Abstract
The genus Ceiba Mill. belongs to the subfamily Bombacoideae (Malvaceae), a paleopolyploid lineage characterized by numerous small chromosomes, which has frequently been reported to have variable intra- and interspecific chromosome numbers. The genus is of Miocene origin, representing a suitable model for studying the relationships between chromosome variability and paleopolyploidy. A comparative cytogenetic analysis of five Ceiba species was undertaken to determine their karyotype variability. New chromosome number counts, chromosome morphological observations, CMA/DAPI double staining, and in situ hybridization (FISH) with 5S and 45S rDNA were performed for five Ceiba species, which represent three out of five main lineages of the genus. Karyotypic data were discussed in the light of molecular phylogenies available for the group. All species showed 2n = 86 and similar karyotypes, composed predominantly of metacentric chromosomes. Two pairs of CMA+/DAPI− bands were located on the short arms of the metacentric chromosomes. The CMA+ bands colocated with 45S rDNA sites, while the 5S rDNA sites were situated in the interstitial regions of other chromosome pairs. Contrary to the intra- and interspecific chromosome number diversity reported for Ceiba species in the literature, our findings suggest chromosome number stability in four of the five lineages within the genus for which data are available. Moreover, our data suggest that karyotypes are evolutionary conserved in the three lineages for which we generated new karyotypic data. Our data, as well as recent cytogenetic reviews of other Bombacoideae genera, indicate numerical stability for these taxa suggesting that counting errors, especially in earlier research, may have overestimated karyotype variability. Chromosome count errors can be attributed to technical difficulties associated with high chromosome numbers, and/or the reduced size of Bombacoideae chromosomes.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Chromosome numbers (2n) are the most reported elements of plant karyotypes, and represent an important tool for understanding the phylogenetic relationships and the genetic mechanisms of evolution at and above the species level (Guerra 2000, 2012). Chromosome counts are especially important for species which are difficult to analyze karyotypes composed of small, numerous, and morphologically similar chromosomes (Ragone 2001; Felix and Guerra 2010; Sousa et al. 2011). In such cases, it is difficult to identify additional structural characteristics of karyotypes, and hence chromosome number counts become the main source of data for cytogenetic analyses. There are frequent reports in the literature of hypervariable and complex chromosome numbers (Lewis 1951; Lewis et al. 1971; Brighton et al. 1973; Assis et al. 2013; Escudero et al. 2015), although it is not always clear if this karyotypic variability is real (e.g., polyploidy, aneuploidy, or disploidy) or simply due to chromosome count errors. Erroneous chromosome counts may represent a significant fraction of the intra- and interspecific variability of reported chromosome numbers (Merxmüller 1970; Guerra 1986; Baum and Oginuma 1994; Marinho et al. 2014). In this context, chromosome recounts, followed by more detailed analyzes of heterochromatin distributions, represent an important tool for determining the real status of karyotypic diversity.
The subfamily Bombacoideae is characterized by small and numerous chromosomes, ranging from 2n = 18 in Bombax insigne Wall. (Sinha and Mazumdar 1993) to 2n = 276 in Eriotheca pubescens (Mart.) Schott & Endl. (Oliveira et al. 1992), with great intra- and interspecific variability (Baker and Baker 1968; Baum and Oginuma 1994; Oginuma et al. 1999; Marinho et al. 2014). Two probable basic chromosome numbers (x) have been suggested for Bombacoideae (x = 44 in Adansonia L., Pachira Aubl., and Pseudobombax Dugand, or x = 46 in Eriotheca Schott & Endl.) (Marinho et al. 2014), but detailed analyzes and cytogenetic revisions are lacking for most genera. A strong phylogenetic signal for higher chromosome numbers (2n = 64–90) has been observed in Bombacoideae compared with other subfamilies of Malvaceae, suggesting a whole genome duplication event in the origin of that subfamily, as well as neopolyploidy events in some lineages (Marinho et al. 2014).
