Abstract
Introduction
Waist-to-height ratio (WHtR) can be effective for the diagnosis of abdominal obesity and the risk of type 2 diabetes. The role of genetic factors in the development of obesity has been broadly recognized. Adiponectin’s level is inversely correlated with body fat percentage and is reduced in obesity and type 2 diabetes. The aim of this study is to investigate the association between WHtR and adiponectin gene polymorphisms in Iranian population.
Methods
This study was conducted on 610 subjects from two Iranian populations. Anthropometric characteristics were measured by routine methods. Blood samples were collected in tubes (3–5 mL) containing EDTA and were stored at 20 °C. After DNA extraction, genotyping was performed using PCR–RFLP technique.
Results
There were statistically significant differences in genotype frequencies of −11391 G/A in centrally obese (WHtR >0.5) and noncentrally obese (WHtR ≤0.5) subjects (P value <0.044). In the former, the frequencies of GG and GA + AA genotypes were 89.4 and 10.6 %, respectively, while the frequencies of GG and GA + AA genotypes were 95.9 and 4.1 %, respectively, in noncentrally obese subjects.
Conclusions
The frequency of GG genotype was significantly increased in subjects with WHtR >0.5 compared to the other group. After adjustment for diabetes, abdominal obesity was significantly associated with the −11391 G/A polymorphism.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Extensive epidemiological studies have shown that obesity is a significant risk factor for several important diseases, including coronary heart disease, stroke, type 2 diabetes, hypertension, some forms of tumors, and musculoskeletal disorders [1]. The prevalence of obesity and overweight in developing countries, including Iran, is increasing [2, 3].
Studies have shown that obesity is more associated with body fat distribution (central obesity) than with general obesity measured by body mass index (BMI). These studies have also suggested a more effective role of obesity in determining one’s health [4]. It has been proposed that waist-to-height ratio (WHtR) can be effective for the diagnosis of abdominal obesity and the risk of type 2 diabetes [5] The use and associated risk factors of WHtR were first suggested in the mid-1990s [6, 7]. According to a study, BMI and WHtR in males and BMI, WHtR, and waist circumference (WC) in females are the best predictors of diabetes incidence in a population [8]. Other studies have also shown that the waist-to-hip ratio (WHR) and WHtR are better anthropometric indices than BMI and waist circumference (WC) [9–11]. In comparison with BMI, WC is more responsive to diet and exercise in that any increase in muscle mass can lead to more noticeable changes in WC and WHtR than in BMI [12]. In fact, the use of WC as the only indicator of abdominal obesity may lead to the overestimation of associated risks in tall people and the underestimation of the risks in short ones [13].
The effectiveness of anthropometric indicators for predicting health risk factors such as cardiovascular disease is related to genetic factors and might vary in different populations. The role of genetic factors in the development of obesity and diabetes has been broadly recognized [14–16].
Adiponectin is an abundant adipose-derived protein whose levels are inversely correlated with body fat percentage [17]. Like so, a reduction in adiponectin level may enhance the probability of obesity [18], type 2 diabetes [19], dyslipidemia [20], and coronary heart disease [21]. Adiponectin plays an important role in different metabolic processes including glucose regulation and fatty acid metabolism [22]. Individuals with visceral adipose tissue have been found to have lower levels of adiponectin [23]. Single nucleotide polymorphisms have been identified in adiponectin gene, including polymorphism at positions −11391 G/A, −11377 G/C, +45 T/G, and +276 G/T [24]. Recent molecular epidemiological studies have evaluated the association between polymorphisms of adipokine genes and the risk of obesity [25]. In addition, several studies have shown that +45 T/G polymorphism is significantly associated with obesity and type 2 diabetes [26].
Established upon the best anthropometric index, this study is the first to evaluate the association between abdominal obesity, type 2 diabetes, and adiponectin. The aim of this study was to examine the association between WHtR and adiponectin gene polymorphisms in an Iranian population.
Materials and methods
This cross-sectional study was conducted on 610 subjects who were selected from Kerman (n = 310) and Tehran populations (n = 300) [27]. The selection of the patients was carried out resting on a hospital-based clinic approach covering a diabetes clinic.
