Abstract
Background
The US Food and Drug Administration (FDA) collects and retains several data sets on post-market drugs and associated adverse events (AEs). The FDA Adverse Event Reporting System (FAERS) contains millions of AE reports submitted by the public when a medication is suspected to have caused an AE. The FDA monitors these reports to identify drug safety issues that were undetected during the premarket evaluation of these products. These reports contain patient narratives that provide information regarding the AE that needs to be coded using standardized terminology to enable aggregation of reports for further review. Additionally, the FDA collects structured drug product labels (SPLs) that facilitate standardized distribution of information regarding marketed medical products. Manufacturers are currently not required to code labels with associated AEs.
Objectives
Approaches for automated classification of reports by preferred terminology could enhance regulatory efficiency. The goal of this work was to assess the suitability of manually annotated FDA FAERS and SPL data sets to be subjected to predictive modeling.
Methods
A recurrent neural network (RNN) was proposed as a proof-of-concept model for automated extraction of preferred AE terminology. A separate RNN was fit and cross-validated on two regulatory data sets with varying properties. First, the researchers trained and cross-validated a model on 325 annotated FAERS patient narratives for a sample of AE terms. A model was then trained and validated on a data set of 100 SPLs.
Results
Model cross-validation results for product labels demonstrated that the model performed at least as well as more conventional models for all but one of the terms selected based on F1-score. Model results for the FAERS data set were mixed.
Conclusions
This work successfully demonstrated a proof-of-concept machine learning approach to automatically detect AEs in several textual regulatory data sets to support post-market regulatory activities. Limited instances of each AE class likely prohibited models from generalizing data effectively. Additional data may permit more robust validation.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Regulatory adverse event reports can be used by the US Food and Drug Administration to identify post-market drug safety trends; however, this requires intensive manual review. |
Machine learning in combination with natural language processing techniques can be used to classify textual report data based on manually annotated training data sets. |
This work successfully demonstrated a proof-of-concept machine learning approach to automatically detect adverse events in several textual regulatory data sets to support post-market regulatory activities. |
1 Introduction
The US Food and Drug Administration (FDA) Adverse Event Reporting System (FAERS) is a database that contains adverse event (AE) reports associated with marketed drugs and supports the FDA’s post-market drug safety efforts (https://open.fda.gov/data/faers/). FAERS receives safety reports voluntarily from drug consumers and health care providers, while manufacturers are required by law to submit safety reports they receive from the public [1]. Despite rigorous clinical trial requirements, new and unknown safety issues may arise in post-market phases for drugs. This can be primarily attributed to the challenges associated with perfectly mimicking post-market conditions during clinical trials [2]. FAERS can help identify previously unknown safety issues and drug–AE associations. While data are plentiful with more than a million reports filed each year, identifying these associations is laborious because the reports require intense manual review [3].
In each report, a summary of the AE that occurred is contained in a patient narrative. The patient narrative contains a free-text description of the event and is coded with preferred terms (PTs) from the Medical Dictionary for Regulatory Activities (MedDRA). By utilizing standardized codes for labeling AE reports, the FDA can identify trends regarding potential safety threats and causal relationships between drugs and AEs [4]. This information could also aid in identifying at-risk patient subpopulations, tracking inappropriate prescription trends, and facilitating continued surveillance over time [5]. The coding is performed manually and is therefore highly labor intensive [6, 7]. Providing automated and standardized support for the labeling of patient narratives could improve reviewer efficiency and is of significant interest to the FDA [3, 8].
In addition to AE narratives, MedDRA has also been applied to structured drug product labels (SPLs). SPLs facilitate distribution of information regarding marketed medical products in a standard format for use in health information systems [9]. Currently, manufacturers are not required to describe AEs using MedDRA terminology in SPLs [3]. Therefore, AE terms must be manually extracted from product labels to provide useful data. The process of manually annotating product labels using preferred MedDRA terminology is laborious [8]. Methods for automating the extraction of AEs from drug labels and mapping these events to MedDRA terminology could be beneficial for regulatory bodies.
Automated detection of AEs from text is an active area of research in this field, with experts exploring many techniques. The simplest approaches have relied on lexicon matching [3, 10,11,12,13,14,15,16] and rule-based systems [3, 6, 9, 15, 17,18,19,20]. While generally reliable and simple to apply, these approaches may have difficulty managing informal language and deciphering complex linguistic relationships. Assorted predictive statistics models (e.g., regression, support vector machine [SVM], decision trees) have been used as well [6, 11, 16, 18, 21,22,23,24,25]. Text and data from many sources have been explored for detection of AEs, including drug labels [3, 9, 10], social media posts [11,12,13, 21], biomedical literature [14, 15, 22,23,24,25,26], web search logs [27], health records [16,17,18,19, 28, 29], and regulatory reports [6, 15, 20, 26]. For a more in-depth review of recent literature, see [30].
