivated European populations of the gilt head sea bream (Sparus aurata). Aquaculture 230:65–80
Aljanabi SM, Martinez I (1997) Universal and rapid salt-extraction of high quality genomic DNA for PCR-based techniques. Nucleic Acids Res 25:4692–4693
An HS, Park JY, Kim MJ (2009) Isolation and characterization of microsatellite markers for the heavily exploited rockfish Sebastes Schlegeli, and cross-species amplification in four related Sebastes spp. Conserv Genet 10:1969–1972
An H, Kim MJ, Park K, Cho K, Bae B, Kim J, Myeong JI (2012) Genetic diversity and Population structure in the heavily exploited Korean Rockfish, Sebastes Schlegeli, in Korea. J World Aquaculture Soc 43:73–83
An HS, Nam MM, Myeong JI, An CM (2014) Genetic diversity and differentiation of the Korean starry flounder (Platichthys stellatus) between and within cultured stocks and wild populations inferred from microsatellite DNA analysis. Mol Biol Rep 41:7281–7292
Araki H, Waples RS, Ardren WR, Cooper B, Blouin MS (2007) Effective population size of steelhead trout: influence of variance in reproductive success, hatchery programs, and genetic compensation between life-history forms. Mol Ecol 16:953–966
Barbour RC, Otahal Y, Vaillancourt RE, Potts BM (2008) Assessing the risk of pollen-mediated gene flow from exotic Eucalyptus globulus plantations into native eucalypt populations of Australia. Biol Conserv 141(4):896–907
Bell JD, Rothlisberg PC, Munro JL, Loneragan NR, Nash WJ et al (eds) (2005) Restocking and stock enhancement of marine invertebrate fisheries. Adv. Mar Biol 49: 1–370
Bell JD, Leber KM, Blankenship HL, Loneragan NR, Masudar R (2008) A new era for restocking, stock enhancement and sea ranching of coastal fisheries resources. Rev Fish Sci 16:1–9
Blankenship HL, Leber KM (1995) A responsible approach to marine stock enhancement. In: American Fisheries Society Symposium Vol. 15, No. 167475, pp. 167–175
Blaxter JHS (2000) The enhancement of marine fish stocks. Adv Mar Biol 38:1–54
Botsford LW, Castilla JC, Peterson CH (1997) The management of fisheries and marine ecosystems. Science 277:509–515
Botstein D, White RL, Skolnick M, Davis RW (1980) Construction of a genetic linkage map in man using restriction fragment length polymorphisms. Am J Hum Genet 32:314–331
Camp EV, Larkin SL, Ahrens RNM, Lorenzen K (2017) Trade-offs between socioeconomic and conservation management objectives in stock enhancement of marine recreational fisheries. Fish Res 186:446–459
Champagnon J, Elmberg J, Guillemain M, Gauthier-Clerc M, Lebreton JD (2012) Conspecifics can be aliens too: a review of effects of restocking practices in vertebrates. J Nat Conserv 20:231–241
Christie MR, Marine ML, French RA, Blouin MS (2012) Genetic adaptation to captivity can occur in a single generation. Proc Natl Acad Sci 109:238–242
Cornuet JM, Luikart G (1996) Description and power analysis of two tests for detecting recent population bottlenecks from allele frequency data. Genetics 144:2001–2014
Crawford NG (2010) SMOGD: software for the measurement of genetic diversity. Mol Ecol Resour 10(3):556–557
DeWoody JA, Avise JC (2000) Microsatellite variation in marine, freshwater and anadromous fishes compared with other animals. J Fish Biol 56:461–473
Dick S, Shurin JB, Taylor EB (2014) Replicate divergence between and within sounds in a marine fish: the copper rockfish (Sebastes caurinus). Mol Ecol 23:575–590
Do C, Waples RS, Peel D, Macbeth GM, Tillett BJ, Ovenden JR (2014) NeEstimator v2: re-implementation of software for the estimation of contemporary effective population size (ne) from genetic data. Mol Ecol Resour 14(1):209–214
Evanno G, Renaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14:2611–2620
Falush D, Stephens M, Pritchard JK (2003) Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics 164(4):1567–1587
Food and Agriculture Organization of the United Nations (FAO) (2022) The state of World fisheries and Aquaculture, 2022. Food and Agriculture Org
