Abstract
Soybean rust, caused by Phakopsora pachyrhizi, is one of the most important foliar diseases threatening soybean production in Uruguay. Knowledge of pathogenic variation among Uruguayan rust populations is necessary to guide development of resistant soybean cultivars in national breeding programs. To assess pathogenic variation,12 P. pachyrhizi isolates were collected from fields across the country over a four-year period and were inoculated onto a set of 12 differential soybean genotypes. All Uruguayan rust isolates (URPs) were highly virulent on differential soybean plants carrying resistance genes Rpp1, Rpp3, and Rpp4. Conversely, all isolates showed resistant reactions on soybean differentials with Rpp1-b and on a line carrying the genes Rpp2, Rpp4, and a Rpp5 allele. The pathogenic variation of the URPs was compared to that of a collection of 157 P. pachyrhizi isolates from other Latin American countries and Japan. All URPs clustered together and with other fungal isolates from South America. Of the seven different pathotypes that were found, four shared identical virulence patterns with isolates from South America, and three were associated with unique virulence patterns, which mainly resulted from the reactions they caused on plants carrying Rpp3, Rpp4 and especially Rpp6. The results indicate that the URPs can overcome the resistance of a larger number of Rpp genes than P. pachyrhizi isolates from neighboring countries. The resistant genes Rpp1-b and Rpp5 proved to be effective against the pathogen and will be introgressed into local soybean lines.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Soybean [Glycine max (L.) Merr.] has become the most important rainfed crop in Uruguay, with hectarage more than tripling in the last decade (DIEA 2016). As the crop has expanded, there have been increasing concerns about diseases affecting its yield. One of the most serious foliar diseases threatening the crop is Asian soybean rust (ASR) caused by Phakopsora pachyrhizi Sydow & Sydow. This pathogen was first reported in Japan in 1902 (Henning 1903), and between 1996 and 2001 it spread throughout different countries in Africa (Levy 2005; Akinsanmi et al. 2001; Pretorius et al. 2001). In 2001, it was first detected in South America, initially in Paraguay and Brazil (Yorinori et al. 2005). It was first found in Argentina in 2003 (Ivancovich 2005; Rossi 2003) and in 2004 in Uruguay (Stewart et al. 2005). Brazil and Argentina, together with the USA are the largest soybean producers in the world (FAOSTAT 2014). Yield losses from 30 to 80% have been observed in South America (Yorinori et al. 2005) and in South Africa (Kawuki et al. 2003; Levy 2005), 25–80% in Mexico (García-Rodríguez et al. 2017) and 43–60% in USA (Mueller et al. 2009; Sikora et al. 2009). In Uruguay, although the disease has been severe in the northeast part of the country, yield losses have not yet been quantified.
Overwintering of P. pachyrhizi requires metabolically active host tissue and temperatures higher than 4 °C. The fungus survives on soybean or on alternative hosts such as kudzu (Pueraria lobata) (Li et al. 2012). According to Pivonia and Yang (2004), Uruguay could be considered a non-overwintering region due to its climate and the absence of kudzu in the country. Thus, the pathogen must be re-introduced into the country every growing season, presumably from Brazil. De Ruyver et al. (2011) showed associations between P. pachyrhizi urediniospores captured in traps and atmospheric patterns favoring wind circulations from the northeast and from northern Argentina during the growing season, which suggests that urediniospores are transported into Uruguay from southern Brazil. This inoculum movement coincides with Uruguayan observations that the first reports of ASR on the crop have always been from the northeastern part of the country, near the Brazilian border, a pattern that has been observed since 2005. Asian Soybean Rust was detected as early as January in 2015 and 2016, and as late as the end of March in 2008, 2009 and 2012 (S. Stewart, unpublished data).
Eight major Rpp resistance genes, and at least five alleles, named Rpp1 - Rpp7 and Rpp1-b, are known to confer resistance to P. pachyrhizi, (Childs et al. 2018; Garcia et al. 2008; Hossain et al. 2015; Li et al. 2012; Ray et al. 2009). The ability of the pathogen to overcome single Rpp genes was reported as early as 1966 for Rpp1 (Bromfield 1984) and in 1978 for Rpp2 (Hartman et al. 2005). The Rpp1 to Rpp4 genes were initially effective in Brazil but were defeated by the pathogen within a few years (Akamatsu et al. 2013; Yorinori 2008). For years Rpp1 and Rpp6 were effective against field populations in southern USA, however in 2012 these genes became ineffective to field populations from north-central Florida (Paul et al. 2013). Pyramiding resistance genes has been proposed as a strategy for durable resistance. Consistently, enhanced resistance in lines possessing the combination of Rpp2 + Rpp4 + Rpp5 was demonstrated by Yamanaka et al. (2013).
