Abstract
The cacao tree surface is a substrate for various microorganisms. However, the species which live on the surface have hardly been described. The aims of this study were to evaluate the diversity of native cacao yeasts and to investigate their possible interaction with Moniliophthora perniciosa. A total of 225 yeast strains were isolated from the surface of leaves, fruits, and flowers of 20 cacao genotypes selected as carriers of resistance genes to M. perniciosa and M. roreri, which are the causal agents of witches’ broom (WB) and frosty pod rot diseases (FPR), respectively. The yeast isolates were molecularly identified based on sequences of the D1/D2 and ITS1/ITS4 regions of the rDNA. A total of 20 taxa distributed into 11 genera was found: Hannaella (23), Cryptococcus (9), Candida (11), Hanseniaspora (4), Kwoniella (1), Occultifur (3), Rhodotorula (17), Pichia (2), Sporobolomyces (1), Wicherhamomyces (1), and Yamadazyma (1). Phylloplane tissues showed the greatest richness: 55% of species and 81.82% of identified genera. Six yeast strains (Candida orthopsilosis—LEV 225; Hanseniaspora uvarum—LEV 162, 210, 211; and Rhodotorula paludigena—LEV 166, 168) exhibited the desired traits for a BCA agent: antibiosis in cocultivation, antibiosis by volatile compounds, germination inhibition, and production of killer toxins. This study is the first to show the great diversity of yeasts found in the cocoa phytobiome and the discovery of at least six microorganisms with excellent potential as BCA against cocoa witches’ broom disease.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Cacao tree is among the most important perennial crops in the world; annual production of dry cocoa beans is estimated at 4834 t (ICCO 2019). Cacao originated in tropical forests of South America (Cuatrecasas 1964) and is commercially exploited by the food industry to produce chocolate, jellies, pulps, and other products in America’s equatorial, Africa, and Asia.
During three decades, Brazil remained as one of the largest cacao-producing countries in the world, reaching a production of 238, 879 t dry cocoa beans in 2020 (IBGE 2020). Currently, Brazil ranks seventh in world production, and Bahia is the main cacao-producing state (ICCO 2019). However, economic factors, climate problems, and especially, the appearance of witches’ broom (WB) disease caused by the fungus Moniliophthora perniciosa (Stahel) Aime and Phillips-Mora led to an enormous socioeconomic crisis in the region, causing Brazil to shift from being a cacao exporter to importer of cocoa beans (Bowers et al. 2001; Marelli et al. 2009; Midlej and Santos 2012; Teixeira et al. 2015).
Since then, efforts have been made to control WB that comes up against pathogen intrinsic factors and environment conditions for this crop cultivation using the so-called cacao–cabruca system or ‘cabruca agroforestry system’ which is characterized by understory tree species in Atlantic Rain Forests (Tavares 1979; Rice and Greenberg 2010; Duarte et al. 2013).
Crop management and application of chemical fungicides alone were not successful to control WB in the cacao crop, but genetic and biological controls have been the most effective alternatives against this disease mainly if they are applied together (Pimenta Neto et al. 2018; Ferraz et al. 2019). In relation to biological control product Tricovab®, based on Trichoderma stromaticum, developed, produced, and distributed by Comissão Executiva do Plano da Lavoura Cacaueira (CEPLAC), it is effective in reducing fungal sporulation. However, production of the fungus is expensive, which reinforces the need for other sustainable biological strategies to control WB (Loguercio et al. 2009; Pomella et al. 2007).
In this context, prospecting yeasts from the cacao tree phytobiome as biological control agents (BCA) seem to be a promising, less expensive and safe alternative (GRAS (generally recognized as safe)) for humans and animals. Yeasts are safe to manipulate and have great competitive potential (Zhang et al. 2007); they release killer toxins (Platania et al. 2012), produce volatile compounds (Bruce et al. 2003), and secrete hydrolytic enzymes that can degrade the phytopathogen cell wall (Masih and Paul 2002; Urquhart and Punja 2002; Lu et al. 2014). In addition, yeasts can withstand environmental changes and stressful conditions (Sui et al. 2015) are are tolerant to pollutants, essential oils, phytoalexins, and some fungicides (Valdebenito-Sanhueza 2000); they can survive saprophytically (Afsah-Hejri 2013) and tolerate adverse conditions (Hu et al. 2015).
Particularly for the cacao tree crop, a worldwide commodity, yeasts studies have been directed to the cacao bean fermentation process (Daniel et al. 2009; Moreira et al. 2013) and to standardize the quality of products for the chocolate manufacture (Leal Jr et al. 2008). Thus, there is a gap regarding the prospection of native yeasts of the cacao tree phytobiome as BCAs to the main pathogens of this crop. With that in mind, this research aimed to isolate yeasts present in the cacao tree phytobiome in the southern region of Bahia State to know their diversity and prospect the most promising ones as BCAs against M. perniciosa, the causal agent of WB.
Material and methods
Sampling site
The samples used were from Ilhéus, a southern coast city of the state of Bahia, Brazil (14°47′20″S, 39°02′58″W and altitude of 52 m), located at 460 km from the state capital, Salvador. Ilhéus has an area of 1584.693 km2, which is part of the 8% of the Atlantic Forest remnants, characterized by tall trees of evergreen leaves and abundance of epiphytes. Cacao trees are grown under the canopy of native trees. The region has an equatorial climate (Köppen–Geiger climate classification), significant rainfall throughout the year, even in the driest month, with average annual temperature of 24.4 °C, and rainfall of 1419 mm. Samples were collected in the rainy period.
The cacao trees selected as substrate for bioprospecting of yeast-like fungi in this study are part of the Cacao Breeding Program of the CEPLAC, and they grow in experimental areas of the Cacao Research Center, Ilhéus (14°46′3.46″S; 39°13′57.31″W). Samples were obtained from 16 cacao genotypes characterized as resistant to Moniliophthora perniciosa, one of them being clone (EET 392), and selected plants of 15 progenies (P213, P225, P232, P238, P243, P245, P247, P253, P258, P278, P282, P284, P285, P300, and P309) (Silva et al. 2010; Pimenta Neto et al. 2018), and four other clones carrying resistance genes to M. roreri (EET 233, EET 75, ICS 95, and UF 233) (Phillips-Mora et al. 2007).
Yeast isolation and preservation
The yeasts were isolated following as described elsewhere (Assis et al. 1999). Five samples from different plant tissues (flower, leaf, and fruit) of each cacao tree genotype were collected and taken to a laboratory. Three samples were randomly collected in the upper, middle, and lower extracts of the outer edges and two in the inner part. Plant parts sampled were not surface sterilized because of the interest in obtaining yeasts that could be colonizing epiphytically. Isolation was made from young healthy plant materials: leaves presenting green petioles, green fruits, and open flowers and without lesions, disease, or presence of insects. One gram of each sample was suspended in 9 mL of saline solution (0.85% NaCl) containing chloramphenicol (50 mg L−1) and sonicated for 15 min in an ultrasonicator (Quimis 0335D). Serial dilutions of the suspended yeast sample were performed in saline solution + chloramphenicol (50 mg L−1). The yeast suspension concentration was defined based on quantification in Neubauer chamber and measurement of optical density by reading in a spectrophotometer (SpectraMax® Paradigm® Multi-Mode Detection Platform, Molecular-Devices Japan, Tokyo, Japan) at 595 nm. Aliquots of 300 μL of each dilution were distributed in sterilized plates containing yeast extract peptone dextrose agar (YEPD) (0.15% yeast extract, 1% neopeptone, 4% dextrose, 2% agar, and 0.05% chloramphenicol). The plates were incubated at 28 °C for five days, and the yeasts were analyzed for morphological characteristics of colonies and cells. One representative sample of each yeast colony was grown in YEPD to obtain pure cultures, which were maintained in tubes containing potato dextrose agar (PDA) (Himedia®) at 4 °C and in 20% glycerol at − 80 °C.
Detection of antifungal activity by diffusible and volatile compounds
The antifungal activity of 172 yeast isolates was evaluated by comparing co-cultivations of four molecularly distinct M. perniciosa isolates (Mp 4009, Mp 4010, Mp 4031, Mp 4145; Patrocínio et al. 2016), according to Mariano et al. (2005). Disks (Ø 5 mm) of culture medium containing fungal structures of M. perniciosa were removed from the peripheral region of the growing isolate and picked at the center of Petri dishes (Ø 9.5 cm) that contained PDA medium (Himedia®). The yeast isolates were placed on plates containing M. perniciosa isolates, bordering the discs deposited in the center of the plate. The plates were incubated for 10 days at 25 °C in a growth chamber. From day six, they were evaluated for inhibition of mycelial growth by daily measuring of colony diameter. The colony diameter was evaluated with a Mitutoyo® (Japan) caliper in two orthogonal measurements. Five replications were made and the experiment design was completely randomized. The control consisted of plates containing culture medium, seeded with the pathogen alone. When the pathogen growth in the control reached the plate edges, the percentages of growth inhibition were calculated according to the formula of Topps and Wain (1957): I% = [(A − B)/A] × 100, I% is the percentage of inhibition, A is the mean growth diameter in the control, and B is the mean diameter growth in the treatment.
