Abstract
Background
Lithium remains the first-line treatment for bipolar disorder (BD), but patients respond to it variably. While a myriad of studies have attributed many genes and signaling pathways to lithium responsiveness, a comprehensive study with an integrated conclusion is still lacking.
Objective
We aim to present an integrated mechanism for the therapeutic actions of lithium in BD.
Methods
First, a list of lithium responsiveness-associated genes (LRAGs) was collected by searching in the literature. Thereafter, gene set enrichment analysis together with gene–gene interaction network analysis was performed, in order to find the cellular and molecular events related to the LRAGs.
Results
Gene set enrichment analyses showed that the chromosomal regions 3p26, 4p21, 5q34 and 7p13 could be novel associated loci for lithium responsiveness in BD. Also, expression pattern analysis of the LRAGs showed their enrichment in adulthood stages and different cell lineages of brain, blood and immune system. Most of the LRAGs exhibited enriched expression in central parts of human brain, suggesting major contribution of these parts in lithium responsiveness. Beside the prediction of several biological processes and signaling pathways related to lithium responsiveness, an interaction network between these processes was constructed that was found to be regulated by a set of microRNAs. Proteins of the network were mainly classified as transcription factors and kinases, which also highlighted the crucial role of glycogen synthase kinase 3β (GSK3β) in lithium responsiveness.
Conclusions
The predicted cellular and molecular events in this study could be considered as mechanisms and also determinants of lithium responsiveness in BD.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
This study presents a comprehensive view of the cellular and molecular actions of lithium in the treatment of bipolar disorder (BD). |
Despite diversities in the biological processes involved in lithium response in BD, this study presents a unique molecular interaction network for the lithium responsiveness phenotype of the disorder. |
Understanding the lithium targets at cellular and molecular levels and elucidating their expression pattern in patients allow clinicians to determine which patients are most likely to respond to lithium. |
1 Introduction
Bipolar disorder (BD) is one of the most severe mental illnesses, causing extreme mood swings between mania and depression [1]. Lithium is primarily used for long-term treatment of BD with the aim to prevent further manic and depressive recurrences. In this indication, lithium remains the first-line treatment of BD [2, 3]. Lithium is also known to substantially reduce the risk of suicide in people with BD and unipolar depression [3]. However, only in one third of BD patients does long-term monotherapy with lithium totally prevent affective episodes [4]. The therapeutic properties of lithium in BD have been the subject of a number of candidate gene studies [5,6,7,8,9] and also that of the International Consortium on Lithium Genetics (ConLiGen) genome-wide associated study (GWAS) [2]. These studies have used several experimental approaches, such as GWAS [2], gene expression [10], and functional-based analyses [11, 12], to find the genes involved in lithium responsiveness. Several GWASs have been published to date but the majority focused on BD in general and not lithium responsiveness [13]. One of the greatest efforts in order to delineate the genetic basis of lithium response has been launched by the ConLiGen, which in its first project has conducted a GWAS of stringently defined response to lithium treatment in BD [2, 14]. However, in the future, ConLiGen may also study the response to lithium treatment in general (e.g., lithium augmentation in the treatment of major depression) [15].
To date, many genetic variants and altered expression of many genes have been found to be associated with lithium responsiveness. A large variety of the associated genes were identified by GWAS analyses, and their association was further confirmed by in vitro or in vivo functional assays. In fact, many of the associated genetic variants reported by GWAS analyses have been examined in cultured cells or model organisms and shown to influence the expression or function of the genes. Such functional analyses confirmed some of the lithium responsiveness-associated genes (LRAGs) as direct lithium targets, such as the phosphatidylinositol (PI) pathway, which is one of the first and most studied cellular processes where lithium plays an inhibitory role [16, 17], and also the glycogen synthase kinase 3β (GSK3β) gene, which is one of the best characterized targets of lithium [18]. Both GSK3β and the genes of the PI pathway were shown to harbor the genetic variants associated with lithium responsiveness in BD [19,20,21]. However, the association of many other genes with lithium responsiveness was confirmed by genomics tools, without any functional studies [22, 23]. Despite the large number of reports on the association of many genes with lithium responsiveness, a single comprehensive study to illustrate how these genes contribute to response to lithium treatment in BD is lacking.