Physiological specialization of the pathogen on soybean has also been known for several decades (Bromfield 1984). Resistance breakdown has been reported around the world (Hartman et al. 2005; Miles et al. 2008) as new pathotypes emerge due to spontaneous mutations, long distance movement (Freire et al. 2008), high evolutionary rates (Langenbach et al. 2016), and selection pressure (McDonald and Linde 2002). Temporal and geographic comparisons of pathogenicity have been made; isolates collected from Africa and South America in 2001 were more virulent than Asian and Australian isolates from the 1970s (Bonde et al. 2006). Moreover, higher virulence was also revealed in the Brazilian P. pachyrhizi populations compared to Japanese populations (Yamanaka et al. 2010).
Management of ASR in Uruguay currently relies heavily on fungicide applications, and although they can be effective if applied in a timely manner, they add production costs and are not environmentally friendly. The use of rust resistant soybean varieties would be the most sustainable and cost-effective disease management approach (Hartman et al. 2005). But, knowledge on the pathogenic variation of the rust populations in the country is necessary to be able to incorporate effective resistance to soybean cultivars in national breeding programs. Therefore, the objective of this study was to characterize URPs to determine the effectiveness of soybean resistance genes and to compare pathogenic variability of URPs with those of populations from neighboring and some other countries.
Materials and methods
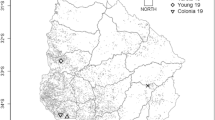
Soybean rust isolates were collected from different soybean plants in individual fields across the country during four growing seasons. Urediniospores from individual fields were harvested in bulk from 30 to 40 leaflets, and these non-purified samples were defined as isolates or “URPs”. Three, four, two and three isolates were obtained in 2014, 2015, 2016 and 2017, respectively (Fig. 1). Urediniospores of P. pachyrhizi were promptly inoculated onto the susceptible variety BRS154 for multiplication purposes. A differential set of 11 soybean varieties, each containing one or three Rpp genes, and a universal susceptible variety, BRS154, were used to evaluate pathogenicity of the URPs (Table 1). Three plants per genotype were grown in growth chambers at 24 °C, and were inoculated at stages V3-V4 each year, immediately after multiplication of rust isolates, following a slightly modified Yamanaka et al. (2017) protocol. Briefly, plants were inoculated with a urediniospore suspension either in mineral oil (Soltrol 170, Phillips Petroleum Co.) or in distilled water with 0.04% Tween 20 to homogeneously cover the leaf surfaces. Plants were maintained in a humid chamber in the dark for 12 h, and later placed in a growth chamber at 24 °C.
Disease evaluations were made two weeks after inoculation on at least one leaflet from two or three plants per genotype. Thirty individual lesions from these leaflets were evaluated for sporulation level (SL) and number of uredinia (NoU) following Yamanaka et al. (2010). SL was rated using a 0 to 3 scale, where 0 is no sporulation and 3 is abundant sporulation (Yamanaka et al. 2017). NoU were counted on each lesion prior to removal of spores with a paint brush. Finally, reaction types were classified as resistant (R), intermediate (M) and susceptible (S) according to Yamanaka et al. (2017) (Table 2).
Cluster analysis for each URP was conducted by transforming R, M and S reaction types into 0, 1, and 2 trinomial characters, respectively. Reactions type patterns of URPs were compared to those of 145 rust isolates from South America (44 from Argentina, 58 from Brazil, 43 from Paraguay), four from Mexico and eight from Japan using the same differential soybean genotypes and methods (Akamatsu et al. 2013, 2017; García-Rodríguez et al. 2017). Raw data for this analysis were provided by the Japan International Research Center for Agricultural Sciences. Distance matrices were prepared by calculating the Euclidean distance between isolates using R software v. 3.0.1 (R Core Team 2015), and the resulting matrices were run in a hierarchical clustering function of the software. A dendrogram based on the unweighted pair group method with arithmetic mean (UPGMA) was also constructed with R software. Approximate unbiased probability values (AU p-values) were calculated for each cluster by 10,000 multiscale bootstrap resampling with the “pvclust” package of R (Suzuki and Shimodaira 2006).