Determination of the producing potential of volatile antifungal compounds
Antifungal action was detected in volatile compounds through the culture technique in split Petri dishes containing PDA medium (Himedia®). Sixty-six yeast isolates previously selected with antifungal activity to M. perniciosa were compared with the four M. perniciosa strains. The yeast isolate was inoculated on one side of the plate, and a disc containing fungal structures of the pathogen was placed on the other side of the plate. The plates were incubated for 10 days at 25 °C in a growth chamber and evaluated daily using a caliper ruler with 0.5 mm accuracy. Five replications were made and the experiment design was completely randomized. The control consisted of plates containing culture medium with only the pathogen. The percentage of growth inhibition was calculated according to Topps and Wain (1957).
Heatmap analysis with yeast cluster for evaluation of diffusible and volatile compounds
A matrix generated for the treatments (yeast isolates) was evaluated using the R 3.6.2 program (R Core Team 2019), which allowed the construction of heatmaps (Gu 2016) using Euclidean distance to evaluate yeast clustering based on the inhibition profile by diffusible and volatile compounds, and volatile compounds alone, in the inhibition of mycelial growth of four M. perniciosa strain.
Determination of potential to inhibit basidiospore germination
The yeast isolates tested were cultured in liquid YEPD medium and incubated at 150 rpm and 28 °C in an orbital shaking incubator (ACBLABOR® AC-830) for 72 h. The secretome or crude extract was obtained based on Coelho et al. (2011), 45 mL of YEPD liquid medium and 5 mL of the standard yeast suspension (108 CFU mL−1) was used for measuring the optical density in a spectrophotometer at 595 nm. The sample was centrifuged at 6500 g for 15 min at 10 °C, separating the supernatant from the yeast cells. The supernatant of the 66 evaluated yeasts was filtered with a sterile syringe and a 0.22-μm Millipore® syringe filter, isolating the remaining cells. The filtrate was stored in a freezer at − 80 °C for the preservation of bioactive molecules until use for the tests to inhibit the germination of M. perniciosa spores. Aliquots of the yeast secretome and basidiospore suspension (106), at a 1 : 1 ratio, were homogenized (together) in 2-mL microtubes and poured in Petri dishes (60 × 15 mm). The experimental design was completely randomized, with two plates divided into quadrants; each quadrant constituted one replication, totaling eight replications (treatments). They were evaluated at 6 h after incubation by counting the number of total and germinated spores in the area, using a 40x objective optical microscope at three distinct points in each quadrant. The basidiospore whose germ tube was greater than or equal to its length was considered germinated and the percentage of spores was calculated for each replication. The size of the germ tube was measured under the microscope with a micrometer adapted to the microscope eyepiece (Mares et al. 2016). The data were transformed [(Sqrt (x + 0.5)] and subjected to analysis of variance (ANOVA) followed by Scott–Knott test (p ≤ 0.01), using R version 3.6.2 environment (R Core Team 2019), with the SK function of the Scott-Knott package (Scott and Knott 1974).
Determination of the killer factor
The killer activity of 66 yeast isolates was investigated by co-cultivating the selected yeasts and reference strains as a killer sensitivity standard. The tests were performed using the sowing technique with YEPD-MB agar (1% yeast extract, 2% glucose, 2% peptone, 2% agar, 0.003% methylene blue, and buffered to pH 4.0 with phosphate citrate buffer 0.5 M) in Petri dishes as described by Sangorrín et al. (2001). The yeasts were resuspended in 0.85% saline solution to a concentration of approximately 5 × 105 CFU mL−1. Then, 100 μL of this cell suspension, corresponding to each of the sensitive strains (Saccharomyces cerevisiae NCYC1006® and Candida glabrata Y55 ATCC90525®), was spread on the YEPD-MB agar medium surface in the Petri dishes (Ø 90 × 15 mm) using a Drigalski spatula. After the suspension dried, the yeast isolates were point-shaped inoculated. Thus, it was possible to inoculate 11 test isolates plus the killer toxin-producing strain in each Petri dish. Three replications were made and the experiment design was completely randomized. The plates were incubated in a BOD chamber at 25 °C for 72 h and evaluated daily. The presence of killer phenotype (K+) was found in isolates that caused a clear inhibition zone with no sensitive yeast growth around the colony, which was delimited by an adjacent halo of blue-stained cells, denoting cell death and killer activity (Young 1987).
Multivariate analysis of antifungal activity
To jointly assess the effect of all variables: antifungal activity, volatile compounds, inhibition of basidiospore germination, and killer activity, a principal component analysis (PCA) analysis was performed. For this, the values of the inhibition of basidiospore germination variable were converted to a binary system and the matrix containing the measurements of those four variables were used in the PCA. The PCA was performed in R version 3.6.2 environment (R Core Team 2019), with the function prcomp (stats package) with the parameters center = TRUE and scale = TRUE. The k-means function (stats package) was applied in the PCA, for partial grouping in 2 clusters. A biplot, with components 1, 2, and 3, was plotted using the function plot3d (package rgl) together with the groupings.
Evaluation of yeast–Moniliophthora perniciosa interaction in liquid culture medium
The yeast Hannaella sp. (LEV 207—107 ufc) was co-cultivated with the pathogen M. perniciosa (Mp 4009: disks of culture medium containing fungal structures) in 125-mL conical flasks containing 22.5 mL of potato dextrose (PD) medium, adding to them 2.5-mL aliquots of the yeast isolate and a disc containing fungal structures of the pathogen, with shaking at 150 rpm at 28 °C in a thermal shaker, forming different types of yeast treatments and reinoculation. The treatments used were T1 = Hannaella sp., T2 = M. perniciosa, T3 = Hannaella sp. + M. perniciosa (simultaneous), T4 = Hannaella sp. + M. perniciosa (simultaneous) + 1 yeast reinoculation (after 120 h), T5 = Hannaella sp. + M. perniciosa (simultaneous) + 2 yeast reinoculation (after 96 and 1920 h), T6 = Hannaella sp. + M. perniciosa (simultaneous) + 3 yeast reinoculation (after 48, 144, and 216 h), T7 = Hannaella sp. + simultaneous M. perniciosa) + 4 yeast reinoculation (after 48, 96, 144, and 192 h), T8 = Hannaella sp. + M. perniciosa (simultaneous) + 1 yeast reinoculation (after 192 h), and T9 = M. perniciosa + Hannaella sp. (after 168 h + 1 reinoculation after 216 h). Samples were collected and vacuum filtered on Whatman® 110 mm Ø filter paper at 10 days after the assay begun, then, quantified for dry biomass on an analytical balance. The dry sample was obtained through dehydration using an oven at 70 °C for 48 h. Glucose consumption was inferred by the quantification of reducing sugars in a DNS (dinitrosalicylic acid) assay (Miller 1959). Aliquots (0.1 mL) of the liquid fraction of each treatment were placed in test tubes and 0.9 mL of distilled water was added, followed by 3 mL of DNS reagent. The mixture was boiled for 5 min and cooled to room temperature, and 7 mL of distilled water was added. The absorbance values of the samples were collected at 540 nm. The absorbance was measured in comparison to a blank sample in a spectrophotometer. The experiment was conducted in a completely randomized design with three replications.
Molecular analyses for yeast isolates identification
Eighty of the yeast isolates, selected by their distinct morpho-physiological characteristics, were molecularly identified. Total DNA was extracted using the UltraClean® Microbial DNA Isolation kit (MoBio, USA), following the manufacturer’s recommendations. DNA integrity and quantity were verified by electrophoresis in 1.0% agarose gel stained with ethidium bromide and viewed under ultraviolet light, followed by photodocumentation and quantification with a Qubit® 2.0 Fluorometer (Invitrogen).
The D1/D2 region of the large subunit (26S) of the rDNA were amplified with the primers NL1 (5′-GCATATCAATAAGCGGAGGAAAAG-3′) and NL4 (5′-GGTCCGTGTTTCAAGACGG-3′) (O’Donnell 1993). Reactions were prepared with the following reagents and concentrations: 60 g of DNA from each sample, 1x of Taq DNA polymerase buffer, 3.7 mM of MgCl2, 0.6 pmol μL−1 of dNTPs, 0.4 pmol μL−1 of each primer, and 2.5 U of Taq DNA polymerase, with final volume adjusted to 50 μL with ultrapure water. Amplification cycles were performed in the Veriti Thermal Cycler PCR (Applied Biosystems), with initial denaturation temperature of 95 °C for 5 min; and 35 amplification cycles of three steps: at 95 °C for 30 s, at 54 °C for 30 s, and at 72 °C for 7 min; and final extension temperature of 72 °C for 10 min. The amplified products were applied in 1% agarose gel, which was used to do electrophoresis, and stained with ethidium bromide, and visualized under ultraviolet light. Then, the amplicons were purified using the PureLink™ PCR Purification kit (Invitrogen), quantified with Qubit® 2.0 Fluorometer (Invitrogen) for subsequent nucleotide identification, using the automatic sequencer ABI-Prism 3500 Genetic Analyzer (Applied Biosystems). The sequences were edited and assembled using the Sequencher 4.1.4 program (Gene Code Corporation). Taxonomic identification of the isolated yeasts was verified through the database GenBank, using the Basic Local Alignment Search Tool (BLASTn) of the NCBI (http://www.ncbi.nlm.nih.gov).