Gao T, Ding K, Song N, Zhang X, Han Z (2018) Comparative analysis of multiple paternity in different populations of viviparous black rockfish, Sebastes schlegelii, a fish with long-term female sperm storage. Marine Biodivers 48:2017–2024
Gold JR, Ma L, Saillant E, Silva PS, Vega RR (2008) Genetic effective size in populations of hatchery-raised red drum released for stock enhancement. Trans Am Fish Soc 137:1327–1334
Goudet J (2001) FSTAT, a program to estimate and test gene diversities and fixation indices, version 2.9.3. http://www2.unil.ch/popgen/softwares/fstat.htm
Grant WS, Jasper J, Bekkevold D, Adkison M (2017) Responsible genetic approach to stock restoration, sea ranching and stock enhancement of marine fishes and invertebrates. Rev Fish Biol Fish 27:615–649
Hagen IJ, Ugedal O, Jensen AJ, Lo H, Holthe E, Bjøru B, Sægrov H, Skoglund H, Karlsson S (2021) Evaluation of genetic effects on wild salmon populations from stock enhancement. ICES J Mar Sci 78(3):900–909
Hamasaki K, Toriya S, Shishidou H, Sugaya T, Kitada S (2010) Genetic effects of hatchery fish on wild populations in red sea bream Pagrus major (Perciformes, Sparidae) inferred from a partial sequence of mitochondrial DNA. J Fish Biol 77:2123–2136
Hedgecock D, Coykendall K (2007) Genetic risks of marine hatchery enhancement: the good, the bad, and the unknown. Ecological and genetic implications of aquaculture activities. Springer, Dordrecht, pp 85–101
Jost L (2008) GST and its relatives do not measure differentiation. Mol Ecol 17(18):4015–4026
Kalinowski ST, Taper ML, Marshall TC (2007) Revising how the computer program CERVUS accommodates genotyping error increases success in paternity assignment. Mol Ecol 16(5):1099–1106
Kitada S (2018) Economic, ecological and genetic impacts of marine stock enhancement and sea ranching: a systematic review. Fish Fish 19(3):511–532
Kitada S, Shishidou H, Sugaya T, Kitakado T, Hamasaki K, Kishino H (2009) Genetic effects of long-term stock enhancement programs. Aquaculture 290(1–2):69–79
Kohlmann K, Kersten P, Flajšhans M (2005) Microsatellite-based genetic variability and differentiation of domesticated, wild and feral common carp (Cyprinus carpio L). Populations Aquaculture 247:253–266
Konovalov DA, Manning C, Henshaw MT (2004) KINGROUP: a program for pedigree relationship reconstruction and kin group assignments using genetic markers. Mol Ecol Notes 4(4):779–782
Laikre L, Schwartz MK, Waples RS (2010) Compromising genetic diversity in the wild: unmonitored large-scale release of plants and animals. Trends Ecol Evol 25:520–529
Leber KM (2004) Marine stock enhancement in the USA: status, trends and needs. Stock Enhancement and Sea Ranching: Developments, Pitfalls and Opportunities: 11–24
Lin L, Lee TM, Shi HT (2023) China should re-evaluate its stance on wildlife release. Proceedings of the National Academy of Sciences, 120(32): e2221766120
Liu Q, Wang Z, Li W, Zhang Q, Liu B, Li M, Liu Y, Tian T, Yan H (2022) Genetic stock identification and adaptability of hatchery-reared black rockfish, Sebastes schlegelii, released into the North Yellow Sea waters. Front Mar Sci 9:800607
Lorenzen K, Beveridge M, Mangel M (2012) Cultured fish: integrative biology and management of domestication and interactions with wild fish. Biol Rev 87:639–660
Lü H, Zhang X, Xi D, Gao T (2014) Use of calcein and alizarin red S for immersion marking of black rockfish Sebastes schlegelii juveniles. Chin J Oceanol Limnol 32:88–98
Luikart G, Cornuet JM (1998) Empirical evaluation of a test for identifying recently bottlenecked populations from allele frequency data. Conserv Biol 12:228–237
Marchant MH, Viens NA, Cook C, Vail TP, Bolognesi MP (2009) The impact of glycemic control and diabetes mellitus on perioperative outcomes after total joint arthroplasty. J Bone Joint Surg 91:1621–1629
McCay DPF, Rowe JJ (2003) Habitat restoration as mitigation for lost production at multiple trophic levels. Mar Ecol Prog Ser 264:233–247
Millar MA, Byrne M (2007) Pollen contamination in Acacia saligna: assessing the risks for sustainable agroforestry. Ecosytems Sustainable Dev VI 106:101–110