Deep learning models have been proposed as a means for automated extraction of AEs from text data. While less interpretable and more computationally expensive than other predictive models, neural networks can provide classifications based on complex contextual associations between features. Deep neural networks, typically in conjunction with word embeddings, such as GloVe [31], have proven useful for the classification of text associated with adverse drug events [6, 14, 22, 23, 25]. Recent advances have made more complex models available as well. Convolutional neural networks (CNNs) learn contextual information about features by including a convolutional and pooling layer in its architecture. While most commonly applied to machine vision tasks, CNNs have been used in research for text classification and AE detection [16]. Recurrent neural networks (RNNs), which are designed to make predictions based on arbitrary-length sequences of input, have been used as well. Because the information conveyed by text is highly dependent on word order, RNNs are highly suited for text classification. Recent implementations for drug AE detection include the use of bidirectional RNNs [12, 13, 19, 24], RNN with an attention mechanism [16, 29], and a RNN-Conditional Random Field ensemble [28].
For this work, the researchers propose an RNN for classification of patient narratives using MedDRA PTs. This proof-of-concept model will demonstrate the suitability of RNNs for the classification of MedDRA PTs given FAERS patient narratives. Further, the model was tested by the researchers on FDA SPLs to demonstrate its utility with text data that is more structured. The focus of this research is on the suitability of the data sets for predictive modeling, not to demonstrate the limits of state-of-the-art machine learning algorithms. Further, the objective was not to benchmark the results of the prediction task for each data set against one another. This proof-of-concept will serve as a building block for additional projects with the goal of aiding narrative coders in standardizing FDA AE data.
2 Methods
In this work, two types of data common in FDA regulatory activities were used to develop and validate a proof-of-concept application of an RNN model to extract PTs. The objective was to use the full documents contained in each data set to predict PT labels. First, the researchers performed training and cross-validation using patient narratives from the FAERS database. Second, a new RNN was fit and cross-validated on a data set of SPLs. Performance for these efforts is reported.
2.1 FAERS Data Set
The FAERS data set used for this project contained 325 event entries that included both patient narratives and PT labels. Each event can have many PT labels associated with it. For example, the following event narrative was contained in the data set:
“A one-year-old female (born in [RED]) experienced an encephalopathy (no etiology for the encephalopathy could be found) with severe hypotonia and epilepsy (exact start date not reported). The epilepsy is partly controlled with valproate sodium, clonazepam and vigabatrin treatment. Her father had been treated with omeprazole for several years (exact start date, dosage and indication not reported, his medical history included a nephrotic syndrome [sic] for which he had been treated for more than two years before the conception). Concomitant medication not précised [sic]. The reporting physician considered this to be a congenital anomaly.’
The PTs associated with this event narrative were ‘epilepsy,’ ‘encephalopathy,’ ‘hypotonia,’ and ‘congenital anomaly.’ These PTs are highlighted in the above excerpt. Note that PTs may not exist in a document in the exact preferred form. The model must therefore be able to detect synonyms of PTs, or collections of words that describe the PT. In all, there were 618 unique PTs in the data set. Many of these PTs occurred sparsely, with most of them occurring only once. In this work, several of the frequently occurring PTs are considered. Narratives can be labeled with more than one PT; however, in this research each PT is treated as an individual, binary classification problem.
Narratives are unstructured and highly variable. The length of narratives can vary considerably, with the shortest containing a single word and the longest containing more than 2000. Narratives are typically informal descriptions of the event and can contain many contextual clues about the nature of the event that do not necessarily use official or predictable vocabulary.
2.2 Structured Drug Product Label (SPL) Data Set
The SPL data set included 100 product labels manually annotated with MedDRA PTs by FDA personnel. The data set was originally provided as part of the FDA Adverse Drug Event Evaluation challenge conducted during 2018–2019, through which researchers worked to develop a tool for automated extraction of AEs from SPLs. Details on the data sets are available in [32].
Each SPL was provided in an XML format, from which relevant text was extracted. Each label is organized into several standardized sections. These sections include Boxed Warnings, Warnings, General Precautions, Warnings and Precautions, and Adverse Reactions [32]. This work focuses exclusively on the Adverse Reaction sections of the labels. This section is used as the primary basis for manual PT annotations. The Adverse Reactions section should contain all adverse reactions that have been attributed to a medication, while other sections of the label may provide additional information regarding the severity of certain reactions, as well as recommendations on how to monitor or treat patients who experience certain adverse reactions [32].