The yeast isolates were subjected to a second sequencing with the ITS region (ITS1 and ITS4 primers), as described by White et al. (1990), to confirm the species to which they grouped or the impossibility of separating the sequences obtained at the species level only with study of the D1/D2 region/domain.
Yeast diversity evaluation
Richness, relative frequency, and similarity were evaluated as ecological attributes. Richness was defined as the total number of species found in each plant organ, in each genotype, and each group of plants characterized as resistant to M. perniciosa and M. roreri. Relative frequency was calculated based on the formula F = n × 100/N, where n is the number of samples in which a species or genus was found and N is the total number of samples in each plant organ or in each resistance standard. The following frequency classes were determined: sporadic (F ≤ 10%), slightly frequent (10 < F ≤ 30%), frequent (30 < F ≤ 70%), and very frequent (F > 70%) (Dajoz 1983). The similarity between plant tissues and resistance standards was measured by the Sorensen Index (Muller-Dombois 1981), S = 2c.100/a + b, were c is the number of yeasts common to two plant tissues or two resistance standards and a + b is the number of yeasts present in the tissues or resistance standards. The formula S = 3d.100/a + b + c was used for the three plant tissues studied, where d is the number of yeasts found in three plant tissues and a + b + c is the number of fungi present in the tissues. Venn diagrams were developed to show the number of taxa shared between communities. The diagrams were generated in the R 3.0.1 program, using the VennDiagram package (Oliveros 2007).
Results
In the present study, 225 yeast isolates were found on the surface of leaves (63.5%), fruits (14.2%), and flowers (22.2%) of cacao tree genotypes (20) selected as carriers of genes of resistance to the witches’ broom (WB) and frosty pod rot (FPR) diseases. The higher number of isolates from leaf blades was due to the greater availability of this plant tissue. Yeasts were found in 100% of the samples. The total density of yeasts varied from 3.2 × 104 CFU g−1 to less than 50 CFU on fruits, from 2.8 × 104 CFU g−1 to less than 50 CFU on flowers, and from 2.0 × 104 CFU g−1 to less than 50 CFU on leaves.
A heatmap analysis was done based on the antifungal action of the 172 yeast isolates native to cacao plants (y axis) on four distinct strains of the pathogen M. perniciosa (Mp) in five evaluation days (6–9 days after pairing) (x axis) and allowed the separation of two groups (clades) (Fig. 1). The yeast isolates inhibited more than 50% mycelial growth of the four pathogens strains evaluated, except for the yeast isolates LEV 136, LEV 019, and LEV 110. These three isolates were part of the yeast clade that presented less inhibition of the pathogen growth (Fig. 1). The yeast isolates LEV 079, 139, 002, 047, 023, 034, 210, 194, 211, 030, 028, 097, 227, 204, 206, 189, 007, 021, and 162 and LEV 212 differed from the other isolates causing the highest percentages of mycelial growth inhibition of the four strains of the pathogen.
The inhibition pattern of each yeast was not uniform in the four strains of M. perniciosa and also between evaluation days (Fig. 1). The rate of mycelial growth of the pathogens was variable when it was grown in the presence of diffusible and volatile compounds. The fungistatic potential of yeast was similar for the pathogens Mp 4010 and Mp 4031, whereas the highest percentages of inhibition were found for the Mp 4009 (Fig. 1).
There was a trend of increasing the percentage of inhibition with time for most of the yeasts evaluated; however, some of them showed constant inhibition throughout the evaluation period for the four strains of the pathogen: LEV 039, 168, 169, 006, 164, 018, 166, 182, 067, 036, 010, 084, 023, 047, 002, 139, 179, 085, 212, and 013 and LEV 202 for Mp 4009; LEV 010, 067, 212, 021, 162, 097, 078, and 194 and LEV 210 for Mp 4010; LEV 001, 018, 006, 212, 084, 002, 139, 162, 021, 097, 078, and 194 and LEV 210 for Mp 4031; LEV 006, 018, 182, 036, 039, 010, 047, and 162 and LEV 021 for the pathogen Mp 4145. Inhibition oscillation was also found in the evaluation days for the yeast isolates LEV 067, 202, 064, 069, 096, 074, 062, 110, and 210 and LEV 211. The yeasts LEV 039, 074, 212, and 096 and LEV 069 showed a trend of decreasing in percentages of inhibition for some M. perniciosa strains (Fig. 1).
The inhibitor potential of mycelial growth by volatile compounds of most yeasts fluctuated, but some remained constant during the evaluation days for some of the four pathogens of the M. perniciosa tested (Fig. 2): LEV 165, 002, 118, 084, 140, 111, 169, 082, 209, 059, and 226 and LEV 088 for the pathogen Mp 4009; LEV 002, 101, 140, 161, 178, 109, 090, 077, 185, and 189 and LEV 225 for the pathogen Mp 4010; LEV 162, 118, 166, 149, 143, 111, 178, 109, 100, and 211 and LEV 080 for the pathogen Mp 4031. Only LEV 162 for the pathogen Mp 4145.
For the evaluation of volatile compounds, two yeast groups (clades) were separated by Euclidean distance and visualized in heatmap (Fig. 2). Twentty-two yeast isolates (33.33%) composed the clade 1 that had the lowest percentages of growth inhibition. However, in this clade yeast isolates, LEV 170, LEV 169, and LEV 082 showed inhibition levels like those of clade 2 for three of the pathogen isolates (Mp 4009, Mp 4010, and Mp 4031). Among the individuals that grouped in clade 2 presents the highest percentages of growth inhibition by volatile compounds, four yeast isolates (LEV 225, 206, and 166 and LEV 092) stand apart in a subgroup (2nd level clade) since they inhibited above 50% the growth of the pathogen isolates Mp 4009 and Mp 4010. Yeasts LEV 074, 210, 089, and 189 and LEV 211 belonging to the second subgroup maintained the same inhibition pattern for the four M. perniciosa strains (Fig. 2). Seven isolates (LEV 002, 084, 162, 189, 206, 210, and 211) in clade 2 inhibited the mycelial growth of M. perniciosa by volatile compounds (Fig. 2) and also the sum of diffusible and volatile compounds (Fig. 1).
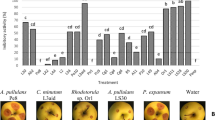
All the filtrates obtained from the cultures of 66 yeast isolates in YEPD medium at 1 : 1 dilution (filtrate : basidospore suspension) inhibited basidiospore germination after six hours of incubation when compared to the control (Fig. 3). From the three groups statistically distinct (p < 0.001) according to the Scott–Knott test, 19 isolates stood out (LEV 049, 063, 065, 093, 101, 105, 111, 147, 149, 166, 168, 169, 185, 204, 207, 210, 211, and 219 and LEV 225) by inhibiting basidiospore germination from 57.5 (LEV 093) to 87.5% (LEV 169) (Fig. 3).
Inhibition of basidiospore germination and capability to produce killer toxins by different isolates of native cacao yeasts. *Non-killer toxin-producing yeasts. The means of inhibition percentages were transformed Sqrt (x + 0.5) and analyzed by Scott–Knott test (p < 0.01). Three groups, statistically distinct, were formed and identified as represented by specific markings on the bars
Only 13 yeast isolates (LEV 049, 077, 78, 88, 090, 092, 101, 106, 109, 118, 143, and 161 and LEV 229) (19.69%) did not produce killer toxins (Fig. 3). Only LEV 049 and LEV 101 were from the group with the highest inhibitions for other parameters evaluated in this study.
In the PCA, significant differences were found for yeast isolates, antibiosis, volatile compounds, germination inhibition, and killer activity. In the biplot distribution (Fig. 4), yeast isolates were scattered on a three-dimensional graph of the first three main components (synthetic variables), which together account for 78.09% of the total variance analysis.
Principal component analysis (PCA) of the relationship between biocontrol mechanisms (real variables), produced by yeast cacao tree isolates when evaluated against Moniliophthora perniciosa. Points are plotted in a color gradient to show data cluster trends by diffusible and volatile compounds, antibiosis assays (AB: light blue), volatile compounds (VC: dark blue), germination inhibition (GI: maroon), and killer activity (KA: yellow). The k-means function (stats package R version 3.6.2) was applied in the PCA, for partial yeast clusters (red point and black point). The biplot with k-means clusters was obtained by plotting three main components (vectors): PC1, PC2, and PC3
The principal component 1 (PC1) represented 31.73% of the total variance; the variable antibiosis (AN) represented the largest variance of this component due to its highest coefficient (auto vector: 0.62). The variables killer activity (KA), volatile compounds (VC), and germination inhibition (GI) presented coefficients of 0.60, 0.48, and 0.06, respectively. GI showed the highest weight (0.85) in the variance of the principal component 2 (PC2), and VC was the major contributor to the variance of the principal component 3 (PC3). PC2 and PC3 accounted for 26.14% and 20.22% of the total variance data, respectively.