Nei M (1978) Estimation of average heterozygosity and genetic distance from a small number of individuals. Genetics 89(3):583–590
Nellemann C (ed) (2009) The environmental food crisis: the environment’s role in averting future food crises: a UNEP rapid response assessment. UNEP/Earthprint
Peakall R, Smouse P (2012) GenAlEx 6.5: genetic analysis in Excel. Population genetic software for teaching and research-an update. Bioinformatics 28:2537–2539
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155(2):945–959
Romo MDMO-V, Aritaki M, Taniguchi N (2006) Pedigree analysis of recaptured fish in the stock enhancement program of spotted halibut Verasper variegates. Fish Sci 72:48–52
Ryman N, Laikre L (1991) Effects of supportive breeding on the genetically effective population size. Conserv Biol 5:325–329
Saitoh K, Rousset F (2008) Genepop: a complete re-implementation of the genepop software for Windows and Linux. Mol Ecol Resour 8:103–106
Schindler D, Hilborn R, Chasco B, Boatright C, Quinn T, Rogers L, Webster M (2010) Population diversity and the portfolio effect in an exploited species. Nature 465:609–613
Szpiech ZA, Jakobsson M, Rosenberg NA (2008) ADZE: a rarefaction approach for counting alleles private to combinations of populations. Bioinformatics 24:2498–2504
Van Oosterhout C, Hutchinson WF, Wills DPM, Shipley P (2004) MICRO-CHECKER: software for identifying and correcting genotyping errors in microsatellite data. Mol Ecol Notes 4:535–538
Wang WQ, Zhang Y, Liu MX, Wu ZH, Wang LJ, Zi J, Guan J, You F (2012) Microsatellite analysis of genetic diversity in four wild stocks of black rockfish Sebastes Schlegeli. Mar Sci 36:10–16
Wang L, Wu Z, Wang Y, Liu M, Song A, Liu H, You F (2020) Genetic assessment of a black rockfish, Sebastes schlegelii, stock enhancement program in Lidao bay, China based on mitochondrial and nuclear DNA analysis. Front Mar Sci 7:94
Waples RS, Hindar K, Karlsson S, Hard JJ (2016) Evaluating the ryman–laikre effect for marine stock enhancement and aquaculture. Curr Zool 62(6):617–627
Ward RD (2006) The importance of identifying spatial population structure in restocking and stock enhancement programmes. Fish Res 80:9–18
Weir BS, Cockerham CC (1984) Estimating F-statistics for the analysis of population structure. Evolution 1984:1358–1370. https://doi.org/10.2307/2408641
Welcomme RL, Bartley DM (1998) An evaluation of present techniques for the enhancement of fisheries. FAO Fisheries Technical Paper (FAO)
Willoughby JR, Christie MR (2019) Long-term demographic and genetic effects of releasing captive-born individuals into the wild. Conserv Biol 33(2):377–388
Xu SB, Tao YF, Yang ZQ, Chu JY (2002) A simple and rapid method used for silver staining and gel preservation. Hereditas 24:335–336
Yoshida K, Nakagawa M, Wada S (2005) Multiplex PCR system applied for analysing microsatellite loci of Schlegel’s black rockfish, Sebastes Schlegeli. Mol Ecol Note 5:416–418
Zhang H, Yanagimoto T, Zhang X, Song N, Gao T (2016) Lack of population genetic differentiation of a marine ovoviviparous fish Sebastes schlegelii in Northwestern Pacific. Mitochondrial DNA 27:1748–1754
Acknowledgements
This research was funded by the Key Research and Development Programs of Shandong Province (No. 2021LZGC029), the National Special Research Fund for Non-Profit Marine Sector (No. 201305043 and 200805069), and the National Marine Genetic Resource Center.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
On behalf of all authors, the corresponding author states that there is no conflict of interest.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic Supplementary Material
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Zhang, Y., Wang, L., Wu, Z. et al. Microsatellite Analysis of the Influence of Sebastes schlegelii Restocking on the Genetic Structure of Stocks in Rongcheng Bay. Thalassas 40, 743–754 (2024). https://doi.org/10.1007/s41208-024-00688-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s41208-024-00688-0