In this study, a list of the genes that were previously attributed to lithium responsiveness in BD was collected and analyzed to elucidate the mechanisms related to these genes. Understanding the molecular mechanisms underlying lithium responsiveness in BD could help clinicians to select the best therapy approach for bipolar patients. Also, clinicians will be assisted by a panel of genetic tests that may successfully predict which patients are most likely to respond to lithium.
2 Methods
2.1 Searches and Dataset
To systematically study the genes involved in lithium response in BD patients, we performed a search in the literature followed by a manual assembly of the gene list. In detail, all publications reporting the genes associated with lithium responsiveness in BD, published before November 2019, were retrieved by a comprehensive electronic search in PubMed, Google Scholar, ScienceDirect and Web of Science, without any restriction in publication language (Fig. 1). The keywords used for this search were ‘lithium responsive genes’, ‘lithium genes and bipolar disorder’, ‘genes and lithium response’, ‘lithium susceptibility genes’, ‘lithium mechanism of action’, and ‘lithium treatment and genes’. In addition, the reference lists of the studies and related publications were searched manually to identify potential publications that were not covered by the electronic search.
After retrieving the results of the initial search (409 records) and removing duplicates, we screened the title and abstract of 151 publications (145 original and 6 review articles) to ensure that they had enough evidence of significant association between their candidate genes and lithium responsiveness. Through reading the full text of 144 published articles (138 original and 6 review articles), 142 articles (136 original and 6 review article) were finally selected for preparation of our gene set. The inclusion criteria for selection of the candidate genes were replicated genetic polymorphisms in different populations of BD patients, and altered gene expression and functional analysis of the genes in cellular or animal models. Here, we named the collected gene set as ‘lithium responsiveness-associated genes’ (LRAGs). The details of the stages of literature searching and study selection are presented in a flowchart in Fig. 1, based on the PRISMA 2009 statement [24].
2.2 Gene Set Enrichment Analysis for Chromosomal Regions
Many human traits might appear due to alterations in particular regions of the genome [25,26,27]. Therefore, it is of great interest to identify the chromosomal regions linked to the phenotype of lithium responsiveness in BD. To this purpose, gene set enrichment analysis for chromosomal regions was performed by the web-based tools Gene Set Enrichment Analysis (GSEA)–Broad Institute [28] and Enrichr [29, 30], using official symbols of the LRAGs as input data (Supplemental file S1, see electronic supplementary material [ESM]). Among all predicted results, the chromosomal regions with p value < 0.05 were considered as significantly associated loci with lithium responsiveness.
2.3 Expression Pattern Analysis of the Genes
To determine which parts of the human body express the highest number of LRAGs, an enrichment analysis for gene expression was performed by Enrichr, based on text mining information of the genes obtained from the ‘Jensen TISSUES’ database. The Jensen TISSUES is a database of gene–tissue associations in human, mouse, rat, and pig that integrates evidence on tissue expression from manually curated literature, proteomics and transcriptomics screens, and automatic text mining [31]. In addition, microarray data of the GSEA–Broad Institute [28] were used for clustering the genes with similar expression patterns across different cell lineages of the human body. To identify the human brain regions that express the LRAGs at the highest level, enrichment analysis was performed in Enrichr using the data from the ‘Allen Brain Atlas’. The Allen Brain Atlas is an online public resource integrating extensive gene expression data, connectivity data, and high resolution neuroanatomical information with powerful search and viewing tools for the adult and developing brain in mice, humans and non-human primates [32, 33].
2.4 Gene Ontology Analysis of the Genes
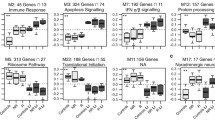
In order to provide an insight into the functional role of the set of LRAGs, Gene Ontology (GO) analysis of both biological processes and signaling pathways was performed by the Database for Annotation, Visualization and Integrated Discovery (DAVID) software [34, 35]. In DAVID results, GO terms with adjusted p value < 0.05 and gene count ≥ 5 were considered as statistically significant enriched biological processes and signaling pathways.