In Bombacoideae, Ceiba Mill. stands out by having chromosome numbers ranging from 2n = 72 to 2n = 88 and by varying at intraspecific level. Ceiba pentandra (L.) Gaertn. has been reported as 2n = 72, 74, 75, 76, 80, 84, 86, and 88 (Heyn 1938; Baker and Baker 1968; Gill et al. 1979; Gibbs et al. 1988). This monophyletic genus (Duarte et al. 2011; Marinho et al. 2014; Carvalho-Sobrinho et al. 2016) comprises 18 tree species distributed mainly in the Neotropics with one disjunct species in West Africa (Gibbs and Semir 2003; Carvalho-Sobrinho and Queiroz 2008). Chorisia Kunth was subsumed into Ceiba based on morphological data (Gibbs et al. 1988; Gibbs and Semir 2003) and subsequently confirmed by molecular (Duarte et al. 2011; Carvalho-Sobrinho et al. 2016) and karyotypic data. The probable basic chromosome number of Ceiba sensu lato is x = 43 (Gibbs and Semir 2003) and the genus probably originated in the Miocene (Marinho et al. 2014), representing a suitable model for studying the relationships between chromosome variability and paleopolyploidy.
Diploidization, a phenomenon common to ancient polyploids (paleopolyploids) that consists of different genetic modifications resulting in a stable polyploid genome (Wolfe 2001), could explain the chromosome variability observed in Ceiba. These diploidizations can occur at both molecular (e.g., silencing, sub- and/or neofunctionalization of genes, elimination of repetitive DNA) and cytogenetic (e.g., aneuploidy, dysploidy, rDNA changes, alterations in genome size) levels (Wolfe 2001; Clarkson et al. 2005; Parisod et al. 2012). Drastic cytogenetic changes can occur causing alterations of chromosome numbers, primarily by dysploidy (decrease or increase of chromosome number due to chromosome rearrangements; not by addition or loss of single chromosomes as in aneuploidy. Therefore, dysploidy is also called pseudoaneuploidy) (Schubert and Lysak 2011).
The present work analyzed the karyotypic diversity of the paleopolyploid genus Ceiba based on chromosome counts, morphology, double staining with the fluorochromes chromomycin A3 (CMA) and 4′,6-diamidino-2-phenylindole (DAPI), and fluorescent in situ hybridization (FISH) with 5S and 45S rDNA. The new chromosome counts were compared to those from the literature to test the existence of intra- and interspecific variability reported for the genus (Baker and Baker 1968; Baum and Oginuma 1994; Oginuma et al. 1999; Gibbs and Semir 2003). These data were interpreted in an evolutionary context based on a molecular phylogeny of Bombacoideae (Marinho et al. 2014) and Ceiba (Carvalho-Sobrinho et al. 2016), which was used to subsidize discussion of chromosome evolution in the group.
Materials and methods
Plant material
The present study was based on examinations of live specimens of five species of Ceiba [C. erianthos (Cav.) K. Schum., C. glaziovii (Kuntze) K. Schum., C. jasminodora (A. St.-Hil.) K. Schum., C. pentandra (L.) Gaertn., and C. speciosa (A. St.-Hil.) Ravenna]. The collection sites and numbers of individuals examined are listed in Table 1. Voucher specimens were deposited in the Universidade Estadual de Feira de Santana Herbarium (HUEFS).
Root tips obtained from seeds or seedlings were pretreated with 8-hydroxyquinoline (0.002 M) for 24 h at 12 °C, fixed in ethanol:acetic acid (3:1; v/v) from two to 24 h at room temperature, and then stored at −20 °C. For conventional staining, the fixed tissues were hydrolyzed in 5 N HCl for 20 min and then washed with distilled water. The meristems were macerated in a drop of 45 % acetic acid and the coverslips subsequently removed in liquid nitrogen. The slides were stained with 2 % Giemsa for 3 min.
Chromosome banding
Fixed root tips were washed in distilled water and digested in a solution of 2 % (w/v) cellulase (Onozuka)/20 % (v/v) pectinase (Sigma) at 37 °C for 90 min. The meristems were macerated in a drop of 45 % acetic acid and the coverslips subsequently removed in liquid nitrogen. For CMA/DAPI double staining, the slides were aged for three days, stained with 10 μl of 0.1 mg mL−1 CMA for 30 min, and then restained with 10 μl of DAPI 2 μg/ml for 60 min (Barros e Silva and Guerra 2010). The slides were mounted in glycerol:McIlvaine buffer pH 7.0 (1:1) and aged for 3 days before analysis. The slides were examined using a Leica DMLB epifluorescence microscope; images were captured with a Cohu CCD video camera using Leica QFISH software, and edited using Adobe Photoshop CS3 version 10.0.