After obtaining informed consent, personal and demographic information for each participant was collected by questionnaires. Waist circumference was measured with a tape measure resting on the distance between the smallest area below the rib cage and the iliac crest with the patient’s lung at the end of expiration [24]. Height was measured using a SECA height meter with an accuracy of 0.1 cm. WHtR was calculated by dividing waist circumference (in cm) by height (in cm) [11]. The diagnosis of diabetes was made based on the American Diabetes Association Criteria [28]. Fasting blood glucose was taken from the participants and blood samples were collected in tubes (3–5 mL) containing EDTA, and were stored in 20 °C. This study is approved by the Ethics Committee of Tehran University of Medical Sciences.
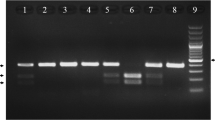
DNA samples in EDTA containing tubes were extracted using salting-out method. The molecular analysis of adiponectin gene +45 T/G polymorphism was examined based on a method described by Schaffler and his colleagues [29]. The molecular analysis of the −11391 G/A polymorphism was performed by PCR–RFLP [30]. Statistical analysis was performed by Chi square, t test, and regression tests using SPSS software version 16. In the univariate model, the association between abdominal obesity and adiponectin gene polymorphisms was investigated. In adjusted models, this association was examined with regard to diabetes.
Result
This study was conducted on 610 subjects from two Iranian populations. 66 % of participants were female and 34 % were male. Table 1 summarizes the mean and standard deviations of age and anthropometric indices of subjects and their association with adiponectin gene polymorphisms. When patients were stratified, there were statistically significant differences between genotype frequencies of −11391 G/A in centrally obese (WHtR >0.5) and noncentrally obese (WHtR ≤0.5) subjects (P value <0.05). However, there were no statistically significant differences between genotype frequencies of +45 T/G in centrally obese (WHtR >0.5) and noncentrally obese (WHtR ≤0.5) subjects (P value = 0.162).
In Table 2, there were no statistically significant differences between genotype frequencies of −11391 G/A in diabetic subjects and non-diabetic subjects (P value = 0.117). In addition, there were no statistically significant differences between genotype frequencies of +45 T/G in diabetic subjects and non-diabetic ones (P value = 0.337). Table 3 shows the association between abdominal obesity and adiponectin gene polymorphisms in logistic regression. As it can be observed in the univariate model, none of these polymorphisms were associated with abdominal obesity. However, after adjustment for diabetes, −11391 G/A polymorphism was significant.
Discussion
The aim of this study was to evaluate the effect of two common polymorphisms of the adiponectin gene and WHtR as indicators of abdominal obesity. The results of this study were based on a study by Esmaillzadeh et al. where the cutoff point of waist-to-height ratio is considered to be 0.5 cm [31]. We found significant associations between −11391 G/A polymorphism and WHtR in the subjects and, after adjustment for diabetes, abdominal obesity was still significantly associated with the −11391 G/A polymorphism.
WHtR had the highest risk-adjusted incidence of diabetes than other anthropometric indicators in all quartiles including BMI, when compared with other central obesity indicators, having significantly lower area under the curve than WHtR [32]. According to a study on a group of men, a strong correlation was established between WHtR and the risk of type 2 diabetes. This study determined WHtR as the best predictor of obesity and diabetes in adult males. This is mainly because the predictive power of anthropometric indicators in each population varies from race to race, which is suggesting that WHtR should be used as a screening indicator [32–34].
Other studies have shown that WHtR is strongly related to visceral adipose tissue while adiponectin is inversely associated with visceral adipose tissue [35, 36]. Previous studies have also reported that WHtR is a stronger predictor of visceral adipose tissue compared to BMI and WC [6, 37]. Thus, WHtR is a strong anthropometric index for predicting adiponectin levels [38].
As reported by Dolley et al., there is a significant association between −11391 G>A polymorphism and WHtR changes, which was evaluated during a 7-year follow-up period. However, this study did not include BMI changes over this time period [39]. Conversely, Fumeron et al. found no association between the −11391 G>A and WHtR at the baseline during a 3-year follow-up period [40]. Furthermore, −11391 G/A polymorphism was associated with higher adiponectin levels in obese children [41].This is supported by a study in Denmark which found relationships between −11391 G/A polymorphism and obesity [42].
Stumvoll et al. showed that, in individuals without familial predisposition to type 2 diabetes, adiponectin polymorphism may moderately increase the risk of obesity and secondarily cause insulin resistance. Furthermore, they suggested that the G allele was significantly associated with obesity [26]. In addition, in the present study, the genotype frequency of GG was higher in centrally obese subjects and this finding could be a facet to consider G allele as a risk factor for abdominal obesity. In 2010, the findings of a study showed that the frequencies of allele A −11391 G/A polymorphism in women and allele G +45 T/G polymorphism in both men and women have a supporting role in weight gain [30].