2.5 Protein Class Identification for the Genes
The PANTHER (Protein ANalysis THrough Evolutionary Relationships) web server [36, 37] was used for enrichment analysis of the LRAGs based on their protein classes. Protein class terms were taken from the PANTHER Protein Class ontology and represent common grouping terms for families or subfamilies of proteins with common functions. Protein class terms may overlap to some degree with GO molecular function terms, but in many cases are distinct [36].
2.6 Construction of Molecular Interaction Network
The Search Tool for the Retrieval of Interacting Genes (STRING) is a useful tool, capable of providing a comprehensive view of the known and predicted interactions and associations between a set of proteins [38]. The STRING online software was used to construct a protein–protein interaction network using the LRAGs of the four significantly enriched signaling pathways (i.e., circadian rhythm, dopaminergic synapse, cholinergic synapse, and morphine addiction) as the input data (Supplemental file S2, see ESM). Within the constructed network, the proteins served as 'nodes' and the links connecting two nodes represent a pairwise protein interaction. The degree of each node corresponds to the number of interactions that the protein is in possession of.
2.7 Prediction of Regulatory microRNAs for Molecular Interaction Network
In order to identify the microRNAs (miRNAs) capable of regulating the molecular interaction network, GSEA analysis for ‘miRNA targets’ was performed by the Broad Institute online tool (https://software.broadinstitute.org/gsea/msigdb/annotate.jsp). This online tool utilizes an algorithm for finding the genes in the network that share the target sites of human miRNAs in their 3′-UTR (https://www.gsea-msigdb.org/gsea/msigdb/collection_details.jsp). The miRNAs with at least two target genes in the network were considered as potential upstream regulators of the network.
3 Results
3.1 Identification of Lithium Responsiveness-Associated Genes (LRAGs)
The search of the literature identified a total of 211 genes that have been found to be associated with lithium response in BD. The list of the genes with their full names is presented in supplemental file S1 (see ESM).
3.2 Chromosomal Regions Linked to Lithium Responsiveness in Bipolar Disorder (BD)
Analysis by the GSEA online tool predicted three chromosomal regions (chr5q34, chr7p13 and chr3p26) as the most associated loci for lithium responsiveness in BD (Fig. 2a). Another similar analysis was performed by the Enrichr software and the same results were obtained (Fig. 2b). However, Enrichr predicted an additional chromosomal region (chr4q21) related to lithium responsiveness, which was not significantly enriched in the GSEA results. The enrichment results of other chromosomal regions were not statistically significant (p ≥ 0.05) (Fig. 2b).
Chromosomal regions related to the LRAGs. a The enrichment results of the GSEA tool (Broad Institute) highlighted three relevant chromosome loci for the LRAGs. b In addition to prediction of chr4q21 as another lithium-associated locus, Enrichr results were consistent with the results of the GSEA. p < 0.05 was considered as statistically significant. The statistically significant results (p < 0.05) are denoted by green shading. GSEA Gene Set Enrichment Analysis, LRAGs lithium responsiveness-associated genes
The LRAGs located on these loci are GABRB2 (gamma-aminobutyric acid type A receptor beta2 subunit), GABRA1 (gamma-aminobutyric acid type A receptor alpha1 subunit), DUSP1 (dual specificity phosphatase 1), and DRD1 (dopamine receptor D1) on the chromosomal region 5q34; ADCY1 (adenylate cyclase 1), IGFBP1 (insulin like growth factor binding protein 1), IGFBP3 (insulin like growth factor binding protein 3), and SEPT7P2 (septin 7 pseudogene 2) on the chromosomal region 7p13; CNTN4 (contactin 4), IL5RA (interleukin 5 receptor subunit alpha), and BHLHE40 (basic helix-loop-helix family member E40) on the chromosomal region 3p26. The enriched LRAGs for the chromosomal region 4q21 were CDKL2 (cyclin dependent kinase like 2), CCNG2 (cyclin G2), G3BP2 (G3BP stress granule assembly factor 2), and C4ORF26 (chromosome 4 open reading frame 26) (data not shown). Taken together, these data suggest the chromosomal loci 5q34, 7p13, 3p26, and 4q21 as the most relevant genomic regions to the phenotype of lithium responsiveness in BD.