The smaller the angle between vectors of the variables, the greater the correlation. AN did not present high correlation as other variables, but it correlated positively with VC and KA, rather than with GI. The negative correlation between VC and GI is shown by their opposite vectorization. The eigenvector coefficients explain the correlation between real variables and synthetic variables; however, VC present significant negative correlation for PC2 and AB and KA for PC3. The relationship between variables (vectors) and treatment distributions (dispersion) is explained by the principal component analysis (PCA) represented in the biplot (Fig. 4).
The yeast isolates that presented the highest means for each variable are shown in Fig. 4 near the apex of the vector, as LEV 169 and LEV 219 for GI. When the isolates showed low means for each variable, they are opposite to the vector of the analyzed variable represented by red spots in Fig. 4.
LEV 210 was in a central position in comparison to the vectors of the four variables evaluated; it was the yeast isolate that stood out with the first, second, and fourth highest means for AB, VC, and GI, and presented the qualitative characteristic KA. The LEV 211 is also located in a central region, close to isolate 210, because it is in the groups of isolates that most inhibited the growth of the pathogen (all variables analyzed). LEV 206 and LEV 189 are between vectors VC and AB, just as LEV166 and LEV 225 are between vectors VC and GI. These isolates belong to the groups that had the highest averages for these respective variables. Although LEV 169 had the highest mean for GI, it was close only to the vector CP2 the component most influenced by this variable. LEV 169 did not provide high mycelial growth inhibition by diffusible and volatile compounds (Fig. 4).
As LEV 207 (Hannaella sp.) was the most prevalent isolate of the genus, it was selected to evaluate the direct interaction between yeast and pathogen and to infer nutrient competition through the determination of reducing sugars. When cultured in liquid medium, LEV 207 inhibited M. perniciosa growth in all the seven treatments, regardless of the number of reinoculation and time of application. The pathogen dry weight of the biomass (DB) was measured after 240 h of cultivation and it was similar for all evaluated treatments. When M. perniciosa was cultivated alone (T2), the DB generated differed significantly from all other treatments (T3 to T9) in co-cultivation with LEV 207 (107 CFU mL−1), thus, was possible to infer the yeast inhibition potential. The minimum inhibition provided was of 95.3% and no significant differences (p < 0.05) were observed in Mp DB among the treatments were LEV 207 was added (T3, T4, T5, T6, and T7) denoting that this yeast isolate does not require reinoculation to increase its ability to inhibit in vitro the pathogen growth. The highest concentration of reducing sugars was found in T8 ((T8 = Hannaella sp. + M. perniciosa (simultaneous) + 1 yeast reinoculation (after 192 h)), differing statistically from all other treatments. T1 and T9 showed no statistical difference between themselves but differed from all other treatments. T2, T3, T4, T5, T6, and T7 did not differ statistically.
Considering the genomic DNA sequencing of 80 of the yeasts isolated from the selected cacao trees, 25% of them were of the phylum Ascomycota, 67.5% Basidiomycota, and 7.5% (six isolates) were undetermined (Table 1). Twenty taxa of 11 genera were found: Candida (6), Cryptococcus (2), Hannaella (4), Hanseniaspora (1), Kwoniella (1), Occultifur (1), Pichia (1), Rhodotorula (1), Sporobolomyces (1), Wicherhamomyces (1), and Yamadazyma (1) (Fig. 1). Most isolates showed > 99% correspondence to the GenBank sequences.
The order Saccharomycetales of the phylum Ascomycota predominated, with the families Pichiaceae and Saccharomycetaceae/Wickerhamomycetaceae. Fungi of the phylum Basidiomycota were identified as belonging to the sub-phyla Agaromycotina and Pucciniomycotina; classes Tremellamycetes, Cystobasidiomycetes, and Microbotryomycetes; orders Tremellales, Cystobasidiales, and Sporidiobolales; and families Tremellaceae and Cystobasidiaceae, apart from isolates whose precise family could not be established in the classification (Incertae sedis).
The yeast isolates with highest frequency were from the order Tremellales, especially of the genus Hannaella (23 isolates) and the taxon Rhodotorula paludigena, with 13 isolates among the sequenced ones (Table 1).
In general, all yeast isolates showed positive results in the in vitro assays for each one of the variables evaluated: mycelial growth inhibition by diffusible compounds, by volatile compounds, production of killer toxins, and inhibition of basisdiospores germination. Some were the best ones for the four tests: Candida orthopsilosis—LEV 225; Hanseniaspora uvarum—LEV 162, 210, and 211; and Rhodotorula paludigena—LEV 166, 168. Some only inhibited basidiospore germination like LEV 169 (R. paludigena), LEV 219 (C. orthopsilosis), and LEV 111 (Cryptococcus heimayensis) and have average behavior in comparison to the other variables. Other yeast isolates as LEV 002 (C. intermedia) and LEV 084, 118 (C. nonsorborphila) performed well in the other three variables but were among those with lower percentage of inhibition of basidiospore germination (Figs. 1, 2, and 3). Some isolates of the species Cryptococcus heimayensis (LEV 206), Occultifur externus (LEV 065), Hannaella oryzae (LEV 093), H. surugaensis (LEV 187), H. coprosmae (LEV 164), H. luteola (LEV 063), Kwoniella mangrovensis (LEV 227), Pichia sporocuriosa (LEV 204), and Wicherhamomyces lynferdii (LEV 047) also presented at least two characters among the ones analyzes, which make them promising biological control agents (Figs. 1, 2, 3, and 4).
Most of the sequenced isolates were obtained from cacao leaves (44%), followed by flowers (27%) and fruits (9%) (Table 2, 3). The richness of yeasts was analyzed on the surface of cacao leaves and 15 taxa (75%) and 9 genera (81.82%) were identified in 44 isolates obtained from the plant substrate. Of the 27 isolates obtained from flowers, 7 taxa (35%) and 7 genera (63.64%) were identified. Nine isolates (11.25%) were obtained from the surface of fruits, and 5 taxa (25%) were identified from 4 different genera (36.36%) (Table 3).
The highest number of different isolates was found in the cacao tree genotypes P232 and P243 (both WB resistant) and EET 233 (FPR resistant), whose richness was 26.32% at species level and 45.45% at genus level. The genotypes P225, P238, and P245 (all WB resistant) presented the lowest richness with only 5.26% species and 9.09% genera among the sequenced isolates (Table 3).
The taxa distribution per frequency class showed predominance of sporadic and slightly frequent taxa in the three plant tissues and all cacao genotypes evaluated. Among the 15 yeast species found on the leaves, 14 showed sporadic frequency and only the species Rhodotorula paludigena (15 isolates) was classified as frequent (Table 4). According to the genus distribution on the leaves per frequency class, 55.56% of the genera were sporadic (Candida, Cryptococcus, Hannaella, Sporobolomyces, Wicherhamomyces, and Yamadazyma), but 33.34% of the genera were characterized as slightly frequent (Hannaella, Cryptococcus, and Candida), and only one genus was classified as frequent (Rhodotorula). Most of the yeast species found on cacao leaves showed sporadic distribution (85.71%), except for Hanseniaspora uvarum, classified as slightly frequent (14.29%). At genus level, four were classified as sporadic (Candida, Occultifur, Pichia, and Rhodotorula), two as slightly frequent (Hannaella, Hanseniaspora), and one as frequent (Cryptococcus). All species found on fruits were classified as slightly frequent, but three genera were classified as slightly frequent (Cryptococcus, Hannaella, Kwoniella) and one genus as frequent (Candida). When the frequency was analyzed by group of resistant cacao tree genotypes, sporadic taxa and species predominated for FPR- and WB-resistant genotypes (Table 5).
The Venn diagram (Fig. 5) made it possible to visualize the intersection of the set of genera present in the different substrates. When evaluated only genotypes with resistance to WB (Fig. 5a and Table 2), three genera were common to all plant tissues (Candida, Hannaella, and Cryptococcus), besides the group of unidentified yeast isolates. However, three genera were identified only on leaf and flower surfaces (Hanseniaspora, Occultifur, and Rhodotorula), three only on leaf surfaces (Yamadazyma, Wicherhamomyces, and Sporobolomyces), and only Pichia on flower surface. For genotypes with resistance to FPR (Fig. 5b and Table 2), the genus Hannaella was identified in all plant tissues, one on fruit and flower surfaces (group of unidentified isolates), one on flower and leaf surfaces (Rhodotorula), two on fruit surface (Candida and Kwoniella), and three on flower surface (Pichia, Hanseniaspora, and Cryptococcus). Seven genera (Hannaella, Rhodotorula, Candida, Pichia, Hanseniaspora, Cryptococcus, and unidentified isolates) were common to the substrates of cacao trees with resistance to WB or FPR, four genera were identified only on genotypes with WB resistant (Yamadazyma, Occultifur, Wicherhamomyces, and Sporobolomyces), and one (Kwoniella) only for those resistant to FPR (Fig. 5c and Table 2). Fifty-four percent of similarity of yeast species was found on cacao leaves and flowers; 35% on leaves and fruits, 27% on flowers and fruits, and 9% species were found on the three plant tissues. The yeast species found in cacao tree genotypes WB resistant were 50% similar to the species found in those resistant to FPR (Fig. 5 and Table 2).