Table 1 contains a summary of the data sets used in this work.
2.3 Model Choice
RNNs are an adaptation of standard neural networks designed to handle sequential data of arbitrary length [33]. This is particularly useful for making predictions based on unstandardized text which can vary in length by word. An RNN is composed of a series of ‘cells,’ where sequential data (e.g., text) is used as input. A series of algebraic operations are applied to the data inside the cell, and the resulting information is passed to the next cell. Within these operations, the RNN cell applies learned weight parameters to a concatenated vector containing the cell input and a ‘hidden state.’ In the case of text classification, a cell input corresponds to a word in a sequence, with the total number of cells equal to the length of the sequence. The hidden state is a vector of user-specified length that is passed from cell to cell and serves to provide ‘memory’ over the sequence to the model. Learned parameters are fit to the data set by comparing the output of the final cell (prediction) to the true value or classification associated with each datapoint. The function that computes the difference between model output and the ground truth is called the loss function. The loss function is typically computed and summed for all datapoints in the training data set, resulting in the total loss. To fit the model, the loss function is minimized, typically using a gradient-based optimization technique, where parameters within each cell are varied to achieve a minimum loss.
The issue with simple RNNs is that they are susceptible to difficulties when training model parameters. This is referred to as vanishing gradients, whereby gradients approach zero during optimization, preventing parameters from updating. This is particularly problematic for large networks [34]. Long Short-Term Memory (LSTM) networks were developed to address this limitation by including mechanisms to prevent vanishing gradients. The core concept behind LSTM is the inclusion of a memory cell that maintains information over long periods of time (elements in a sequence) and nonlinear gating units that regulate the flow of information in and out of the memory cell [35]. Gated Recurrent Unit (GRU) [36] (pre-print article) networks are a more recent development that take the memory cell and gating concept of LSTM, but reduce the number of required gating units, and therefore reduce the number of parameters. GRU has performed similarly to LSTM, but with reduced computational burden [37]. Because this work was to serve as a proof-of-concept, GRUs were used, providing benefits of the LSTM model while also reducing computational resources and the amount of time required.
2.4 Preprocessing Tasks
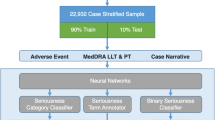
Figure 1 displays a high-level diagram of the preprocessing tasks detailed in Sect. 2.4 (left side), and the experimental validation procedures detailed in 2.5 (right side).
It is typical to perform several standard natural language preprocessing (NLP) tasks to format text for use in machine learning models. First, text data was tokenized, a process in which bodies of text are split into individual words (tokens). Next, unwanted text is filtered. Punctuation, nonalphabetic words, and stop words were removed. Stop words are common words that provide little information in prediction tasks, such as ‘what,’ ‘where,’ ‘is,’ ‘are,’ ‘a,’ and ‘the.’ Tokens were then lemmatized, which transforms words into their base morphological form. For example, ‘mice’ would become ‘mouse’, and ‘eating’ would become ‘eat.’ All-natural language processing tasks were performed using the Natural Language Toolkit (NLTK) in the Python programming language (https://www.nltk.org/).
For use in an RNN, data must be translated into a sequence format. In sequence format, each token in each data point becomes a unique feature or column entry. Rows are padded with zeros to the length of the longest sequence. Keras, a Python library that provides the building blocks for developing deep learning models, provides many features and resources to facilitate model building [38]. Keras was used to format data into sequence format for RNN model input and is also used for model definition and training in the steps outlined below.
To train a statistical model, text must be translated into a numerical format. One of the simplest models for creating real vectors from text is bag-of-words. This model characterizes documents in a textual data set as vectors, where each entry of the vector typically corresponds to the frequency of unique words present across the entire data set. Words are also typically assigned a weight based on their frequency across documents [39]. The limitation of this and similar approaches is that information contained by word order is lost. Additionally, there is no way to compare lexically similar words. For example, comparing the words ‘cat’ and ‘lion’ will result in the same value as comparing ‘cat’ and ‘automobile’, even though the former are obviously more related. This also means the model will not be able to handle words not encountered in training data [40].
Word embeddings are vector representations of words generated with the goal of representing linguistic similarity mathematically, and are commonly used in text classifications [41]. Word embeddings allow algebraic operations to be performed on words such that linguistic meaning is preserved. Most word embeddings are derived from massive corpora using unsupervised or semi-supervised machine learning and dimensionality reduction techniques. Word2vec [42] (pre-print article) and GloVe [31] are popular algorithms that have seen significant use in recent years for NLP tasks.