3.3 The Link of Human Body Parts to Lithium Responsiveness in BD
Expression pattern analysis of the LRAGs using Enrichr and GSEA tools showed that adult tissues (the tissues that specifically correspond to the adult life stage), ganglia, and the immune system are the most related parts of the human body to lithium responsiveness, by expressing the highest number of LRAGs (Fig. 3a). These data also showed that neural and immune systems are the most related organs to the LRAGs. Additional to Enrichr, investigations for finding the expression pattern of LRAGs across the human body were performed by GSEA (Broad Institute). Results showed that LRAGs are mostly expressed in the brain (~ 31.23%) and blood/immune system (~ 15.28%) (Fig. 3b). The results also showed an expression pattern in which the brain-enriched genes are downregulated in blood and immune cells, while the blood- and immune system-enriched genes exhibit their lowest expression levels in the brain cells (Fig. 3b). These data suggest that the brain is the main part of the human body associated with response to lithium therapy in BD.
Expression pattern of the LRAGs. a Text mining (by Enrichr, Jensen TISSUES) was applied to identify an association between the LRAGs, human tissues, and developmental time. b A total of 211 LRAGs were analyzed (by GSEA, Broad Institute) based on their expression pattern across different cell lineages of human tissues. Data showed the enrichment of most LRAGs in brain, blood, and immune system cells. Red and blue colors in the heatmap denote high and low expression levels, respectively. (c) The human brain regions wherein the LRAGs are mostly enriched, profiled by the Allen Brain Atlas (in Enrichr). Brain regions are denoted as distinct numbers that correspond to the significance order based on their p value of analysis (bar plots). GSEA Gene Set Enrichment Analysis, LRAGs lithium responsiveness-associated genes
Given the highest enrichment score for the brain, in the next step, the expression pattern of the LRAGs across different regions of the human brain was analyzed. Results showed that LRAGs are mostly enriched in the regions located in the central part of human brain; these brain regions were (1) external globus pallidum, (2) subpallidum, (3) paracentral nucleus, (4) putamen, (5) caudate nucleus, (6) striatum dorsal region, (7) dorsal region of pallidum, (8) interanterodorsal nucleus, (9) intralaminar nuclei, and (10) interanteromedial nucleus of the thalamus (Fig. 3c). These data suggest a pivotal role for the central regions of the brain in the therapeutic effects of lithium in BD.
3.4 Cellular Components Related to Lithium Responsiveness in BD
Results of the GO analysis for cellular components showed that ~ 75% of the LRAGs (Adj. p < 0.05) encode for the proteins that are physically located in plasma membrane or cytosol of the cells (Table 1). These data suggest that a significant number of LRAGs could drive intracellular signaling pathways by their encoded proteins.
3.5 Biological Processes Related to the LRAGs
Since most of the LRAGs are membrane-bound or cytosolic proteins (Table 1), they may participate in some intracellular molecular networks. GO analysis of biological processes for these genes showed their significant enrichment (Adj. p < 0.01) in ‘circadian rhythm’, ‘circadian regulation of gene expression’, ‘regulation of circadian rhythm’, ‘regulation of cell cycle’, ‘signal transduction’, ‘intrinsic apoptotic signaling pathway in response to endoplasmic reticulum stress’, and ‘negative regulation of glucocorticoid receptor signaling pathway’ (Table 2). Most LRAGs were enriched for circadian rhythm-related processes (rows 1–3; 18.5% of the analyzed genes) and signal transduction (15.2% of the analyzed genes). Further analysis of the genes of the ‘signal transduction’ process showed the specific enrichment of IMPA1 (inositol monophosphatase 1), IMPA2 (inositol monophosphatase 2), INPP1 (inositol polyphosphate-1-phosphatase), and PLCG1 (phospholipase C gamma 1) genes in PI signaling pathway (Table 2; bold-faced genes) and enrichment of GNB1 (G protein subunit beta 1), PDE10A (phosphodiesterase 10A), PDE11A (phosphodiesterase 11A), PDE4D (phosphodiesterase 4D), and PDE8A (phosphodiesterase 8A) genes in morphine addiction pathway (Table 2; underlined bold-faced genes). These data suggest the prominent role of the PI signaling system and morphine addiction in lithium responsiveness in BD.