Venn diagram of the number of taxa in common among the different cacao plant tissues of witches’ broom (WB) (a), or frost pod rot (FPR) (b) resistant and the ones only found in each kind of genotypes and cocoa plant tissues (c). In the intersections of the diagram are common taxa to the communities present in the different cacao plant tissues
Discussion
Biological control is one of the most effective components of the integrated management program for WB control in Brazil. However, a single biological control product is registered for this crop in the country, Tricovab® formulated from Trichoderma stromaticum (BRASIL 2020). The effectiveness and action of Tricovab® are influenced by strong seasonality and conditioned by meteorological factors and environmental conditions of the crop area (Loguercio et al. 2009). It is known that yeasts are capable of colonizing different habitats (Sampaio et al. 2007), competing for space and nutrients. They occur naturally in the microbiome and act as BCA to pathogens of different crops (Bettiol 1997; Ferraz et al. 2019). But there is almost no information about yeasts as biocontrol agents in cacao tree crop. In this study, we isolated 225 yeasts from tissues of leaf, flowers, and cacao pods and studied their diversity and the capability of some to be BCAs for M. perniciosa, the agent of cacao WB disease. We are using the generic term yeasts, to represent both yeast-like fungi (develops in the mycelial form) and species that develop exclusively through the yeast phase.
Since improved genetic materials for WB resistance are of primordial importance to control the disease this work strategically collected and identified yeasts coexisting in 16 cacao tree genotypes with known resistance (horizontal, quantitative, and polygenic) to M. perniciosa (Pimenta Neto et al. 2018). Yeasts were also collected from four clones indicated as resistant to M. roreri (Phillips-Mora et al. 2007) the agent of frosty pod rot (FPR), an A1 quarantine pest (BRASIL 2018) threatening the Brazilian cacao tree crop. Many genotypes are selected for being resistant against plant diseases. Some of these phenotypes may be favored by beneficial microorganisms which interactions improve desirable phenotypic characteristics (Smith et al. 1997; Ab Rahman et al. 2018). In addition, plant surfaces which have natural ‘openings’ are easier targets for M. perniciosa (Sena et al. 2014), and these characteristics have been used by bioprospection.
In general, all yeast isolates obtained from the cacao tree phytobiome proved to have one or more of the skills evaluated in the in vitro tests performed to characterize them as antagonists to M. perniciosa (Figs. 1, 2, 3, and 4) (Zhang et al. 2007; Bruce et al. 2003; Platania et al. 2012). They show different levels of competition with the pathogen, ability to produce diffusible and volatile compounds, release killer toxins (with exception of 13 isolates; Fig. 3) and inhibit basidiospore germination.
Our results indicate the yeast preference for different plant tissues and genotypes groups (Tables 2 and 3, Fig. 5). Four genera occurred only in the plant tissues of genotypes resistant to WB, two genera only in genotypes resistant to FPR, and seven genera were common to both genotypes (Fig. 5). The yeasts preference for some genotypes may be influenced by anatomic and morphophysiological characteristics of the plant tissues or the availability of nutrients (Leveau and Lindow 2001). Yeasts preferentially colonize substrates which are rich in carbohydrates (Sampaio et al. 2007), such as the carposphere, spermosphere, and antosphere as well as the phyllosphere (Sláviková et al. 2009). Of the total yeasts identified, the majority (58.11% Table 2) was isolated from leaves, which represents an important and abundant cacao tree biomass (Siswanto et al. 2009). Other 32.43% were isolated from flowers and 9.46% from pods, result that supports the characteristics of survival and adaptation from yeasts.
In general, there was a predominance of yeasts belonging to the Basidiomycete class compared to Ascomycetes (Table 1). Rhodotorula paludigena was the predominant species corresponding to 34.1% of the isolates obtained from cacao leaves. This species has already been found on the surface of pod, leaves and in aquatic environments such as swamps and ocean (Sampaio 2011); however, this is the first report of its occurrence on leaves, flowers and pods of cacao tree. This species acts as biocontrol microorganism for post-harvest diseases, such as gray mold in tomatoes caused by Botryts cinerea (Wang et al. 2010), green mold in Ponkan mandarin, caused by Penicilium digitatum (Lu et al. 2014) and in oranges (Lu et al. 2013). It is also associated with soil suppressiveness and growth promotion (El-Tarabily and Sivasithamparam 2006). R. paludigena was isolated from the phytobiome of six of the genotypes resistant to WB and two of those resistant to FPR (Table 2).
The taxa Rhodotorula paludigena, Cryptococcus heimaeyensis, Hanseniaspora uvarum, and Candida orthopsilosis presented the best results for one or more studied variables: mycelial growth inhibition by diffusible compounds (Fig. 1), by volatile compounds (Fig. 2), spore germination inhibition (Fig. 3), and Killer toxin production (Fig. 3). Among these four species, R. paludigenum grows at 30 °C, but not at 35 °C (Sampaio 2011); Cryptococcus heimaeyensis does not grow at 30 °C (Fonseca 2011); H. uvarum grows at 30 °C; rare isolates grow at 34 °C and does not grow at 37 °C (Cadez and Smith 2011); however, the species Candida orthopsilosis grows at 37 °C and 42 °C (Lachance et al. 2011); thus, the latter have the potential to grow in human body. The genus Candida brings together species widely used to control plant diseases (Droby et al. 2003; Zhang et al. 2014); it also comprises species that participate in a complex of human pathogens (Bertini et al. 2013). Although C. orthopsilosis has been found in clinical samples on all continents, it is also found in vegetables (Lachance et al. 2011), as in tomato fruits (Robl et al. 2014), sugar cane leaves (Limtong et al. 2014), and in Piper sp. (Kanti et al. 2013).
Cryptococcus heimaeyensis showed the ability to colonize all the cacao tree surfaces sampled: leaf, flower, and pod (Table 2). The literature describes its isolation from several environments, such as soil near volcano (Vishniac 2002), flower nectar (Herzberg et al. 2002), alluvial forest soil in Austria (Wuczkowski and Prillinger 2004), and leaves (Vishniac 2002). New species of the genus Cryptococcus, such as C. foliicola and C. taibaiensis (Wang et al. 2011) were described to inhabit plant tissues.
This study is the first report of occurrence of the species R. paludigenum, Occultifur externus, Sporolomyces roseus, Cryptococcus tabaiensis, Candida heimaeyensis, Hannaella luteola, H. pagnoccae, H. oryzae, and H. coprosmae on the surface of leaves, flowers, and pod from cacao tree (TableS 1 and 2). Members of the phylum Basidiomycota, Tremellales clade, are used as biocontrol of plant diseases, as among them Cryptococcus albidus (Tian et al. 2002), C. laurentii (Tian et al. 2007; Wang and Bai 2008), C. flavus, H. sinensis, and R. palludigenum (Nutaratat et al. 2014). Hannaela was the most frequent genus in cacao tree phytobiome with 23 isolates, representing 42.6% of the identified isolates within the Basidiomycetes. However, only seven of these isolates were specifically identified belonging to four species (Table 1). From this genus, LEV 63 (H. luteola), 93 (H. oryzae), and Lev 10, 147, and 207 (Hannaela sp.) were among the yeast isolates that most inhibited M. perniciosa basidiospore germination and release killer toxins (Fig. 3). Lev 207 also produced diffusible and volatile compounds that induced high mycelial growth inhibition (Fig. 1). Among the nonspecifically identified isolates of this genus it may be possible to find new species for science.
Three of the Hanseniaspora uvarum isolates (LEV 210, 211, and 162) stood out for growth inhibition (Figs. 1 and 2) and spore germination (Fig. 3) of M. perniciosa, in addition to being killer toxin producers (Fig. 3). Several yeasts secrete killer toxins, which allows establishment in the environment, inhibiting the growth of other organisms, through damage to the plasma membrane (Santos and Marquina 2004; Santos et al. 2009), cell wall damage by the inhibition of β-glucans synthesis (Marquina et al. 2002; Peng et al. 2010), blocking the cell cycle (Klassen et al. 2004) and the calcium uptake (Gage et al. 2001), and triggering DNA damage and inducing apoptosis (Klassen and Meinhardt 2005). Killer toxins could be also conjugated with other biocidal biomolecules for the formation of bionanohydro agroparticles (Baker et al. 2017).
Hanseniaspora uvarum is widely used in the grape’s fermentation for wine production (Escalante et al. 2011; Moreira et al. 2011) and enzyme production for industrial interest (López et al. 2016). In addition, it is used to control post-harvest diseases (Janisiewicz et al. 2010) through the production of killer toxins (Schmiti and Neuhausen 1994) and volatile compounds (Moreira et al. 2011). Although H. uvarum has already been reported as part of the microbiota present in the fermentation of cacao hybrid beans (Moreira et al. 2013), this is the first report of this species that inhabits the surface of cacao leaves and flowers.