In this work, GloVe pretrained word embeddings were utilized. Past researchers have used GloVe successfully for similar AE detection tasks [28]. GloVe uses a log-bilinear regression model to fit weight vectors to words based on the probability of word–word co-occurrence in a large text corpus. The resulting word vectors, or word embeddings, exhibit contextual information with relation to one another—in essence, quantifying the relatedness of words [31]. GloVe can be used to fit new custom word embeddings given a large corpus. GloVe also has pretrained embeddings that can be repurposed for new tasks. In this work, a GloVe word embedding trained on Common Crawl (https://commoncrawl.org/) data was used. The embedding was trained on 42 billion tokens, has a vocabulary of 1.9 million words, and contains 300-dimension word vectors. For the FAERS data set, the GloVe word embedding covered 92.9% of the contained words. For the SPL data sets combined, 82.7% was covered. For the annotated SPL data set, 93.8% was covered.
2.5 Model Experiments
The first devised test was to develop an RNN model to classify FAERS patient narrative entries by FDA PTs. A 5-layer deep GRU network was defined using Keras in Python 3.6. To evaluate model performance, several of the highest prevalence PTs were selected for validation. The PTs included, their frequency, and lengths of the processed text in the data set are shown in Table 2.
K-fold cross-validation is a technique to estimate the performance of a model on new data and is performed on labeled training data. This technique splits the data into k ‘folds,’ trains the model on k-1 folds, and tests the model on the remaining fold. This is repeated until each fold has been left out and tested on once. The results are then averaged. The folding procedure is often repeated several times to get unique combinations of datapoints, usually noted as r × k-fold cross-validation, where r is the number of repeats.
We performed 5 × 5-fold stratified cross-validation for ‘drug interaction’ and ‘drug ineffective;’ 4-fold stratified cross-validation was used for other PTs with lower frequencies to ensure several positive cases were present in the validation set. To manage the relative unbalanced nature of the data set, minority classes were randomly oversampled to the size of the majority class. Initial tests indicated overfitting may be an issue, so dropout was incorporated into the model [43]. Machine learning models are trained by minimizing some loss function comparing model-predicted labels with the true labels. Optimization of model parameters is dependent on the parameters of the optimization routine, referred to as hyperparameters [44]. Hyperparameter tuning was performed to improve the model training performance. Model parameters are shown in Table 3. Binary cross-entropy was used for the loss function.
The focus of this work was to assess the given data sets as candidates for modeling with sequence-based architecture neural networks, not necessarily optimizing the model for the data. Therefore, the choice of five hidden layers was an arbitrary choice given the proof-of-concept nature of this work. Further, while a bidirectional architecture was considered for the model, a standard uni-directional RNN was opted for to demonstrate the simplest case benchmark for future improvement.
To demonstrate a predictive classifier, it is common to benchmark the results against other conventional classifiers [28, 29]. Two simpler classifiers were used for validation as well as for comparison. Logistic regression and support vector machine with a radial basis function as the kernel function were used. In both cases, the average word embedding for each narrative was used as input.
Metrics used to evaluate performance were recall, precision, and F1-score, of which the latter is typically the preferred metric in NLP for evaluating model performance [45]. Accuracy can provide misleading results when classes are imbalanced if detection of the minority class is valued more than the majority, such as the case with the data sets in this paper. For model validation, training continued for 20 epochs or until validation F1 did not improve for 10 epochs. Medians and interquartile ranges for results are reported. Multiple comparisons using Mood’s median test with Bonferroni correction were performed for each PT between each model for the F1-score.
In the second test, the goal was to evaluate how well the same type of model (RNN) would classify SPLs based on relevant PTs. The researchers performed validation using the 100 annotated product labels. We performed 5 × 5-fold stratified cross-validation for several high prevalence PTs, as well as for several low prevalence PTs to serve as a comparison to the narrative prediction performance. The included PTs, their frequency, and the length of the processed text in the data set are shown in Table 4. Model performance was benchmarked against logistic regression and SVM with a radial basis function. Model results were compared again using Mood’s median test for post-hoc comparisons.
3 Results
Discussed here are the proof-of-concept results for the FAERS data set and SPL data set PT classification tasks.
3.1 FAERS Patient Narrative Classification
Reported in Table 5 are the median and interquartile range (IQR) for validation F1-score, recall, and precision for the FAERS data cross-validation. Superscripts are used to signify multiple comparisons results. The RNN only outperformed the other models for ‘drug interaction’ but not significantly so for either. The RNN performed significantly worse for ‘acute kidney injury’ and ‘seizure.’