Diversity, defined as pathotype richness within South American countries, was estimated by the Gleason index (Sanders 1968), and was calculated using the formula: Hg = (n-1)/ln (N), where n is the number of pathotypes and N the number of rust isolates per country. This index is less sensitive to differences in population size due to the use of a logarithmic scale (Groth and Roelfs 1987).
Results
No mixed reactions, such as TAN and RB, on the soybean genotypes where observed for any of the isolates studied. All URPs were highly virulent on the susceptible variety BRS154, resulting in the development of the expected tan-colored lesions and the development of many uredinia per lesion, ranging from 4.1 to 7.7, with abundant sporulation (SL = 3) (Tables 2 and 3). Susceptible reactions types to all URPs were also observed on PI 200492, PI 587886, PI 462312, PI 416764 and PI 459025 carrying the Rpp1, Rpp3 and Rpp4 genes, respectively, resulting in high NoU means ranging from 2.8 to 4.5, and high SL means ranging from 2.4 to 3.0. On the contrary, resistant reaction types resulted on PI 587880A, PI 594767A and NO6–12-1, which carry the genes Rpp1b and Rpp2 + Rpp4 + Rpp5, with NoU means ranging from 0 to 0.025, and SL means ranging from 0 to 0.25. Rpp1b was the only resistant gene effective against all URPs.
Additionally, 10 URPs were virulent and two had intermediate reactions on PI 567102B (Rpp6). On the other hand, all URPs, except URP-3 and URP-9, were avirulent on PI 200526 carrying the Rpp5 allele. URP-9 was the only isolate virulent on PI 200526 (Rpp5) (Table 3). Two and five of the 12 URPs caused susceptible and intermediate reactions, respectively, on PI 230970 carrying Rpp2. The highest pathotypic variability of the URPs was observed on soybean differentials PI 230970 and PI 200526, carrying Rpp2 and Rpp5, respectively, whose reaction types ranged from resistance to susceptibility (Tables 3).
Seven pathotypes were defined based on the reaction types observed on the differential soybean plants when challenged with the 12 URPs studied, and were named PT1 to PT7 (Table 3). Pathotypes were distinguished due to the reaction of only three (PI 230970, PI 200526, PI 567102B) of the 12 soybean differentials. The pathotypes PT1, PT2, PT3, PT5, and PT7, corresponding to URP-9, URP-3, URP-11, URP-5 and URP-2, respectively, were unique within 12 URPs tested. On the other hand, the other two pathotypes were represented by three or more URPs, PT4 included four (URP-1, URP-6, URP-8, URP-12) and PT6 included three (URP-4, URP-7, URP-10) P. pachyrhizi isolates.
A dendrogram based on the reactions of the 12 soybean differentials to the seven Uruguayan pathotypes (PT1-PT7) compared to the reactions of 157 fungal isolates previously evaluated by the same methodology was obtained (Fig. 2). All Uruguayan pathotypes grouped together in a large cluster (AU p value = 99) separately from both Japanese and Mexican isolates. Four of the Uruguayan pathotypes (PT2, PT4, PT5 and PT7), represented by seven URPs, clustered together with isolates from Argentina, Brazil and Paraguay. Isolate URP-2 had a reaction type pattern on the soybean differentials identical to those of the Argentinian isolate APM4–3 and the Paraguayan isolate PMA6–5 and grouped together in PT7. URP-3 had a reaction type pattern identical to those of Brazilian and Paraguayan isolates BS03–1, PMA9–1 and PNC1–1, and grouped together in PT2. The reaction type pattern of the four isolates of PT4 (URP-1, URP-6, URP-8 and URP-12) were identical to that of the Brazilian isolate BCW10–5. The reaction type pattern of URP-5 (PT5) was identical to those of Argentinian APM16–8, APM2–3, ANW10–3 and Paraguayan PSI15–1, PSI13–1 isolates. The other five URPs represented three unique pathotypes in South America (URP-9 as PT1, URP-11 as PT3, and URP-4, URP-7, and URP-10 as PT6) (Fig. 2). Additionally, URP-9 (PT1) and URP-11 (PT3) sub-clustered separately from the other isolates mainly due to virulence differences on soybean differentials PI 230970 (Rpp2), PI 200526 (Rpp5) and PI 567102B (Rpp6).