In a non-diabetic Korean population, an association was found between +45 T/G polymorphism and serum levels of adiponectin, as well as obesity and insulin resistance [43]. These results are in contrast to the findings of our study. Moreover, a study conducted in Italy demonstrated significant associations between +276 and +45 T/G polymorphisms and insulin resistance and other parameters such as weight gain [44].
In a study of Indian population, Ranjith et al. found no association between G allele of SNP +45 and obesity, while +45 T/G genotype was only associated with low WC. This study found that the +45 T allele has a lower frequency compared to the +45 G allele [45]. Nevertheless, in Arnaiz-Villena’s study, the +45 T allele was reported to have a higher frequency [46].
In this study we concluded that there is significant association between the −11391 G/A polymorphism and abdominal obesity.
It is recommended that future studies be replicated in other populations and also other polymorphisms of adiponectin need to be considered.
Abbreviations
- WHtR:
-
Waist-to-height ratio
- WC:
-
Waist circumference
- BMI:
-
Body mass index
References
Jeffery RW, Sherwood NE (2008) Is the obesity epidemic exaggerated? No. Br Med J 336(7638):245. doi:10.1136/bmj.39458.495127
Hajian-Tilaki KO, Heidari B (2007) Prevalence of obesity, central obesity and the associated factors in urban population aged 20–70 years, in the north of Iran: a population-based study and regression approach. Obes Rev 8(1):3–10. doi:10.1111/j.1467-789X.2006.00235.x
de Azeredo Passos VM, Barreto SM, Diniz LM, Lima-Costa MF (2005) Type 2 diabetes: prevalence and associated factors in Brazilian community—the Bambuí health and aging study. Sao Paulo Med J 123(2):66–71
Lee J, Chen L, Snieder H et al (2010) Heritability of obesity related phenotypes and association with adiponectin gene polymorphisms in the chinese national twin registry. Ann Hum Genet 74(2):146–154. doi:10.1111/j.1469-1809.2010.00565.x
Schneider HJ, Klotsche J, Silber S, Stalla GK, Wittchen H-U (2011) Measuring abdominal obesity: effects of height on distribution of cardiometabolic risk factors risk using waist circumference and waist-to-height ratio. Diabetes Care 34(1):e7. doi:10.2337/dc10-1794
Ashwell M, Lejeune S, McPherson K (1996) Ratio of waist circumference to height may be better indicator of need for weight management. Br Med J 312(7027):377. doi:10.1136/312.7027.377
Lee JS, Aoki K, Kawakubo K, Gunji A (1995) A study on indices of body fat distribution for screening for obesity. J Occup Health 37(1):9–18
Tulloch-Reid MK, Williams DE, Looker HC, Hanson RL, Knowler WC (2003) Do measures of body fat distribution provide information on the risk of type 2 diabetes in addition to measures of general obesity? Comparison of anthropometric predictors of type 2 diabetes in Pima Indians. Diabetes Care 26(9):2556–2561. doi:10.2337/diacare.26.9.2556
Lin W-Y, Lee L-T, Chen C-Y et al (2002) Optimal cut-off values for obesity: using simple anthropometric indices to predict cardiovascular risk factors in Taiwan. Int J Obes 26(9):1232–1238. doi:10.1038/sj.ijo.0802040
Lorenzo C, Serrano-Ríos M, Martínez-Larrad MT et al (2007) Which obesity index best explains prevalence differences in type 2 diabetes mellitus? Obesity 15(5):1294–1301. doi:10.1038/oby.2007.151
Mi SQ, Yin P, Hu N et al (2013) BMI, WC, WHtR, VFI and BFI: which indictor is the most efficient screening index on type 2 diabetes in Chinese community population. Biomed Environ Sci 26(6):485–491. doi:10.3967/0895-3988.2013.06.009
Hadaegh F, Shafiee G, Azizi F (2009) Anthropometric predictors of incident type 2 diabetes mellitus in Iranian women. Ann Saudi Med 29(3):194–200. doi:10.4103/0256-4947.51788
Hsieh SD, Muto T (2006) Metabolic syndrome in Japanese men and women with special reference to the anthropometric criteria for the assessment of obesity: proposal to use the waist to-height ratio. Prev Med 42(2):135–139. doi:10.1016/j.ypmed.2005.08.007