Some species of the Saccharomycetaceae family obtained in this study were identified for the first time in the phyllosphere, carposphere, and antosphere from cacao tree: Yamadazyma triangularis (syn. Pichia triangularis), Candida polymorpha, C. sorbosivorans, C. nonsorbophila, C. saopaulonensis, C. parapsilosis, and C. orthopsilosis; as well as Pichia sporocuriosa, which currently belong to the Pichiaceae family. Some of these species were reported in the literature as the biocontrol agent of plant diseases such as Candida saopaulonensis controlling stem rot and damping-off in cowpea (Tenório et al. 2019), as well as P. sporocuriosa (Péter et al. 2000) and Wickerhamomyces anomalus (Hashem et al. 2014).
Also, these evaluated yeasts presented the main characteristics for the selection of putative BCA - Biological Control Agents (Ferraz et al. 2019), attributed to the mechanisms of biocontrol such as the production of lytic enzymes (Urquhart and Punja 2002) and killer toxins (Cabral et al. 2009; Perez et al. 2016) and spore germination inhibition (Lutz et al. 2012). In addition to the possibility of inducing or producing phytohormones (Sun et al. 2014), biofilm formation (Li et al. 2015) and being excellent competitors for nutrients and space, which allow their stabilization in hostile environments, exposed to variations in temperature and humidity (Barbieri and Carvalho 2001; Lahlali et al. 2011; Zhao et al. 2011).
This paper comprises an unprecedented study showing for the first time the great yeast diversity existing in the cacao tree phytobiome with some new assignments of species for this crop and Bahia State. The essential in vitro tests to screen for putative BCAs allowed to find 21 yeast isolates belonging to 17 taxa as promising BCA against M. perniciosa, the agent of cacao Witches’ broom disease. Six of them (Candida orthopsilosis—LEV 225; Hanseniaspora uvarum—LEV 162, 210, and 211; and Rhodotorula paludigena—LEV 166, 168) performed among the best ones for all essential qualities of a BCA agent now tested. Thus, it opens the path to cover the lack of new BCA agents to control the most important cacao tree disease in Brazil.
Data availability
The data that support the findings of this study are available from the corresponding author on request.
References
Ab Rahman SFS, Singh E, Pieterse CMJ, Schenk PM (2018) Emerging microbial biocontrol strategies for plant pathogens. Plant Science 267:102–111
Afsah-Hejri L (2013) Saprophytic yeasts: effective biocontrol agents against Aspergillus flavus. International Food Research Journal 20:3403–3409
Assis SMP, Mariano RLR, Michereff SJ, Silva G, Maranhão EAA (1999) Antagonism of yeasts to Xanthomonas campestris pv. campestris on cabbage phylloplane in field. Revista de Microbiologia 30:191–195
Baker S, Volova T, Prudnikova SV, Satish S, Prasad N (2017) Nanoagroparticles emerging trends and future prospect in modern agriculture system. Environmental Toxicology and Pharmacology 53:10–17
Barbieri R, Carvalho IF (2001) Coevolução de plantas e fungos fitopatogênicos. Revista Brasileira de Agrociência 7:79–83
Bertini A, De Bernardis F, Hensgens LA, Sandini S, Senesi S, Tavanti A (2013) Comparison of Candida parapsilosis, Candida orthopsilosis, and Candida metapsilosis adhesive properties and pathogenicity. International Journal of Medical Microbiology 303:98–103
Bettiol W (1997) Biocontrole na filosfera: problemas e perspectivas. Revisão Anual de Patologia de Plantas 5:59–97
Bowers JH, Bailey BA, Hebbar PK, Sanogo S, Lumsden RD (2001) The impact of plant diseases on world chocolate production. Plant Health Progress 2:12. Available at: https://doi.org/10.1094/PHP-2001-0709-01-RV. Accessed on December 18, 2020
BRASIL. Ministério da Agricultura, Pecuária e Abastecimento. Instrução normativa 39. Oct 2018. Available at: www.in.gov.br/materia/-/asset_publisher/Kujrw0TZC2Mb/content/id/43460217/do1-2018-10-02-instrucao-normativa-n-39-de-1-de-outubro-de-2018-43460055. Accessed on April 8, 2020
BRASIL. Ministério da Agricultura, Pecuária e Abastecimento. AGROFIT: consulta de produtos formulados. Available at: www.agrofit.agricultura.gov.br/agrofit_cons/principal_agrofit_cons. Accessed on: 08 April 8, 2020
Bruce A, Douglas S, Susan V, Ron EW (2003) Effect of volatiles from bacteria and yeast on the growth and pigmentation of sapstain fungi. International Biodeterioration & Biodegradation 51:101–108
Cabral AS, De Carvalho PMB, Pinotti T, Hagler AN, Mendonça-Hagler LCS, Macrae A (2009) “Killer” yeasts inhibit the growth of the phytopathogen Moniliophthora perniciosa, the causal agent of witches’ broom disease. Brazilian Journal of Microbiology 40:108–110
Cadez N, Smith MT (2011) Hanseniaspora Zikes (1912). In: Kurtzman CP, Fell JW, Boekoult T (eds) The yeast, a taxonomic study, 5th edn. The Netherlands. Elsevier Academic Press, Amsterdam, pp 421–434
Coelho AR, Nóbrega GMA, Pagnocca FC, Hoffmann FL, Harada K, Hirooka EY (2011) Avaliação do potencial antagônico de leveduras, visando biocontrole de deterioração por Penicillium expansum. Semina: Ciência Agrárias 32:1879–1892
Cuatrecasas J (1964) Cacao and its allies: a taxonomic revision of the genus Theobroma. Contributions from the United States National Herbarium 35:379–614
Dajoz R (1983) Ecologia geral. Rio de Janeiro. Brasil, Editora Vozes
Daniel HM, Vrancken G, Takrama JF, Camu N, De Vos P, De Vuyst L (2009) Yeast diversity of Ghanaian cocoa bean heap fermentations. FEMS Yeast Research 9:774–783
Droby S, Wisniewski M, El Ghaouth A, Wilson C (2003) Influence of food additives on the control of postharvest rots of apple and peach and efficacy of the yeast-based biocontrol product aspire. Postharvest Biology and Technology 27:127–135
Duarte SH, de Andrade CC, Ghiselli G, Maugeri F (2013) Exploration of Brazilian biodiversity and selection of a new oleaginous yeast strain cultivated in raw glycerol. Bioresource Technology 138:377–381
El-Tarabily KA, Sivasithamparam K (2006) Potential of yeasts as biocontrol agents of soil-borne fungal plant pathogens and as plant growth promoters. Mycoscience 47:25–35
Escalante WE, Rychtera M, Melzoch K, Sakoda BH, Polo EQ, Cervantes ZL, Casavilca VS, Quilca GC (2011) Actividad fermentativa de Hanseniaspora uvarum y su importancia en la producción de bebidas fermentadas. Revista de la Sociedad Venezolana de Microbiología 31:57–63
Ferraz P, Cássio F, Lucas C (2019) Potential of yeasts as biocontrol agents of the phytopathogen causing cacao witches’ broom disease: is microbial warfare a solution? Frontiers in Microbiology 10:1–13
Fonseca Á (2011) Cyptococcus Vuillemin (1901). In: Kurtzman CP, Fell JW, Boekoult T (eds) The yeast, a taxonomic study, 5th edn. The Netherlands. Elsevier Academic Press, Amsterdam, pp 1661–1737
Gage MJ, Bruenn J, Fischer M, Sanders D, Smith TJ (2001) KP4 fungal toxin inhibits growth in Ustilago maydis by blocking calcium uptake. Molecular Microbiology 41:775–785
Gu Z (2016) Complex heatmaps reveal patterns and correlations in multidimensional genomic
Hashem M, Alamri SA, Hesham AE, Al-Qahtani FMH, Kilany M (2014) Biocontrol of apple blue mould by new yeast strains: Cryptococcus albidus KKUY0017 and Wickerhamomyces anomalus KKUY0051 and their mode of action. Biocontrol Science and Technology 24
Herzberg M, Fischer R, Titze A (2002) Conflicting results obtained by RAPD-PCR and large-subunit rDNA sequences in determining and comparing yeast strains isolated from flowers: a comparison of two methods. International Journal of Systematic and Evolutionary Microbiology 52:1423–1433
Hu H, Xu Y, Lu H, Xiao R, Zheng X, Yu T (2015) Evaluation of yeasts from Tibetan fermented products as agents for biocontrol of blue mold of Nashi pear fruits. Journal of Zhejiang University-Science b (biomedicine & biotechnology) 16:275–285
IBGE. Sistema IBGE de Recuperação Eletrônica (SIDRA) Available at: www.sidra.ibge.gov.br/bda/agric/. Accessed on: April 07. 2020
ICCO Quarterly bulletin of cocoa statistics, Vol. XLV, No. 1, Cocoa year 2018/19. 2019
Janisiewicz WJ, Kurtzman CP, Buyer JS (2010) Yeasts associated with nectarines and their potential for biological control of brown rot. Yeast 27:389–398