Frequencies of resistant reaction types on soybean genotypes with different Rpp genes varied depending on the geographic origin of the P. pachyrhizi isolate (Fig. 3). Japanese isolates were less virulent than isolates from South American countries, including Uruguay. Major reaction differences were seen on PI 200492 (Rpp1), PI 462312 and PI 416764 (Rpp3) and PI 567102B (Rpp6) which were 100% resistant to isolates from Japan, as opposed to 0% for URPs. Additionally, frequency of resistant reaction types on soybean differentials PI 462312 and PI 416764 (Rpp3), PI 459025 (Rpp4), and PI 567102B (Rpp6) ranged from 32 to 66% for isolates from other South American countries (Argentina, Brazil and Paraguay), while no resistant reaction was found for URPs. The most noticeable difference between South American countries with regard to resistant reaction frequencies was observed for PI 567102B (Rpp6), which ranged from 0% for Uruguayan to 93% for Paraguayan isolates (data not shown). One-hundred percent resistance effectiveness was observed for single gene Rpp1-b (PI 587880A and PI 594767A) and for the pyramided line No6–12-1 carrying Rpp2, Rpp4 and a Rpp5 allele. Furthermore, the latter was the only soybean genotype that was resistant to more than 98% of the ASR isolates from all countries studied.
No clear correlation between clustering and geographic origin was noticed for the set of URPs studied (Fig. 2). On the contrary, three (URP- 4, URP-7, and URP-10) out of the four isolates from 2015 clustered together (PT6) denoting similarities according to collection date, at least for that particular year. URP-8, which was the fourth isolate collected in 2015, differed from the three previously mentioned URPs because its reaction type on PI 230970 (Rpp2) was M instead of S (Fig. 2; Table 3). Conversely, PT4, the dominant pathotype in Uruguay, included rust isolates from 2014, 2015 and 2017 originating from different parts of the country (Fig. 3).
The pathotype richness of URPs was the lowest when compared to isolates from other South American countries (Table 4). The richness of URPs was 3.6 to 4.7 times lower than populations from Argentina and Brazil, respectively.
Discussion
A wide virulence spectrum was found in URPs, as four (Rpp1, Rpp3, Rpp4 and Rpp6) out of the seven Rpp genes were no longer effective against the pathogen. The frequency of resistant reactions conferred by these genes to isolates from the other South American countries ranged from 4 to 66%, denoting a narrower virulence (S reaction type on fewer Rpp genes) when compared to URPs. These results suggest that the virulence spectrum of the Uruguay population is wider (S reaction type on more Rpp genes) than that of the neighboring countries, and certainly broader than that of Japanese isolates, corroborating findings of previous studies with Brazilian and Japanese isolates (Yamanaka et al. 2010).
Temporal and geographic comparisons of pathogenicity among P. pachyrhizi populations have previously been made (Bonde et al. 2006; Akamatsu et al. 2017). In contrast to our results, the Rpp1 and Rpp3 genes were found to be effective in several other countries, such as Australia, Hawaii, India and Japan (Kato 2017). Although Rpp1 is still useful in Japan and the USA, there are previous reports of soybean genotypes carrying this gene that exhibit susceptible reactions to P. pachyrhizi isolates collected between 1993 and 1997 in Japan (Kato 2017; Yamanaka et al. 2010) and in north-central Florida (Paul et al. 2015). As in Mexico (García-Rodríguez et al. 2017), Rpp4 was ineffective in our study, although it is still effective to more than half of the isolates from other South American countries. Effectiveness of PI 567102B carrying Rpp6 has been tested (Miles et al. 2008; Paul et al. 2015), but its future as a source of resistance has already been jeopardized by results from Tanzania where half of the isolates induced susceptible reactions (Murithi et al. 2017). In contrast, both lines carrying Rpp1-b which were previously reported as ineffective to P. pachyrhizi isolates from Vietnam (Pham et al. 2010), the USA (Paul et al. 2015), and Mexico (García-Rodríguez et al. 2017), conferred complete resistant to all fungal isolates from Uruguay and to almost all isolates from South America, which is in agreement with reports obtained in Nigeria (Twizeyimana et al. 2009).
All URPs clustered together and with other rust isolates from South America. More than half of the URPs exhibited virulence patterns that were identical to pathotypes from that continent. This result was expected, as the pathogen is known to enter Uruguay via neighboring countries every year (De Ruyver et al. 2011). URPs shared reaction patterns with isolates from Argentina, Brazil and Paraguay on lines with Rpp1, Rpp2, Rpp5, Rpp1-b, and on the pyramided line carrying Rpp2, Rpp4, and a Rpp5 allele.