Mehrab-Mohseni M, Tabatabaei-Malazy O, Hasani-Ranjbar S et al (2011) Endothelial nitric oxide synthase VNTR (intron 4 a/b) polymorphism association with type 2 diabetes and its chronic complications. Diabetes Res Clin Pract 91(3):348–352. doi:10.1016/j.diabres.2010.12.030
Rankinen T, Zuberi A, Chagnon YC et al (2006) The human obesity gene map: the 2005 update. Obesity 14(4):529–644. doi:10.1038/oby.2006.71
Bazzaz JT, Shojapoor M, Nazem H et al (2010) Methylenetetrahydrofolate reductase gene polymorphism in diabetes and obesity. Mol Biol Rep 37(1):105–109. doi:10.1007/s11033-009-9545
Chandran M, Phillips SA, Ciaraldi T, Henry RR (2003) Adiponectin: more than just another fat cell hormone? Diabetes Care 26(8):2442–2450. doi:10.2337/diacare.26.8.2442
Arita Y, Kihara S, Ouchi N et al (1999) Paradoxical decrease of an adipose-specific protein, adiponectin, in obesity. Biochem Biophys Res Commun 257(1):79–83. doi:10.1016/j.bbrc.2012.08.024
Hotta K, Funahashi T, Arita Y et al (2000) Plasma concentrations of a novel, adipose-specific protein, adiponectin, in type 2 diabetic patients. Arterioscler Thromb Vasc Biol 20(6):1595–1599. doi:10.1161/01.ATV.20.6.1595
Weyer C, Funahashi T, Tanaka S et al (2001) Hypoadiponectinemia in obesity and type 2 diabetes: close association with insulin resistance and hyperinsulinemia. J Clin Endocrinol Metab 86(5):1930–1935
Ouchi N, Kihara S, Arita Y et al (1999) Novel modulator for endothelial adhesion molecules: adipocyte-derived plasma protein adiponectin. Circulation 100(25):2473–2476. doi:10.1161/01.CIR.100.25.2473
Díez JJ, Iglesias P (2003) The role of the novel adipocyte-derived hormone adiponectin in human disease. Eur J Endocrinol 148(3):293–300
Hanley AJG, Bowden D, Wagenknecht LE et al (2007) Associations of adiponectin with body fat distribution and insulin sensitivity in nondiabetic Hispanics and African-Americans. J Clin Endocrinol Metab 92(7):2665–2671. doi:10.1210/jc.2006-2614
Hasani-Ranjbar S, Amoli MM, Tabatabaei-Malazy O et al (2012) Effect of adiponectin gene polymorphisms on waist circumference in patients with diabetes. J Diabetes Metab Disord 11(1):14. doi:10.1186/2251-6581-11-14
Yu Z, Han S, Cao X, Zhu C, Wang X, Guo X (2012) Genetic polymorphisms in adipokine genes and the risk of obesity: a systematic review and meta-analysis. Obesity 20(2):396–406. doi:10.1038/oby.2011.148
Stumvoll M, Tschritter O, Fritsche A et al (2002) Association of the T-G polymorphism in adiponectin (Exon 2) with obesity and insulin sensitivity: interaction with family history of type 2 diabetes. Diabetes 51(1):37–41. doi:10.2337/diabetes.51.1.37
Bazzaz JT, Nazari M, Nazem H et al (2010) Apolipoprotein e gene polymorphism and total serum cholesterol level in Iranian population. J Postgrad Med 56(3):173–175. doi:10.4103/0022-3859.68629
Genuth S, Alberti KGMM, Bennett P et al (2003) Follow-up report on the diagnosis of diabetes mellitus. Diabetes Care 26(11):3160–3167. doi:10.2337/diacare.26.11.3160
Schäffler A, Büchler C, Müller-Ladner U et al (2004) Identification of variables influencing resistin serum levels in patients with type 1 and type 2 diabetes mellitus. Horm Metab Res 36(10):702–707. doi:10.1055/s-2004-826015
Tabatabaei-Malazy O, Hasani-Ranjbar S, Amoli MM et al (2010) Gender-specific differences in the association of adiponectin gene polymorphisms with body mass index. Rev Diabetic Stud 7(3):241–246. doi:10.1900/RDS.2010.7.241
Esmaillzadeh A, Mirmiran P, Azizi F (2006) Comparative evaluation of anthropometric measures to predict cardiovascular risk factors in Tehranian adult women. Public Health Nutr 9(1):61–69. doi:10.1079/PHN2005833
Hadaegh F, Zabetian A, Harati H, Azizi F (2006) Waist/height ratio as a better predictor of type 2 diabetes compared to body mass index in Tehranian adult men—a 3.6-year prospective study. Exp Clin Endocrinol Diabetes 114(6):310–315. doi:10.1055/s-2006-924123