Kanti A, Sukara E, Latifah K, Sukarno N, Boundy-Mills K (2013) Indonesian oleaginous yeasts isolated from Piper betle and P. nigrum. Mycosphere 4:1015–1026
Klassen R, Meinhardt F (2005) Induction of DNA damage and apoptosis in Saccharomyces cerevisiae by a yeast killer toxin. Cellular Microbiol 7:393–401
Klassen R, Teichert S, Meinhardt F (2004) Novel yeast killer toxins provoque S-phase arrest and DNA damage checkpoint activation. Molecular Microbiology 53:263–273
Lachance MA, Boekhout T, Scorzetti G, Fell JW, Kurtzman CP (2011) Candida Berkhout (1923). In: Kurtzman CP, Fell JW, Boekoult T (eds) The yeast, a taxonomic study, 5th edn. The Netherlands. Elsevier Academic Press, Amsterdam, pp 987–1278
Lahlali R, Hamadi Y, El Guilli M, Jijakli MH (2011) Efficacy assessment of Pichia guilliermondii strain Z1, a new biocontrol agent, against citrus blue mould in Morocco under the influence of temperature and relative humidity. Biological Control 56(3):217–224
Leal GA Jr, Gomes LH, Efraim P, Tavares FCA, Figueira A (2008) Fermentation of cacao (Theobroma cacao L.) seeds with a hybrid Kluyveromyces marxianus strain improved product quality attributes. FEMS Yeast Research 8:788–798
Leveau JH, Lindow SE (2001) Appetite of an epiphyte: quantitative monitoring of bacterial sugar consumption in the phyllosphere. Proceedings of the National Academy of Science of The United States of America 98:3446–3453
Li X, Yu VY, Lin Y, Chomvong K, Estrela R, Park A, Liang LM, Znameroski EA, Feehan J, Kim SR, Jin Y, Glass NL, Cate JHD (2015) Expanding xylose metabolism in yeast for plant cell wall conversion to biofuels. Elife 4:1–16
Limtong S, Kaewwichian R, Youngmanitchai W, Kawasaki H (2014) Diversity of culturable yeast in phylloplane of sugarcane in Thailand and their capability to produce índole-3-acetic acid. World Journal Microbiology and Biotechnology 30:1785–1796
Loguercio LL, Santos LS, Niella GR, Miranda RAC, Souza JT, Collins RT, Pomella AWV (2009) Canopy-microclimate effects on the antagonism between Trichoderma stromaticum and Moniliophthora perniciosa in shaded cacao. Plant Pathology. 58:1104–1115
López S, Mateo JJ, Maicas S (2016) Screening of Hanseniaspora strains for the production of enzymes with potential interest for winemaking. Fermentation 1:1–16
Lu L, Ye C, Guo S, Sheng K, Shao L, Zhou T, Yua T, Zheng X (2013) Preharvest application of antagonistic yeast Rhodosporidium paludigenum induced resistance against postharvest diseases in mandarin orange. Biological Control 67:130–136
Lu H, Lu L, Zeng L, Fu D, Xiang H, Yu T, Zheng X (2014) Effect of chitin on the antagonistic activity of Rhodosporidium paludigenum against Penicillium expansum in apple fruit. Postharvest Biology and Technology 92:9–15
Lutz MC, Lopes CA, Rodriguez ME, Sosa MC, Sangorrín MP (2012) Efficacy and putative mode of action of native and commercial antagonistic yeasts against postharvest pathogens of pear. International Journal of Food Microbiology. 164:166–172
Marelli JP, Maximova SN, Gramacho KP, Kang S, Guiltinan MJ (2009) Infection biology of Moniliophthora perniciosa on Theobroma cacao and alternate solanaceous hosts. Tropical Plant Biology 2:149–160
Mares JH, Gramacho KP, Santos EC, Santiago AS, Silva EMA, Alvim FC, Pirovani CP (2016) Protein profile and protein interaction network of Moniliophthora perniciosa basidiospores. BMC Microbiology 16:120
Mariano RLR, Gomes AMA, Assis SMP, Silveira EB (2005) Mecanismos de ação de bactérias promotoras de crescimento de plantas. In: Mariano RLR, Silveira EB (Eds.) Manual de práticas em fitobacteriologia. 2a. Ed. Recife, PE. UFRPE. pp. 143–157
Marquina D, Santos A, Peinado JM (2002) Biology of killer yeasts. International Microbiology 5:65–71
Masih EI, Paul B (2002) Secretion of β-1,3-glucanases by the yeast Pichia membranifaciens and its possible role in the biocontrol of Botrytis cinerea causing grey mold disease of the grapevine. Current Microbiology 44:391–395
Midlej RR, Santos AM (2012) Economia do Cacau. In: VALLE RR (ed) Ciência, Tecnologia e Manejo do Cacaueiro. Brasília, DF. Ceplac, pp 655–672
Miller GL (1959) Use of dinitrosalicylic acid reagent for determination of reducing sugar. Analytical Chemistry 31:426–428
Moreira N, Moreira N, Pina C, Mendes F, Couto JA, Hogg T, Vasconcelos I (2011) Volatile compounds contribution of Hanseniaspora guilliermondii and Hanseniaspora uvarum during red wine vinifications. Food Control 22:662–667
Moreira IMV, Pedrozo MGC, Duarte MWF, Dias DR, Schwan RF (2013) Microbial succession and the dynamics of metabolites and sugars during the fermentation of three different cocoa (Theobroma cacao L.) hybrids. Food Research International 54:9–17
Muller-Dombois D (1981) Ecological measurements and microbial populations. In: Wicklow DT, Carroll GC (eds) The fungal community: its organization and role in the ecosystem. USA. Marcel Derker, New York, pp 173–184
Nutaratat P, Srisuk N, Arunrattiyakorn P, Limtong S (2014) Plant growth-promoting traits of epiphytic and endophytic yeasts isolated from rice and sugar cane leaves in Thailand. Fungal Biology 118:683–194
O’Donnell K (1993) Fusarium and its near relatives. In: Reynolds DR, Taylor JW (Eds.) The fungal holomorph: Mitotic, Meiotic and Pleomorphic Speciation. Fungal Systematics. Wallingford, UK: CAB International. pp. 225–233
Oliveros JC (2007) Venny. An interactive tool for comparing lists with Venn diagrams. Available at: http://bioinfogp.cnb.csic.es/tools/venny/index.html. Accessed on December 18, 2020
Patrocínio NGRB, Ceresini PC, Gomes LIS, Resende MLV, Mizubuti ESG, Gramacho KP (2016) Population structure and migration of the witches’ broom pathogen Moniliophthora perniciosa from cacao and cultivated and wild solanaceous hosts in southeastern Brazil. Plant Pathology 66:900–911
Peng Y, Chi Z, Wang X, Li J (2010) β-1,3-Glucanase inhibits activity of the killer toxin produced by the marine-derived yeast Williopsis saturnus WC91-2. Marine Biotechnology 12:479–485
Perez MF, Contreras L, Garnica NM, Zenoff MVF, Farias ME, Sepulveda M, Ramallo J, Dib JR (2016) Native killer yeasts as biocontrol agents of postharvest fungal diseases in lemons. PLoS One 11:1–21
Péter G, Tornai-Lehoczki J, Dlauchy D, Vitányi G (2000) Pichia sporocuriosa sp. nov. a new yeast isolated from rambutan. Antonie Van Leeuwenhoek 77:37–42
Phillips-Mora W, Aime MC, Wilkinson MJ (2007) Biodiversity and biogeography of the cacao (Theobroma cacao) pathogen Moniliophthora roreri. Plant Pathology 56:911–922
Pimenta Neto AA, Laranjeira D, Pires JL, Luz EDMN (2018) Selection of cocoa progenies for resistance to witches’ broom. Tropical Plant Pathology 43:1–8
Platania C, Restucciab C, Muccillib S, Cirvilleri G (2012) Efficacy of killer yeasts in the biological control of Penicillium digitatum on Tarocco orange fruits (Citrus sinensis). Food Microbiology 30:219–225
Pomella AWV, Souza JT, Niella GR, Bateman RP, Hebbar PK, Loguercio LL, Lumsden RD (2007) Trichoderma stromaticum for management of witches' broom of cacao in Brazil. In: Vincent C, Goettel MS, Lazarovits G (eds) Biological control: a global perspective. CAB International, Wallingford, p 210
R Core Team (2019) R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. Available at: https://www.R-project.org/. Accessed on April 12, 2020
Rice RA, Greenberg R (2010) Cacao cultivation and the conservation of biological diversity. Ambio: A Journal of the Human Environment 29:167–173
Robl D, Thimoteo SS, Souza GCCF, Beux MR, Dalzoto PR, Pinheiro RL, Pimentel IC (2014) Occurrence of Candida orthopsilosis in Brazilian tomato fruits (Lycopersicum esculentum mill.). Brazilian Journal of Microbiology 45:105–109
Sampaio JP (2011) Rhodotorula Harrison (1928). In: Kurtzman CP, Fell JW, Boekhout T (eds) The yeasts, a taxonomic study. 5th Ed. vol. 3. Elsevier, New York, USA, pp 1873–1927
Sampaio A, Sampaio JP, Leão C (2007) Dynamics of yeast populations recovered from decaying leaves in a nonpolluted stream: a 2-year study on the effects of leaf litter type and decomposition time. FEMS Yeast Research 7:595–603