Fluorescent in situ hybridization (FISH)
In order to localize the rDNA sites, a 500-bp 5S rDNA clone (D2) of Lotus japonicus (Regel) K. Larsen labeled with Cy3-dUTP (Amersham) and a 6.5-kb 18S-5.8S-25S clone (R2) of Arabidopsis thaliana (L.) Heynh. labeled with digoxigenin-11-dUTP were used as probes (Pedrosa et al. 2002). Both labeling techniques were performed using nick translation. The 45S rDNA probe was detected with sheep anti-digoxigenin FITC conjugate (Roche), and amplified with rabbit anti-sheep FITC conjugate (Dako). FISH was performed as described by Pedrosa et al. (2002), with small modifications. The hybridization mix contained 50 % formamide (v/v), 10 % dextran sulfate (w/v), 2 × SSC, and 5 ng μL−1 of each probe. The slides were denaturated at 75 °C for 3 min, and the final stringency of hybridization was ca. 76 %. Images of the best cells were captured as described above.
Phylogenetic relationships of Ceiba
The phylogenetic relationships of Ceiba presented in Fig. 13 were based on Carvalho-Sobrinho et al. (2016). The cladogram originated from Bayesian inference based on nuclear (ETS, ITS) and plastid (matK, trnL-trnF, trnS-trnG) DNA characters and was used to interpret chromosome number variations in the genus. CorelDRAW® version X7 software was used to draw the tree topology and plot chromosome data.
Results and discussion
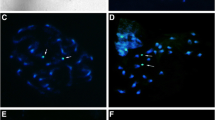
Bombacoideae is karyotypically characterized by having high and hypervariable chromosome numbers, often making interpretations of karyotypic variations difficult (Baker and Baker 1968; Baum and Oginuma 1994; Oginuma et al. 1999; Gibbs and Semir 2003). Our data confirm the predominance of 2n = 86 for Ceiba and did not confirm the intraspecific chromosome number variability previously reported mainly for C. pentandra (Baker and Baker 1968; Baum and Oginuma 1994; Oginuma et al. 1999; Gibbs and Semir 2003). The five species of Ceiba analyzed showed 2n = 86, similar symmetric karyotypes, and small chromosomes (~1.50 µm) which was shown predominantly metacentric (Figs. 1–2). All the five species showed semireticulated interphase nuclei with only one visible nucleolus. Among the seven species with previous chromosome counts, C. insignis (Kunth) P.E. Gibbs & Semir (2n = 72, 80, 86, and 88; Cristobal 1967; Baker and Baker 1968; Sareen and Kumari 1973; Morawetz 1986; Gibbs et al. 1988) and C. pubiflora (A. St.-Hil.) K. Schum. (2n = 86; Gibbs et al. 1988) were not analyzed in the present work (Table 1). Intraspecific sampling ranged from three individuals in C. glaziovii to 11 individuals in C. pentandra and revealed similar karyotypes in all individuals. No intraspecific variability was detected.
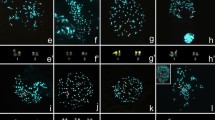
Cytogenetic reanalyses of other Bombacoideae genera, Adansonia (Baum and Oginuma 1994) and Eriotheca (Marinho et al. 2014), also revealed stable karyotypes, refuting the numeric variability previously reported for these genera (Baker and Baker 1968; Baum and Oginuma 1994). Divergences in intra- and interspecific chromosome counts can be attributed to technical difficulties, high chromosome numbers, and/or the small size of the Bombacoideae chromosomes. Specifically, secondary chromosome constrictions, when positively stained using conventional stains, can resemble extra chromosomes and confuse the observer, leading to imprecise determinations of chromosome numbers (Guerra et al. 1997). The high frequency of four distended satellites in Ceiba that stain positively with Giemsa can explain the 2n = 88 count for C. pentandra (Gill et al. 1979). The CMA/DAPI banding allowed us to identify these satellites and precisely characterize the karyotypes. Other sources of chromosome miscounts include aneusomatic mitosis (Gibbs et al. 1988), taxonomic identification errors (Merxmüller 1970; Guerra 1986; 1990), and taxon sampling issues (Guerra 1986). Double staining with CMA and DAPI revealed the presence of CMA+/DAPI− bands on the short arm of two metacentric pairs in C. erianthos (Fig. 3), C. glaziovii (Fig. 5), C. jasminodora (Fig. 7), C. pentandra (Fig. 9), and C. speciosa (Fig. 11).