Table 6 contains the results of the multiple comparisons analysis. The largest differences in performance were observed for ‘acute kidney injury,’ with the RNN performing significantly worse than the other models.
3.2 SPL Classification
Reported in Table 7 are the median and IQR for validation F1-score, recall, and precision for the SPL training data cross-validation. The RNN performed significantly better than the other models for the low occurrence PTs and only performed significantly worse in the case of ‘diarrhea’ compared with logistic regression, but with a very small effect size.
Table 8 contains the results of the multiple comparisons analysis. Differences in performance were largest between the RNN and the other models for ‘arrythmia.’
4 Discussion
The following is a discussion of the model results and their implications as a proof-of-concept.
4.1 FAERS Patient Narrative Classification Validation
These results provide evidence that, with some additional model optimization and more data, the RNN model could assist in automated extraction of AE data from FAERS patient narratives. Overall, however, the given patient narrative data set did not generalize well enough for the RNN model to reliably predict the selected FDA PTs. The generally poor results likely can be attributed to a combination of small sample size, the short length of some narratives, and the unstructured nature of the text. Oversampling of minority classes did aid in overcoming the imbalance issues; however, the poor F1 scores suggest that positive cases may be too few and text may be too variable to generalize effectively, and thus the model was prone to overfitting (particularly for the RNN). For drug ineffective, acute kidney injury, and seizure, the contents of narratives were extremely short. In one case, for seizure, the text was one word after processing. It is very unlikely that these extremely short narratives were correctly predicted in any of the cases.
For the patient narrative classification task, results were mixed between models. Results indicated that for ‘drug interaction’ the RNN had the best performance. For the other terms, the simpler models performed better. The difference in performance may be due to the nature of the terms themselves. ‘Acute kidney injury’ and ‘seizure’ point to specific ailments with distinct terminology that may have resulted in a relatively linear influence on the average word embedding. ‘Drug interaction’ and ‘drug ineffective’ describe more abstract concepts and may require a more complex model to recognize their linguistic signal. That said, the RNN did not perform significantly better than the other models for any term so it is difficult to infer the generalizability of this type of data without a larger sample.
4.2 Structured Product Label Classification Validation and Testing
Overall model performance was significantly better for SPL validation, likely due to the structured nature of the product label text and the more balanced class distribution. The RNN model performed at least as well as the simpler models for all terms except ‘diarrhea’ and performed significantly better than both models for ‘malaise’, ‘flushing,’ ‘atrial fibrillation,’ and ‘arrythmia.’ That the deep learning model performed similar to the simpler models for the high frequency terms and better for the low frequency terms suggests that the ability to detect complex linguistic relationships can help overcome class imbalance issues.
The discrepancy between the RNN’s ability to extract low frequency terms from the FAERS data versus the SPL data is not entirely understood. It may be that the structured and comprehensive nature of product labels contains information that the patient narratives do not. The RNN may be able to extract more predictive value from word order from the SPLs than the patient narratives due to the structured nature. Additionally, SPLs contain comprehensive information regarding possible drug adverse reactions. Patient narratives generally do not contain as many references to medical terminology that may help a model develop associations. RNNs are well suited to identify this type of complex co-occurring medical terminology associated with certain AEs.
4.3 Limitations and Implications
This work suggests that deep learning models, specifically RNNs, can be used to extract AEs as preferred terminology from SPLs at least as well as, and in some cases better than, other standard predictive models. This work was limited in that validation results only demonstrated model effectiveness for a small subset of PTs. Future work should focus on optimizing the current model more rigorously, as well as exploring additional model architectures. A model that produced consistent results across PTs would increase the usefulness of SPLs for discovering drug–AE associations.
Another limitation of this work occurred with respect to data preprocessing. Non-alphabetic words were removed from text prior to fitting models. In hindsight, this may have removed valuable information from the SPL data set, as some PT terminology relies on mixed alpha-numeric terminology (e.g., HLA-B*1502-positive). This was less of a concern for the FAERS data set, which typically contained less technical language. Future work should verify the influence of this terminology in adverse drug event text data for similar prediction tasks.
In this work, each PT prediction task was treated as an individual, binary classification problem, and a separate model was fit in each case. While we could have integrated the prediction tasks into a single architecture as multiple binary classification problems, oversampling the data for minority classes can be more complicated in this case. This ultimately shouldn't significantly influence the results, however if the goal was to classify a much larger sample of PTs, the used approach would be time-prohibitive.