It should be noted that only one isolate of the URPs caused a susceptible reaction on Rpp5 (PI 200526) and three URPs resulted to be unique to the South American population. The uniqueness of those URPs in South America was mainly due to the reaction of soybean genotypes with Rpp3, Rpp4, and specially Rpp6. With regard to Rpp6, 66% of South American isolates induced resistant reactions on plants with this relatively newly identified gene (Li et al. 2012), whereas it was not effective against any isolate from Uruguay. Susceptible reactions of lines carrying this gene were first observed with a few isolates from Brazil and Paraguay in 2013 (data not shown).
No association between pathotypic clustering and geographic origin was noticed among isolates from Uruguay, which was suspected considering the small size of the country, and results from previous studies with South American populations (Akamatsu et al. 2013). In Mexico, pathotypic differentiation was observed among isolates collected 75 km apart. The Mexican population studied formed a cluster separated from all isolates from South America, which was caused mainly by their different virulence on soybean genotypes carrying Rpp1-b (García-Rodríguez et al. 2017). Mexican P. pachyrhizi isolates could be considered as an intermediate population whose virulence patterns partially resemble those from North America and partially those from South America.
In Uruguay, highly resistant plants carrying the Rpp1-b allele will be a useful source for breeding cultivars resistant to ASR, in contrast to the situation in Mexico, the USA and Vietnam (García-Rodríguez et al. 2017; Paul et al. 2015; Pham et al. 2010). The Rpp5 allele from PI 200526 is also a good alternative since it is still effective against 83% of the URPs. Besides these two, no other Rpp gene is useful to confer resistance to the fungal population in Uruguay. Nevertheless, pyramiding multiple Rpp genes into one cultivar could provide durable resistance to the Uruguayan pathotypes, as is the case of the resistance line No6–12-1, carrying Rpp2, Rpp4, and an Rpp5 allele, so far proven to be resistant in Argentina, Brazil, Paraguay, Mexico and Japan.
References
Akamatsu H, Yamanaka N, Yamaoka Y, Soares RM, Morel W, Ivancovich AJG, Bogado AN, Kato M, Yorinori JT, Suenaga K (2013) Pathogenic diversity of soybean rust in Argentina, Brazil and Paraguay. Journal of Genetic Plant Pathology 79:28–40
Akamatsu H, Yamanaka N, Soares RM, Ivancovich AJG, Lavilla MA, Bogado AN, Morel W, Scholz R, Yamaoka Y, Kato M (2017) Pathogenic variation of South American Phakopsora pachyrhizi populations isolated from soybeans from 2010 to 2015. Japan Agricultural Research Quarterly 51:221–232
Akinsanmi O, Lapido J, Oyrkan P (2001) First report of soybean rust (Phakopsora pachyrhizi) in Nigeria. Plant Disease 85:97
Bonde MR, Nester SE, Austin CN, Stone CL, Frederick RD, Hartman GL, Miles MR (2006) Evaluation of virulence of Phakopsora pachyrhizi and P. meibomiae isolates. Plant Disease 90:708–716
Bromfield KR (1984) Soybean rust. Monograph No. 11. American Phytopathological Society, St. Paul, Minnesota
Childs SD, King ZR, Walker DR, Harris DK, Pedley KF, Buck JW, Boerma R, Li Z (2018) Discovery of a seventh Rpp soybean rust resistance locus in soybean accession PI 605823. Theoretical and Applied Genetics 131:27–41
De Ruyver R, de Souza J, Bischoff SA, Formento N (2011) Circulación atmosférica asociada a los casos de aparición de esporas de Roya de la soja en Paraná, Argentina. Meteorologica 36
DIEA, Anuario estadístico Agropecuario (2016). Available at: http://www.mgap.gub.uy/unidad-ejecutora/oficina-de-programacion-y-politicas-agropecuarias/publicaciones/anuarios-diea/anuario2016. Accessed on January, 2018
FAOSTAT (2014) FAO statistical database. Food and Agriculture Organization of the United Nations, Rome. Available at: http://faostat.fao.org/beta/en/#data/QC. Accessed on December 2017