Distribution of CMA+ bands and rDNA sites in the chromosome complements of five Ceiba species. CMA/DAPI-stained metaphase showing CMA+ bands (arrows) in C. erianthos (3), C. glaziovii (5), C. jasminodora (7), C. pentandra (9), and C. speciosa (11). In situ hibridization with 5S (arrowheads) and 45S (arrows) rDNA probes in C. erianthos (4), C. glaziovii (6), C. jasminodora (8), C. pentandra (10), and C. speciosa (12). White arrows point to smaller CMA+/DNAr 45S bands, and white arrowheads point to 5S rDNA sites. Scale bar in (3) = 10 µm
In situ hybridization revealed the presence of two pairs of 45S rDNA sites colocalizing with those terminal CMA+/DAPI− bands. In some metaphases, the 45S rDNA sites were distended, making it difficult to identify their host chromosomes (Figs. 1–2, 10). The metaphasic distentions of 45S sites were often associated with circular structures similar to a nucleolus (Figs. 1–2). Baker (1964) reported a notable cytological characteristic of Bombacoideae in the persistence of the nucleolus during cell division. Nucleoli usually disappear during metaphase, but merely shrink in size in Bombacoideae. As the telophase nuclei reform their membranes, the remains of the old nucleolus are excluded and eventually degraded in the cytoplasm. Our CMA/DAPI staining/in situ hybridization with 45S rDNA indicated that structures similar to nucleoli could persist during metaphase, although additional epigenetic and cytochemical investigations will be needed to better understand the nature of these structures.
All the species of Ceiba analyzed showed just one pair of 5S rDNA sites (Figs. 4, 6, 8, 10, 12) suggesting that 5S sites (different from 45S rDNA sites) undergo diploidization, supporting the idea of chromosome stability. The presence of one pair of 5S rDNA sites has been reported for other Malvaceae subfamilies, although associated with low chromosome numbers and diploidy in Malvoideae (Gossypium; Gan et al. 2013), Byttnerioideae (Theobroma L.; Dantas and Guerra 2010), and Grewioideae (Corchorus L.; Begum et al. 2013). Chromosome number stability in Ceiba is further supported by both heterochromatin and rDNA site distribution data. Given the origin of Ceiba in the Miocene (~16 million years ago; Marinho et al. 2014), the absence of variations in the number and/or distribution of heterochromatin blocks and rDNA sites is remarkable. Many chromosome polymorphisms, however, have been reported in more recent polyploids, such as Gossypium hirsutum L. (2 million years; Hanson et al. 1996), Nicotiana tabacum L. (4.5 million years; Kovarik et al. 2004), and Hepatica Mill. species (4 million years; Weiss-Schneeweiss et al. 2007), especially in rDNA sites. It is striking that no rDNA changes have been observed in such a genus as Ceiba compared the many polymorphisms observed in more recent genera.
Our chromosome counts revealed a marked karyotypic stability in Ceiba, allowing us to challenge reported numeric chromosome variability at both intra- and interspecific levels. This newly generated cytomolecular data support a scenario of karyotypic stability represented by the presence of two pairs of 45S rDNA sites and one 5S rDNA site. The phylogenetic conservation of the numbers of these markers suggests that this karyotype is evolutionary stable based on the phylogeny of Ceiba (Carvalho-Sobrinho et al. 2016) and represents a putative synapomorphy for the genus (Fig. 13). The presence of just one pair of 5S rDNA sites suggests a process of diploidization compatible with the paleopolyploid origin of Bombacoideae, while the presence of two pairs of 45S rDNA sites reveals a distinct tendency. The newly generated karyotypic data for Ceiba along with cytogenetic reviews of other genera (Baum and Oginuma 1994; Marinho et al. 2014) reveal numerical stability for Bombacoideae, and suggest that counting errors, especially in earlier chromosome counts, overestimated the karyotype variability of the subfamily. Errors in chromosome counts can be attributed to technical difficulties, high chromosome numbers, and/or the small sizes of Bombacoideae chromosomes.