3.6 Signaling Pathways Related to the LRAGs
Results of the GO analysis of signaling pathways showed that LRAGs are mostly involved in ‘circadian rhythm’, ‘dopaminergic synapse’, ‘cholinergic synapse’, and ‘morphine addiction’ (Adj. p < 0.01) (Table 3). As shown in Table 3, the highest enrichment score (lowest adj. p-value) was observed for the pathway of circadian rhythm, which is consistent with the data of enrichment analysis for biological processes (Table 2).
To determine the site of action of the LRAGs in signaling pathways, the schematic representation of each pathway was obtained from the Kyoto Encyclopedia of Genes and Genomes (KEGG) database and the LRAGs of the pathways were marked by red asterisks (Supplemental figs. S1, S2 and S3, see ESM).
In addition, gene set enrichment analysis for protein classes revealed that most of the LRAGs in these pathways (Table 3; rows 1–4) are transcription factors (7 out of 32 genes; ~ 21.9%) and kinases (4 out of 32 genes; 12.5%) (Fig. 4). These data suggest that the transcription factors and kinases of the signaling pathways are the main targets of lithium in treatment of BD.
Categorization of the LRAGs based on their protein classes by PANTHER tool. a Analysis of the LRAGs of circadian rhythm, dopaminergic synapse, cholinergic synapse, and morphine addiction showed that the greatest number of them are classified in the transcription factor (21.9%) and protein kinase (12.5%) classes. The enriched genes of these two classes are represented by their official names. b All the enriched protein classes with the number of enriched genes for each class are presented. As shown, most genes are classified as transcription factor (TF) and kinase, marked by asterisks. LRAGs lithium responsiveness-associated genes, PANTHER Protein ANalysis THrough Evolutionary Relationships
3.7 MicroRNA-Mediated Regulation of the Interaction Network of LRAGs
Given the involvement of four signaling pathways in lithium responsiveness in BD, possible gene–gene interactions between these pathways were examined. Results revealed that the four signaling pathways (circadian rhythm, dopaminergic synapse, cholinergic synapse, and morphine addiction) are highly connected to each other through sharing their proteins. In fact, we found that ARNTL (aryl hydrocarbon receptor nuclear translocator like), CREB1 (cAMP responsive element binding protein 1), and GSK3β proteins link the circadian rhythm pathway to the pathways of dopaminergic and cholinergic synapses (Fig. 5a, b). The link between the signaling pathways of dopaminergic/cholinergic synapses and morphine addiction is mediated by GNB1, DRD1, and ADCY1 proteins (Fig. 5a, c).
Molecular interaction network to illustrate the key components of the signaling pathways involved in lithium response in BD. STRING predicted the protein–protein interaction network, wherein the four predicted lithium-related signaling pathways are tightly connected to each other. a Dopaminergic and cholinergic synapse pathways, b circadian rhythm, c morphine addiction. Also, a set of interacting miRNAs were predicted by GSEA online software, to regulate the network. Seed sequences of the miRNAs and their targets are shown by nucleotide sequences and solid lines, respectively. BP bipolar disorder, GSEA Gene Set Enrichment Analysis, miRNAs microRNAs, STRING Search Tool for the Retrieval of Interacting Genes
Further investigations in order to find the regulatory miRNAs for this molecular interaction network showed that a set of miRNAs could regulate the network via targeting at least two genes of the network (Fig. 5). The predicted regulatory miRNAs were miR-30a, miR-30b, miR-30c, miR-30d, miR-493, miR-448, miR-302c, miR-505, miR-494, miR-181a, miR-181b, miR-181c, miR-181d, miR-101, miR-203, and miR-502. The families of miR-30 and miR-181 have their specific seed sequences, shared between their members. The seed sequence of the miR-30 family is ‘TGTTTAC’ and that of the miR-181 family is ‘TGAATGT’ (Fig. 5).