While model performance was less ideal for the patient narrative data, it should not be invalidated as a candidate for prediction using this type of model. The maximum number of samples for a single class was only 14, so it is not overly surprising that it did not generalize well. Manual annotation of patient narratives is an arduous task that requires specialized knowledge of MedDRA. Future work should focus on streamlining acquisition of additional data and evaluating new word embedding techniques as the field advances. Bidirectional Encoder Representations from Transformers (BERT), for example, is a technique that uses contextual information bidirectionally in the construction of word embeddings [46] (pre-print article). This allows BERT to express multiple meanings for words that appear identical based on context. BioBERT, an embedding model trained on a large corpus of biomedical text, could be especially useful in the pharmacovigilance domain [47].
5 Conclusions
Automated extraction of AEs using standardized terminology could aid in streamlining regulatory processes and discovering new drug–AE associations. Extracting events in real time from post-market patient narratives would be especially useful for detecting new safety issues and protecting public health. While model performance was mixed, especially for underrepresented PTs in the FAERS data set, this work provides evidence that well-represented terms can reliably be determined using an RNN. The evaluations of unannotated SPL predictions provided further support for this finding. Machine learning has the potential to increase the efficiency of discovering safety issues associated with pre- and post-market drugs from textual data. A concerted effort should be made to increase the amount of available annotated data such that these models can continue to be developed and optimized.
References
US Food and Drug Administration. Questions and Answers on FDA’s Adverse Event Reporting System (FAERS) [Internet]. FDA; 2019 [cited 2021 Oct 6]. Available from: https://www.fda.gov/drugs/surveillance/questions-and-answers-fdas-adverse-event-reporting-system-faers
Harpaz R, Perez H, Chase HS, Rabadan R, Hripcsak G, Friedman C. Biclustering of adverse drug events in the FDA’s spontaneous reporting system. Clin Pharmacol Ther. 2011;89:243–50.
Ly T, Pamer C, Dang O, Brajovic S, Haider S, Botsis T, et al. Evaluation of natural language processing (NLP) systems to annotate drug product labeling with MedDRA terminology. J Biomed Inform. 2018;83:73–86.
Spiker J, Kreimeyer K, Dang O, Boxwell D, Chan V, Cheng C, et al. Information visualization platform for postmarket surveillance decision support. Drug Saf. 2020;43:905–15.
Karahoca A. Data mining applications in engineering and medicine. London: IntechOpen Limited; 2012.
Hatim Q, Rosario L, Almario EEN, Worthy KM, Sabo TM, McRae E, et al. Modeling and Text Analysis to Empower FAERS Adverse Event Assessment [Internet]. 2018. Available from: https://phuse.s3.eu-central-1.amazonaws.com/Archive/2018/Connect/US/Raleigh/PAP_AB09.pdf
Ramesh BP, Belknap SM, Li Z, Frid N, West DP, Yu H. Automatically recognizing medication and adverse event information from food and drug administration’s adverse event reporting system narratives. JMIR Med Inform. 2014;2: e3022.
Bayer S, Clark C, Dang O, Aberdeen J, Brajovic S, Swank K, et al. ADE eval: an evaluation of text processing systems for adverse event extraction from drug labels for pharmacovigilance. Drug Saf. 2020;44:83–94.
Pandey A, Kreimeyer K, Foster M, Dang O, Ly T, Wang W, et al. Adverse Event extraction from Structured Product Labels using the Event-based Text-mining of Health Electronic Records (ETHER) system: Health Informatics Journal [Internet]. 2018 [cited 2019 Oct 24];25. Available from: https://journals.sagepub.com/doi/pdf/https://doi.org/10.1177/1460458217749883
Kusch MKP, Zien A, Hachenberg C, Haefeli WE, Seidling HM. Information on adverse drug reactions–proof of principle for a structured database that allows customization of drug information. Int J Med Inform. 2019;103970.
Nikfarjam A, Sarker A, O’Connor K, Ginn R, Gonzalez G. Pharmacovigilance from social media: mining adverse drug reaction mentions using sequence labeling with word embedding cluster features. J Am Med Inform Assoc. 2015;22:671–81.
Cocos A, Fiks AG, Masino AJ. Deep learning for pharmacovigilance: recurrent neural network architectures for labeling adverse drug reactions in Twitter posts. J Am Med Inform Assoc. 2017;24:813–21.
Xie J, Liu X, Dajun ZD. Mining e-cigarette adverse events in social media using Bi-LSTM recurrent neural network with word embedding representation. J Am Med Inform Assoc. 2017;25:72–80.
Aronson AR, Lang F-M. An overview of MetaMap: historical perspective and recent advances. J Am Med Inform Assoc. 2010;17:229–36.