Phylogenetic relationships according to Carvalho-Sobrinho et al. (2016) and chromosome numbers (2n) of Ceiba Mill. (Bombacoideae, Malvaceae). Idiogram showing sites of 5S rDNA (black) and 45S rDNA/CMA+/DAPI− bands (gray with stripes)
References
Assis FNM, Souza BCQ, Medeiros Neto E, Pinheiro F, Silva AEB, Felix LP (2013) Karyology of the genus Epidendrum (Orchidaceae: Laeliinae) with emphasis on subgenus Amphiglottium and chromosome number variability in Epidendrum secundum. Bot J Linn Soc 172:329–344
Baker HG (1964) Opportunities for evolutionary studies in the tropics. Taxon 13:121–126
Baker HG, Baker I (1968) Chromosome numbers in the Bombacaceae. Bot Gaz 129:294–296
Barros e Silva AE, Guerra M (2010) The meaning of DAPI bands observed after C-banding and FISH procedures. Biotech Histochem 85:115–125
Baum DA, Oginuma K (1994) A review of chromosome numbers in Bombacaceae with new counts for Adansonia. Taxon 43:11–20
Begum R, Zakrzewski F, Menzel G, Weber B, Alam SS, Schmidt T (2013) Comparative molecular cytogenetic analyses of a major tandemly repeated DNA family and retrotransposon sequences in cultivated jute Corchorus species (Malvaceae). Ann Bot 112:123–134
Brighton CA, Mathew B, Marchant CJ (1973) Chromosome counts in the genus Crocus (Iridaceae). Kew Bull 28:451–464
Carvalho-Sobrinho JG, Queiroz LP (2008) Ceiba rubriflora (Malvaceae: Bombacoideae), a new species from Bahia, Brazil. Kew Bull 63:649–653
Carvalho-Sobrinho JG, Alverson WS, Alcantara S, Queiroz LP, Mota AC, Baum DA (2016) Revisiting the phylogeny of Bombacoideae (Malvaceae): novel relationships, morphologically cohesive clades, and a new tribal classification based on multilocus phylogenetic analysis. Mol Phylogenet Evol. doi:10.1016/j.ympev.2016.05.006
Clarkson JJ, Lim KY, Kovarik A, Chase MW, Knapp S, Leitch AR (2005) Long-term genome diploidization in allopolyploid Nicotiana section Repandae (Solanaceae). New Phytol 168:241–252
Cristobal CL (1967) Cromosomas de Malvales. Kurtziana 4:139–142
Dantas LG, Guerra M (2010) Chromatin differentiation between Theobroma cacao L. and T. grandiflorum Schum. Genet Mol Biol 33:94–98
Duarte MC, Esteves GL, Salatino MLF, Walsh KC, Baum DA (2011) Phylogenetic analyses of Eriotheca and related genera (Bombacoideae, Malvaceae). Syst Bot 36:690–701
Escudero M, Maguilla E, Loureiro J, Castro M, Castro S, Luceño M (2015) Genome size stability despite high chromosome number variation in Carex gr. laevigata. Am J Bot 102:1–6
Felix LP, Guerra M (2010) Variation in chromosome number and the basic number of subfamily Epidendroideae (Orchidaceae). Bot J Linn Soc 163:234–278
Gan Y, Liu F, Chen D, Wu Q, Qin Q, Wang C, Li S, Zhang X, Wang Y, Wang K (2013) Chromosomal locations of 5S and 45S rDNA in Gossypium genus and its phylogenetic implications revealed by FISH. PLoS ONE 8:e68207
Gibbs PE, Semir J (2003) A taxonomic revision of the genus Ceiba Mill. (Bombacaceae). Ann Jard Bot Madrid 60:259–300
Gibbs PE, Semir J, da Cruz ND (1988) A proposal to unite the genera Chorisia Kunth with Ceiba Miller (Bombacaceae). Notes Roy Bot Gard Edinburgh 45:125–136
Gill BS, Bir SS, Singhal VK (1979) Reports. In: Love A (ed) IOPB chromosome number reports LXV. Taxon 28:630
Guerra M (1986) Citogenética de angiospermas coletadas em Pernambuco, I. Braz J Genet 9:21–40
Guerra M (1990) A situação da citotaxonomia de angiospermas nos trópicos e, em particular, no Brasil. Acta Bot Bras 4:75–86
Guerra M (2000) Chromosome number variation and evolution in monocots. Monocots 127-136
Guerra M (2012) Cytotaxonomy: the end of childhood. Plant Biosyst 146:703–710
Guerra M, Pedrosa A, Silva AEB, Cornélio MTM, Santos KGB, Soares Filho WS (1997) Chromosome number and secondary constriction variation in 51 accessions of a Citrus germoplasm bank. Brazil J Genet 20:489–496