4 Discussion
The main indication for lithium is long-term administration for the prevention of manic and depressive recurrences in BD. However, long-term monotherapy with lithium totally prevents the affective episodes in only one third of BD patients [4]. In order to improve the treatment of patients with BD, identification of the factors associated with response and lack of response to lithium is of great importance. In this study, we aimed to define an integrated mechanism related to lithium responsiveness in BD, using a list of previously reported LRAGs. Since the associations of these genes were previously validated by several experimental approaches such as GWAS, gene expression analysis, and functional annotations, they could be a reliable input for downstream analyses. In the first analysis we showed that 5q34, 7p13, 3p26, and 4q21 are the genomic loci most associated with lithium responsiveness. Despite reporting many chromosomal loci associated with the etiology of BD, reports on the chromosomal loci associated with lithium responsiveness in BD are rare. However, the most replicable evidence relates to 21q21.1 [2]. Of the predicted loci in our study, 5q31-34 [39, 40], 7p13 [41], and 4p21 [42] were previously linked to the etiology of BD without any reported association with lithium responsiveness. While the 3p26 locus has been shown to be associated with major depression [43] and schizophrenia [44], it appeared in only one GWAS of BD [45]. However, its neighboring locus (3p24-23) was reported to be associated with lithium responsiveness, harboring two reproducible variants within the glutamate decarboxylase like 1 (GADL1) gene [46].
Expression pattern analysis of the LRAGs showed their expression enrichment in adulthood stage, and within the brain, blood, and immune system. Analyzing the LRAGs based on their expression pattern across different human cell lineages delineated two distinct clusters: first, the ‘brain-enriched genes’ with highest expression in brain and lowest expression in blood and immune system; second, the ‘brain-depleted genes’ with lack or low level of expression in brain cells but enrichment in blood and immune system cells (Fig. 3a, b). These data suggest that most of the LRAGs are expressed in the brain, blood, and immune system. This may also suggest that lithium accumulates in the brain, blood, and immune system to exert its therapeutic effect.
In order to determine which parts of the human brain express the LRAGs, a gene set enrichment analysis was performed and the results emphasized the central regions, suggesting the crucial role of these regions in lithium response of bipolar patients (Fig. 3c). Other studies also showed that lithium accumulates in the neurogenic parts of the brain (especially in the hippocampus, subventricular zone, and striatum) to induce cell proliferation [47]. In fact, at its therapeutic doses (0.6–1.2 mM), lithium appears to accumulate in the brain structures containing a higher proportion of cell bodies in general and in neurogenic regions in particular [48, 49]. However, there is no such evidence for other parts of the brain (such as the brain cortex) [50]. Apart from the nervous system, lithium has been found to affect immune cells [51]. Since there is no empirical evidence for use of lithium in immune system disorders [51], this effect could be considered as a new mechanism for lithium response in BD.
We also found that most of the LRAGs encode for proteins that are localized in the plasma membrane or cytoplasm. With such proximities in subcellular localization, we hypothesized that these genes could participate in some intracellular signaling processes. Interestingly, our analysis showed that LRAGs are mainly involved in circadian rhythm, cell cycle regulation, drug response, and apoptotic processes. Consistent with our results, circadian rhythm dysfunction has been widely demonstrated as a trait marker for BD [52, 53]. Therefore, the effectiveness of lithium therapy in BD could be in part attributed to its modulatory role on circadian rhythm. Enrichment of cell cycle and apoptosis for the analyzed LRAGs highlights the effects of lithium on these processes. Previous studies also showed that lithium can either stimulate or inhibit cell proliferation [54]. Through inducing cell cycle [55, 56] and inhibiting apoptosis [57, 58], lithium can stimulate proliferation of hippocampal stem/progenitor cells to inhibit the apoptosis of neurons and peripheral blood cells, which often occurs in BD [59, 60]. However, other cell types could be subjected to the anti-proliferative effect of lithium; hence, normal development will be disrupted (especially at prenatal stages) [55].