Sangorrín MP, Zajonskovsky IE, Lopes CA, Rodríguez ME, Giraudo de van Broock MR, Caballero AC (2001) “Killer” behavior in wild wine yeasts associated with Merlot and Malbec type musts spontaneously fermented from northwestern Patagonia (Argentina). Journal of Basic Microbiology 41:105–113
Santos A, Marquina D (2004) Killer toxin of Pichia membranifaciens and its possible use as a biocontrol agent against grey mould disease of grapevine. Microbiology 150:2527–2534
Santos A, San Mauro M, Bravo E, Marquina D (2009) PMKT2, a new killer toxin from Pichia membranifaciens, and its promising biotechnological properties for control of the spoilage yeast Brettanomyces bruxellensis. Microbiology 155:624–634
Schmiti MJ, Neuhausen F (1994, 1765-1772) “Killer” Toxin-secreting double-stranded RNA mycoviruses in the yeasts Hanseniaspora uvarum and Zygosaccharomyces bailii. Journal of Virology
Scott RJ, Knott M (1974) A cluster analysis method for grouping mans in the analysis of variance. Biometrics 30:507–512
Sena K, Alemanno L, Gramacho KP (2014) The infection process of Moniliophthora perniciosa in cacao. Plant Pathology 63:1272–1281
Silva SDVM, Luz EDMN, Pires JL, Yamada MM, Santos Filho LP (2010) Parent selection for cocoa resistance to witches’broom. Pesquisa Agropecuária Brasileira 45:680–685
Siswanto S, Hartati RS, Prastowo B (2009) Biomass of cocoa and sugarcane. IOP Conference Series: Earth and Environmental Science 65:1–12
Sláviková E, Vadkertiová R, Vránová D (2009) Yeasts colonizing the leaves of fruit trees. Annals of Microbiology 59:419–424
Smith KP, Handelsman J, Goodman RM (1997) Modeling dose-response relationships in biological control: partitioning host responses to the pathogen and biocontrol agent. Phytopathology 87:720–729
Sui Y, Wisniewski M, Droby S, Liua J (2015) Responses of yeast biocontrol agents to environmental stress. Applied and Environmental Microbiology 81:2968–2975
Sun P, Fang W, Shin L, Wei J, Fu S, Chou J (2014) Indole-3-acetic acid-producing yeasts in the phyllosphere of the carnivorous plant Drosera indica L. PLoS One 9:1–22
Tavares LHD (1979) História da Bahia, 6a edn. São Paulo, Brasil. Ática
Teixeira PJ, Thomazella DP, Pereira GA (2015) Time for chocolate: current understanding and new perspectives on cacao witches’broom disease research. PLoS Pathogens. Online
Tenório DA, Medeiros EV, Lima CS, Silva JM, Barros JA, Neves RP, Laranjeira D (2019) Biological control of Rhizoctonia solani in cowpea plants using yeast. Tropical Plant Pathology. Online
Tian SP, Fan Q, Xu Y, Liu HB (2002) Biocontrol efficacy of antagonist yeasts to gray mold and blue mold on apples and pears in controlled atmospheres. Plant Disease 86:848–853
Tian SP, Yao HJ, Deng X, Xu XB, Qin GZ, Chan ZL (2007) Characterization and expression of β-1,3-glucanase genes in jujube fruit induced by the microbial biocontrol agent Cryptococcus laurentii. Phytopathology 97:260–268
Topps JH, Wain RL (1957) Investigation on fungicides. III. The fungi toxicity of 3-and 5- alkyl salicylanilide and P-chloronilines. Annals of Applied Biology 45:506–511
Urquhart EJ, Punja ZK (2002) Hydrolytic enzymes and antifungal compounds produced by Tilletiopsis species, phyllosphere yeasts that are antagonists of powdery mildew fungi. Canadian Journal of Microbiology 48:219–229
Valdebenito-Sanhueza RMV (2000) Leveduras para o biocontrole de fitopatógenos. In: Melo IS, Azevedo JL (eds) Controle biológico. Jaguariúna, DF, Embrapa Meio Ambiente, pp 41–56
Vishniac HS (2002) Cryptococcus tephrensis, sp. nov. and Cryptococcus heimaeyensis, sp. nov.; new anamorphic basidiomycetous yeast species from Iceland. Canadian Journal of Microbiology 48:463–467
Wang QM, Bai FY (2008) Molecular phylogeny of basidiomycetous yeasts in the Cryptococcus luteolus lineage (Tremellales) based on nuclear rRNA and mitochondrial cytochrome b gene sequence analyses: proposal of Derxomyces gen. nov. and Hannaella gen. nov., and description of eight novel Derxomyces species. FEMS Yeast Research 8:799–814
Wang Y, Yu T, Xi J, Yu D, Wang J, Zheng X (2010) Biocontrol of postharvest gray mold of cherry tomatoes with the marine yeast Rhodosporidium paludigenum. Biological Control 53:178–182
Wang YF, Tang F, Xi J, Yu T, Wang J, Azhati R, Zheng XD (2011) A combination of marine yeast and food additive enhances preventive effects on postharvest decay of jujubes (Zizyphus jujuba). Food Chemistry 125:835–840
White TJ, Bruns T, Lee S, Taylor JW (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ (eds) PCR protocols: a guide to methods and applications. Academic Press Inc, New York, NY, pp 315–322
Wuczkowski M, Prillinger H (2004) Molecular identification of yeasts from soils of the alluvial forest national park along the river Danube downstream of Vienna, Austria (“Nationalpark Donauauen”). Microbiological Research 159:263–275
Young TW (1987) “killer” yeasts. In: Rose AH, Harrison JS (Eds.). The yeasts. v. 2. London: Academic Press. pp. 131–164
Zhang HY, Wang L, Dong Y, Jiang S, Cao J, Meng R (2007) Postharvest biological control of gray mold decay of strawberry with Rhodotorula glutinis. Biological Control 40:287–292
Zhang D, Lu CG, Zhang TT, Spadaro D, Liu DW, Liu WC (2014) Candida pruni sp. nov. is a new yeast species with antagonistic potential against brown rot of peaches. Archives of Microbiology 196:525–530
Zhao K, Sun LX, Ma X, Li XL, Wang X, Ping WX, Zhou DP (2011) Improved taxol production in Nodulisporium sylviforme derived from inactivated protoplast fusion. African Journal of Biotechnology 10:4175–4182
Acknowledgments
The authors thank the Fundação de Amparo à Pesquisa do Estado da Bahia (FAPESB) for funding the project and granting scholarships; the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES) and the Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) for granting scholarships; and the Phytopathology Graduate Program of the Universidade Federal Rural de Pernambuco (UFRPE), the Comissão Executiva do Plano da Lavoura Cacaueira (CEPLAC), the Universidade Estadual de Santa Cruz (UESC), and the Universidade Federal do Recôncavo da Bahia (UFRB) for providing the infrastructure for this study.
Author information
Authors and Affiliations
Contributions
All authors contributed to the idea and design of the study. Material preparation, data collection and analysis were carried out by Antônio Alves Pimenta Neto, Tacila Ribeiro Santos, Elizabeth Amélia Alves Duarte, Thiago Alves Santos de Oliveira, Edson Mário de Andrade Silva, Ana Paula Trovatti Uetanabaro, Andréa Miura da Costa, Edna Dora Martins Newman Luz, and Delson Laranjeira. The first draft of the manuscript was written by Antônio Alves Pimenta Neto and all authors commented on it as previous versions of the manuscript. All authors heard and approved the final manuscript.
Conceptualization: Antônio Alves Pimenta Neto and Edna Dora Martins Newman Luz
Methodology: Antônio Alves Pimenta Neto, Tacila Ribeiro Santos, Elizabeth Amélia Alves Duarte, Thiago Alves Santos de Oliveira
Formal analysis and investigation: Antônio Alves Pimenta Neto, Elizabeth Amélia Alves Duarte, Thiago Alves Santos de Oliveira, Edson Mário de Andrade Silva
Acquisition of financing: Edna Dora Martins Newman Luz
Writing–preparation of the original draft: Antônio Alves Pimenta Neto
Writing–review and editing: Antônio Alves Pimenta Neto, Edna Dora Martins Newman Luz, Elizabeth Amélia Alves Duarte, Thiago Alves Santos de Oliveira, Edson Mário de Andrade Silva
Supervision: Edna Dora Martins Newman Luz, Ana Paula Trovatti Uetanabaro, Andréa Miura da Costa, Delson Laranjeira
Corresponding author
Ethics declarations
Conflict of interest
On behalf of all authors, the corresponding author states that there is no conflict of interest.
Additional information
The results presented are part of the doctoral work of the first author Antônio Alves Pimenta Neto.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Neto, A.A.P., Santos, T.R., Duarte, E.A.A. et al. Yeasts associated with aerial parts of Theobroma cacao L. in southern Bahia, Brazil, as prospective biocontrol agents against Moniliophthora perniciosa. Trop. plant pathol. 46, 109–128 (2021). https://doi.org/10.1007/s40858-020-00418-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40858-020-00418-w