Hanson RE, Islam-Faridi MN, Percival EA, Crane CF, Ji Y, McKnight TD, Price HJ (1996) Distribution of 5S and 18S–28S rDNA loci in a tetraploid cotton (Gossypium hirsutum L.) and its putative diploid ancestors. Chromosoma 105:55–61
Heyn ANJ (1938) Du nombre de chromosomes chez quelques vegetaux en culture aux Indes Neerlandaises (Coffea, Ceiba, Oryza, Derris et Palaquium). Ann Jard Bot Buitenzorg 48:103–120
Kovarik A, Matyasek R, Lim KY, Skalická K, Koukalova B, Knapp S, Leitch AR (2004) Concerted evolution of 18–5.8–6S rDNA repeats in Nicotiana allotetraploids. Biol J Linn Soc 82:615–625
Lewis H (1951) Origin of supernumerary chromosomes in natural populations of Clarkia elegans. Evolution 5:142–157
Lewis WH, Royce LO, Terry JL (1971) Multiple genotypes in individuals of Claytonia virginica. Science 172:564–565
Marinho RC, Mendes-Rodrigues C, Balao F, Ortiz PL, Yamagishi-Costa J, Bonetti AM, Oliveira PE (2014) Do chromosome numbers reflect phylogeny? New counts for Bombacoideae and a review of Malvaceae sI. Am J Bot 101:1456–1465
Merxmüller H (1970) Biosystematics: still alive. Taxon 19:140–145
Morawetz W (1986) Remarks on karyological differentiation patterns in tropical woody plants. Plant Syst Evol 152:49–100
Oginuma K, Alverson WS, Baum DA (1999) A cytological study of three genera of neotropical Bombacaceae (clades Bombacoideae and Malvoideae). Acta Phytotax Geobot 50:173–178
Oliveira PE, Gibbs PE, Barbosa AA, Talavera S (1992) Contrasting breeding systems in two Eriotheca (Bombacaceae) species of the Brazilian cerrados. Plant Syst Evol 179:207–219
Parisod C, Mhiri C, Lim KY, Clarkson JJ, Chase MW, Leitch AR, Grandbastien MA (2012) Differential dynamics of transposable elements during long-term diploidization of Nicotiana section Repandae (Solanaceae) allopolyploid genomes. PLoS One 7:e50352
Pedrosa A, Sandal N, Stougaard J, Schweizer D, Bachmair A (2002) Chromosomal map of the model legume Lotus japonicus. Genetics 161:1661–1672
Ragone D (2001) Chromosome numbers and pollen stainability of three species of Pacific Island breadfruit (Artocarpus, Moraceae). Am J Bot 88:693–696
Sareen TS, Kumari S (1973) Reports. In: Love A (ed) IOPB chromosome number reports XLII. Taxon 22:651-652
Schubert I, Lysak MA (2011) Interpretation of karyotype evolution should consider chromosome structural constraints. Trends Genet 27:207–216
Sinha ARP, Mazumdar LR (1993) New chromosome counts of an endemic species of Andamans. Broteria Genet 14:157–160
Sousa A, Barros e Silva AE, Cuadrado A, Loarce Y, Alves MV, Guerra M (2011) Distribution of 5S and 45S rDNA sites in plants with holokinetic chromosomes and the “chromosome field” hypothesis. Micron 42:625–631
Tijo JH (1948) Notes on nucleolar conditions in Ceiba pentandra. Hereditas 34:204–208
Weiss-Schneeweiss H, Schneeweiss GM, Stuessy TF, Mabuchi T, Park JM, Jang CG, Sun BY (2007) Chromosomal stasis in diploids contrasts with genome restructuring in auto-and allopolyploid taxa of Hepatica (Ranunculaceae). New Phytol 174:669–682
Wolfe KH (2001) Yesterday’s polyploids and the mystery of diploidization. Nat Rev Genet 2:333–341
Acknowledgments
The authors wish to thank the Brazilian agencies, the Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) and the, Fundação de Amparo à Ciência e Tecnologia de Pernambuco (FACEPE process APQ-2008-2.02/12) for their financial supports. A.W.L.O and JGCS thank the CNPq for the awards of Masters and postdoctoral (process 158916/2014-0) fellowships, respectively.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Figueredo, A., de L. Oliveira, Á.W., Carvalho-Sobrinho, J.G. et al. Karyotypic stability in the paleopolyploid genus Ceiba Mill. (Bombacoideae, Malvaceae). Braz. J. Bot 39, 1087–1093 (2016). https://doi.org/10.1007/s40415-016-0296-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40415-016-0296-5