Lear SA, Chen MM, Frohlich JJ, Birmingham CL (2002) The relationship between waist circumference and metabolic risk factors: cohorts of European and Chinese descent. Metab Clin Exp 51(11):1427–1432. doi:10.1053/meta.2002.34042
Molarius A, Seidell JC (1998) Selection of anthropometric indicators for classification of abdominal fatness—a critical review. Int J Obes 22(8):719–727. doi:10.1038/sj.ijo.0800660
Lee S, Bacha F, Gungor N, Arslanian SA (2006) Racial differences in adiponectin in youth: relationship to visceral fat and insulin sensitivity. Diabetes Care 29(1):51–56. doi:10.2337/diacare.29.01.06.dc05-0952
Asayama K, Hayashibe H, Dobashi K et al (2003) Decrease in serum adiponectin level due to obesity and visceral fat accumulation in children. Obes Res 11(9):1072–1079
Wu H-Y, Xu S-Y, Chen L-L, Zhang H-F (2009) Waist to height ratio as a predictor of abdominal fat distribution inmen. Chin J Physiol 52(6):441–445. doi:10.4077/CJP.2009.AMH064
Lee S, Kuk JL, Hannon TS, Arslanian SA (2008) Race and gender differences in the relationships between anthropometrics and abdominal fat inyouth. Obesity 16(5):1066–1071. doi:10.1038/oby.2008.13
Dolley G, Bertrais S, Frochot V et al (2008) Promoter adiponectin polymorphisms and waist/hip ratio variation in a prospective French adults study. Int J Obes 32(4):669–675. doi:10.1038/sj.ijo.0803773
Fumeron F, Aubert R, Siddiq A et al (2004) Adiponectin gene polymorphisms and adiponectin levels are independently associated with the development of hyperglycemia during a 3-year period: the epidemiologic data on the insulin resistance syndrome prospective study. Diabetes 53(4):1150–1157. doi:10.2337/diabetes.53.4.1150
Bouatia-Naji N, Meyre D, Lobbens S et al (2006) ACDC/adiponectin polymorphisms are associated with severe childhood and adult obesity. Diabetes 55(2):545–550. doi:10.2337/diabetes.55.02.06
Tankó LB, Siddiq A, Lecoeur C et al (2005) ACDC/adiponectin and PPAR-gamma gene polymorphisms: implications for features of obesity. Obes Res 13(12):2113–2121
Jang Y, Chae JS, Koh SJ et al (2008) The influence of the adiponectin gene on adiponectin concentrations and parameters of metabolic syndrome in non-diabetic Korean women. Clin Chim Acta 391(1–2):85–90. doi:10.1016/j.cca.2008.02.011
Menzaghi C, Ercolino T, Paola RD et al (2002) A haplotype at the adiponectin locus is associated with obesity and other features of the insulin resistance syndrome. Diabetes 51(7):2306–2312. doi:10.2337/diabetes.51.7.2306
Ranjith N, Pegoraro RJ, Shanmugam R (2011) Obesity associated genetic variants in young Asian Indians with the metabolic syndrome and myocardial infarction. Cardiovasc J Afr 22(1):25–30. doi:10.5830/CVJA-2010-036
Arnaiz-Villena A, Fernández-Honrado M, Rey D et al (2013) Amerindians show association to obesity with adiponectin gene SNP45 and SNP276: population genetics of a food intake control and ‘thrifty’ gene. Mol Biol Rep 40(2):1819–1826. doi:10.1007/s11033-012-2236-1
Acknowledgments
Implementation of this study was sponsored by Tehran University of Medical Sciences (Endocrinology and Metabolism Research Center).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests.
Ethical standard
The study has been approved by ethical committee of Tehran University of Medical Sciences.
Informed consent
Informed consents have been obtained from all patients participating in the study.
Additional information
S. Hasani-Ranjbar and M. M. Amoli equally contributed as corresponding authors.
Rights and permissions
About this article
Cite this article
Payab, M., Amoli, M.M., Qorbani, M. et al. Adiponectin gene variants and abdominal obesity in an Iranian population. Eat Weight Disord 22, 85–90 (2017). https://doi.org/10.1007/s40519-016-0252-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40519-016-0252-1