Gurulingappa H, Toldo L, Rajput AM, Kors JA, Taweel A, Tayrouz Y. Automatic detection of adverse events to predict drug label changes using text and data mining techniques. Pharmacoepidemiol Drug Saf. 2013;22:1189–94.
Tran T, Kavuluru R. Predicting mental conditions based on “history of present illness” in psychiatric notes with deep neural networks. J Biomed Inform. 2017;75:S138–48.
Chazard E, Ficheur G, Bernonville S, Luyckx M, Beuscart R. Data mining to generate adverse drug events detection rules. IEEE Trans Inf Technol Biomed. 2011;15:823–30.
Casillas A, Pérez A, Oronoz M, Gojenola K, Santiso S. Learning to extract adverse drug reaction events from electronic health records in Spanish. Expert Syst Appl. 2016;61:235–45.
Wunnava S, Qin X, Kakar T, Sen C, Rundensteiner EA, Kong X. Adverse drug event detection from electronic health records using hierarchical recurrent neural networks with dual-level embedding. Drug Saf. 2019;42:113–22.
Botsis T, Jankosky C, Arya D, Kreimeyer K, Foster M, Pandey A, et al. Decision support environment for medical product safety surveillance. J Biomed Inform. 2016;64:354–62.
Sarker A, Gonzalez G. Portable automatic text classification for adverse drug reaction detection via multi-corpus training. J Biomed Inform. 2015;53:196–207.
Wang C-S, Lin P-J, Cheng C-L, Tai S-H, Kao Yang Y-H, Chiang J-H. Detecting Potential Adverse Drug Reactions Using a Deep Neural Network Model. J Med Internet Res [Internet]. 2019;21. Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6381404/
Tafti AP, Badger J, LaRose E, Shirzadi E, Mahnke A, Mayer J, et al. Adverse drug event discovery using biomedical literature: a big data neural network adventure. JMIR Med Inform. 2017;5: e51.
Huang D, Jiang Z, Zou L, Li L. Drug–drug interaction extraction from biomedical literature using support vector machine and long short term memory networks. Inf Sci. 2017;415–416:100–9.
Tafti AP, Behravesh E, Assefi M, LaRose E, Badger J, Mayer J, et al. bigNN: An open-source big data toolkit focused on biomedical sentence classification. 2017 IEEE International Conference on Big Data (Big Data). Boston: IEEE; 2017. p. 3888–96.
Xu R, Wang Q. Large-scale combining signals from both biomedical literature and the FDA Adverse Event Reporting System (FAERS) to improve post-marketing drug safety signal detection. BMC Bioinform. 2014;15:17.
White RW, Wang S, Pant A, Harpaz R, Shukla P, Sun W, et al. Early identification of adverse drug reactions from search log data. J Biomed Inform. 2016;59:42–8.
Dai H-J, Su C-H, Wu C-S. Adverse drug event and medication extraction in electronic health records via a cascading architecture with different sequence labeling models and word embeddings. J Am Med Inform Assoc [Internet]. 2019 [cited 2019 Oct 23]; Available from: https://academic.oup.com/jamia/advance-article/doi/https://doi.org/10.1093/jamia/ocz120/5537181
Chu J, Dong W, He K, Duan H, Huang Z. Using neural attention networks to detect adverse medical events from electronic health records. J Biomed Inform. 2018;87:118–30.
Harpaz R, Callahan A, Tamang S, Low Y, Odgers D, Finlayson S, et al. Text mining for adverse drug events: the promise, challenges, and state of the art. Drug Saf. 2014;37:777–90.
Pennington J, Socher R, Manning C. GloVe: Global Vectors for Word Representation. Proceedings of the 2014 Conference on Empirical Methods in Natural Language Processing (EMNLP) [Internet]. Doha, Qatar: Association for Computational Linguistics; 2014 [cited 2020 Aug 10]. p. 1532–43. Available from: https://www.aclweb.org/anthology/D14-1162
Demner-Fushman D, Shooshan SE, Rodriguez L, Aronson AR, Lang F, Rogers W, et al. A dataset of 200 structured product labels annotated for adverse drug reactions. Sci Data. 2018;5: 180001.
Ackerson JM, Dave R, Seliya N. Applications of Recurrent Neural Network for Biometric Authentication & Anomaly Detection. Information. Multidisciplinary Digital Publishing Institute; 2021;12:272.
Sherstinsky A. Fundamentals of recurrent neural network (RNN) and long short-term memory (LSTM) network. Physica D. 2020;404: 132306.
Greff K, Srivastava RK, Koutník J, Steunebrink BR, Schmidhuber J. LSTM: a search space odyssey. IEEE Trans Neural Netw Learn Syst. 2017;28:2222–32.