Freire MCM, de Oliveira LO, de Almeida AMR, Schuster I, Moreira MA, Liebenberg MM, Mienie CMS (2008) Evolutionary history of Phakopsora pachyrhizi (the Asian soybean rust) in Brazil based on nucleotide sequences of the internal transcribed spacer region of the nuclear ribosomal DNA. Genetics and Molecular Biology 31:920–931
Garcia A, Calvo ES, de Souza Kiihl RA, Harada A, Hiromoto DM, Vieira LG (2008) Molecular mapping of soybean rust (Phakopsora pachyrhizi) resistant genes: discovery of novel locus and alleles. Theoretical and Applied Genetics 117:545–553
García-Rodríguez JC, Morishita M, Kato M, Yamanaka N (2017) Características patogénicas de la roya asiática de la soja (Phakopsora pachyrhizi) en México. Revista Mexicana de Fitopatología 35:338–349
Groth JV, Roelfs AP (1987) The concept and measurement of phenotypic diversity in Puccinia graminis on wheat. Phytopathology 77:1395–1399
Hartman GL, Miles MR, Frederick RD (2005) Breeding for resistance to soybean rust. Plant Disease 89:664–666
Hartwig EE (1986) Identification of a fourth major gene conferring resistance to soybean rust. Crop Science 26:1135–1136
Hartwig EE, Bromfield KR (1983) Relationships among three genes conferring speci fic resistance to rust in soybeans. Crop Science 23: 237–239
Henning P (1903) Some new Japanese Uredinale. IV. Hedwigia 42:107–108
Hossain MM, Akamatsu H, Morishita M, Mori T, Yamaoka Y, Suenaga K, Soares RM, Bogado AN, Ivancovich AJG, Yamanaka N (2015) Molecular mapping of Asian soybean rust resistance in soybean landraces PI 594767A, PI 587905 and PI 416764. Plant Pathology 64:147–156
Ivancovich AJG (2005) Soybean rust in Argentina. Plant Disease 89:667–668
Kato M (2017) Review: effectiveness of resistance genes to the soybean rust pathogen Phakopsora pachyrhizi. Japan Agricultural Research Quarterly 51:199–207
Kawuki RS, Adipala E, Tukamuhabwa P (2003) Yield losses associated to soybean rust (Phakopsora pachyrhizi Syd.) in Uganda. Phytopathology 151:7–12
Langenbach C, Campe R, Beyer SF, Mueller AN, Conrath U (2016) Fighting Asian soybean rust. Frontiers in Plant Science 7:797
Levy C (2005) Epidemiology and chemical control of soybean rust in Southern Africa. Plant Disease 89:669–674
Li S, Smith JR, Ray JD, Frederick RD (2012) Identification of a new soybean rust resistant gene in PI 567102B. Theoretical and Applied Genetics 125:133–142
McDonald BA, Linde C (2002) Pathogen population genetics, evolutionary potential and durable resistance. Annual Review of Phytopathology 40:349–379
Miles MR, Morel W, Ray JD, Smith JR, Frederick RD, Hartman GL (2008) Adult plant evaluation of soybean accessions for resistance to Phakopsora pachyrhizi in the field and greenhouse in Paraguay. Plant Disease 92:96–105
Mueller TA, Miles MR, Morel W, Marois JJ, Wright DL, Kemerait RC, Levy C, Hartman GL (2009) Effect of fungicide and timing of application on soybean rust severity and yield. Plant Disease 93:243–248
Murithi HM, Haudenshield JS, Beed F, Mahuku G, Joosten MHAJ, Hartman GL (2017) Virulence diversity of Phakopsora pachyrhizi isolates from East Africa compared to a geographically diverse collection. Plant Disease 101:1194–1200
Paul C, Hartman GL, Marois JJ, Wright DL, Walker DR (2013) First report of Phakopsora pachyrhizi overcoming soybean genotypes with Rpp1 and Rpp6 rust resistance genes in field plots in United States. Plant Disease 97:1379
Paul C, Frederick RD, Hill CB, Hartman GL, Walker DR (2015) Comparison of pathogenic variation among Phakopsora pachyrhizi isolates collected from United States and international locations, and identification of soybean genotypes resistant to U.S. isolates. Plant Disease 99:1059–1069
Pham TA, Hill CB, Miles MR, Nguyen BT, Vud TT, Vuong TD, VanToai TT, Nguyen HT, Hartman GL (2010) Evaluation of soybean for resistance to soybean rust in Vietnam. Field Crops Research 117:131–138
Pivonia S, Yang XB (2004) Assessment of the potential year-round establishment of soybean rust throughout the world. Plant Disease 88:523–529
Pretorius ZA, Kloppers FJ, Frederick RD (2001) First report of soybean rust in South Africa. Plant Disease 85:1288
R Core Team (2015) R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna. http://www.r-project.org/. Accessed June 2018
Ray JD, Morel W, Smith JR, Frederick RD, Miles MR (2009) Genetic and mapping of plant rust resistance in soybean PI587886 and PI587880A. Theoretical and Applied Genetics 119:271–280
Rossi RL (2003) First report of Phakopsora pachyrhizi, the causal organism of soybean rust in the province of Misiones, Argentina. Plant Disease 87:102
Sanders HL (1968) Marine benthic diversity: a comparative study. American Naturalist 102:243–282
Sikora EJ, Delaney DP, Delaney MA, Lawrence KS, Pegues M (2009) Evaluation of sequential fungicide spray programs for control of soybean rust. Plant Health Progress Available at: https://www.plantmanagementnetwork.org/php. Accessed January 2018 10:23
Stewart S, Guillin E, Diaz L (2005) First report of soybean rust caused by Phakopsora pachyrhizi in Uruguay. Plant Disease 89:909–909
Suzuki R, Shimodaira H (2006) Pvclust: an R package for assessing the uncertainty in hierarchical clustering. Bioinformatics 22:1540–1542
Twizeyimana M, Ojiambo PS, Sonder K, Ikotun T, Hartman GL, Bandyopadhyay R (2009) Pathogenic variation of Phakopsora pachyrhizi infecting soybean in Nigeria. Phytopathology 99:353–361
Yamanaka N, Yamaoka Y, Kato M, Lemos NG, Andre L, dos Santos JVM, Benitez ER, Abdelnoor RV, Soares RM, Suenaga K (2010) Development classification criteria for resistance to soybean rust and differences in virulence among Japanese and Brazilian rust populations. Tropical Plant Pathology 35:153–162
Yamanaka N, Lemos NG, Uno M, Akamatsu H, Yamaoka Y, Abdelnoor RV, Braccini AL, Suenaga K (2013) Resistance to Asian soybean rust in soybean lines with the pyramided three Rpp genes. Crop Breeding and Applied Biotechnology 13:75–82
Yamanaka N, Hossain M, Yamaoka W (2015) Molecular mapping of Asianso soybean rust resistance in Chinese and Japanese soybean lines, Xiao Jing Huang, Himeshirazu, and Iyodaizu B. Euphytica 205:311–324
Yamanaka N, Kato M, Akamatsu H, Yamaoka Y (2017) Laboratory manual for studies on soybean rust resistance (version 23). Japan International Research Center of Agricultural Sciences (JIRCAS). Available at: http://www.jircas.affrc.go.jp/english/manual/soybean_rust/JIRCAS_manual_soybean_rust_v23.pdf. Accessed on March 2018
Yorinori JT (2008) Soybean germplasms with resistance and tolerance to Asian rust and screening methods. In: Kudo H, Suenaga K, Soares RM, Toledo A (eds) JIRCAS Working Report No 58: Facing the challenge of soybean rust in South America. Japan International Research Center for Agricultural Sciences (JIRCAS), Tsukuba, pp 70–87
Yorinori JT, Paiva WM, Frederick RD, Costamilan LM, Bertagnolli PF, Hartman GE, Godoy CV, Nune J Jr (2005) Epidemics of soybean rust (Phakopsora pachyrhizi) in Brazil and Paraguay from 2001 to 2003. Plant Disease 89:675–677
Acknowledgments
We are grateful to the Brazilian Agricultural Research Corporation (Embrapa) for providing seeds of differential soybean varieties, except for No6-12-1. This research was partly funded by JIRCAS research project “Development of technologies for the control of migratory plant pests and transboundary diseases.”
Author information
Authors and Affiliations
Corresponding author
Additional information
Section Editor: Marcelo A. Carmona
Publisher’s note
Springer nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Stewart, S., Rodríguez, M. & Yamanaka, N. Pathotypic variation of Phakopsora pachyrhizi isolates from Uruguay. Trop. plant pathol. 44, 309–317 (2019). https://doi.org/10.1007/s40858-018-0269-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40858-018-0269-2