Cho K, van Merrienboer B, Gulcehre C, Bahdanau D, Bougares F, Schwenk H, et al. Learning Phrase Representations using RNN Encoder-Decoder for Statistical Machine Translation. arXiv:14061078 [cs, stat] [Internet]. 2014 [cited 2020 Aug 25]; Available from: http://arxiv.org/abs/1406.1078
Dey R, Salem FM. Gate-variants of Gated Recurrent Unit (GRU) neural networks. 2017 IEEE 60th International Midwest Symposium on Circuits and Systems (MWSCAS). Boston, MA, USA: IEEE; 2017. p. 1597–600
Ketkar N. Introduction to Keras. In: Ketkar N, editor. Deep Learning with Python: A Hands-on Introduction [Internet]. Berkeley, CA: Apress; 2017 [cited 2020 Aug 25]. p. 97–111. Available from: https://doi.org/10.1007/978-1-4842-2766-4_7
Soumya G, Joseph S. Text classification by augmenting bag of words (BOW) representation with co-occurrence feature. IOSR-JCE. 2014;16:34–8.
Rudkowsky E, Haselmayer M, Wastian M, Jenny M, Emrich Š, Sedlmair M. More than bags of words: sentiment analysis with word embeddings. Commun Methods Meas Routledge. 2018;12:140–57.
Fan B, Fan W, Smith C, Garner H “Skip.” Adverse drug event detection and extraction from open data: A deep learning approach. Information Processing & Management. 2020;57:102131.
Mikolov T, Sutskever I, Chen K, Corrado G, Dean J. Distributed Representations of Words and Phrases and their Compositionality. arXiv:13104546 [cs, stat] [Internet]. 2013 [cited 2020 Aug 24]; Available from: http://arxiv.org/abs/1310.4546
Labach A, Salehinejad H, Valaee S. Survey of Dropout Methods for Deep Neural Networks. arXiv:190413310 [cs] [Internet]. 2019 [cited 2020 Sep 11]; Available from: http://arxiv.org/abs/1904.13310
SAS® Visual Data Mining and Machine Learning 8.2: Deep Learning Programming Guide. Cary, NC: SAS Institute Inc.; 2017. Available from: https://analyticsconsultores.com.mx/wp-content/uploads/2019/03/SAS-VDMML-8.2-Deep-Learning-Programming-Guide-SAS-2017.pdf
Derczynski L. Complementarity, F-score, and NLP Evaluation. Proceedings of the Tenth International Conference on Language Resources and Evaluation (LREC’16) [Internet]. Portorož, Slovenia: European Language Resources Association (ELRA); 2016 [cited 2020 Aug 27]. p. 261–6. Available from: https://www.aclweb.org/anthology/L16-1040
Devlin J, Chang M-W, Lee K, Toutanova K. BERT: Pre-training of Deep Bidirectional Transformers for Language Understanding. arXiv:181004805 [cs] [Internet]. 2019 [cited 2021 Feb 21]; Available from: http://arxiv.org/abs/1810.04805
Lee J, Yoon W, Kim S, Kim D, Kim S, So CH, et al. BioBERT: a pre-trained biomedical language representation model for biomedical text mining. Bioinformatics. 2019;btz682.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Funding
This manuscript was supported by the US Food and Drug Administration (FDA), an agency within the Department of Health and Human Services (HHS), as part of a financial assistance award (FAIN) totaling US$60,617 with 8% funded by FDA/HHS and US$55,617 and 92% funded by nongovernment source(s). The contents are those of the author(s) and do not necessarily represent the official views of or an endorsement by either FDA/HHS or the US Government.
Conflicts of Interest
The authors declare that they have no conflict of interest.
Ethics Approval
Not applicable.
Consent to Participate
Not applicable.
Consent for Publication
Not applicable.
Availability of Data and Material
Not applicable.
Code Availability
Not applicable.
Authors’ Contributions
BMK—Conception and design of research; acquisition, analysis, and interpretation of data; writing and revision of final manuscript. QH—Conception and design of research; interpretation of data; critical revision of final manuscript. MV-C—Conception and design of research; interpretation of data; critical revision of final manuscript. All authors have read and approved the final submitted manuscript and agree to be accountable for the work.
Rights and permissions
About this article
Cite this article
Knisely, B.M., Hatim, Q. & Vaughn-Cooke, M. Utilizing Deep Learning for Detecting Adverse Drug Events in Structured and Unstructured Regulatory Drug Data Sets. Pharm Med 36, 307–317 (2022). https://doi.org/10.1007/s40290-022-00434-y
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40290-022-00434-y