Consistent with other studies, the components of the PI signaling pathway were enriched in our study. In fact, of the six reported components of this pathway, four genes (IMPA1, IMPA2, PLCG1, and INPP1) were enriched as the main players of the signal transduction process (Table 2). This pathway was previously known as the most important intracellular signaling system related to lithium responsiveness in BD [61, 62]. Another enriched biological process for the LRAGs was ‘response to drug’, suggesting that lithium affects this process, which is shared between most therapeutic reagents. This data may also explain the synergistic or antagonistic effects of drugs when combination therapy is used for treatment of BD [63]. Another enriched biological process for the LRAGs was ‘Negative regulation of glucocorticoid receptor signaling pathway’, previously linked to both pathogenesis and lithium response of BD [64,65,66]. This pathway is a common target for many mood stabilizers [67]. Of note, the enrichment of a particular biological process does not necessarily imply that its activation (rather than suppression) is in favor with lithium responsiveness. For example, there is evidence that shows the induction of the glucocorticoid signaling pathway could interfere with the proliferative effect of lithium on hippocampal neurons [68].
Regarding the signaling pathways of lithium responsiveness, the crucial role of dopaminergic and cholinergic systems are evident in the etiology of BD [69]. In fact, hyperactivation of dopaminergic and cholinergic synapses, respectively, underlie the mania and depression states in BD [70, 71]. Pharmacological studies showed that an efficient mood stabilizer for BD is one that could suppress the activity of both dopaminergic [72] and cholinergic [73] synapses, thereby, both manic and depressive episodes could be controlled. With this capability, lithium is suggested as a potent drug for prevention of manic and depressive episodes in BD.
Another signaling pathway related to lithium responsiveness was morphine addiction that showed a crosstalk with the dopamine pathway (Fig. 5). Studies have shown that the morphine pathway activates dopaminergic synapses, which is not useful in the treatment of BD [71]. By counteracting the negative effect of the morphine pathway [74], lithium could be an efficient treatment for BD.
When we analyzed the LRAGs of the enriched signaling pathways, we found that a significant number of the LRAGs in these pathways are transcription factors (21.9%) and kinases (12.5%). Among these LRAGs, the strongest experimental supports were reported for NR1D1 (nuclear receptor subfamily 1 group D member 1) and GSK-3β [75]. Overall, the four predicted signaling pathways harbor several LRAGs at their critical points (i.e., receptors, kinases, and transcription factors), enabling lithium to effectively modulate the pathways to treat BD. Therefore, patients with different expression of these LRAGs may respond to lithium variably.
While we predicted four distinct significant pathways related to lithium responsiveness, we hypothesize that coordinated activity of these pathways may mediate lithium therapeutic effects. Therefore, we sought to find possible links between the pathways. We found that these pathways can form an interaction network via sharing several proteins. Moreover, we sought to find miRNAs capable of targeting several genes of the network. Such miRNAs not only could regulate the whole network as master regulators, but also they have the criteria to be a predictive value for discriminating lithium responders from non-responders [76]. Of the predicted miRNAs, miR-181b [77], miR-181c [77], miR-494 [78, 79], miR-30a [80], miR-30c [80], and miR-203a [79] were previously linked to lithium responsiveness in BD. miR-493, miR-302c, miR-505, and miR-101 were not previously linked to lithium responsiveness, but there is evidence for their association with the etiology of BD [81].
In addition, there is no evidence for the association of miR-448 and miR-502, either in the etiology or in lithium responsiveness of BD, and this is the first time they have been suggested to be lithium-responsive miRNAs. miR-502 is known to be highly expressed in lymphoid organs; therefore, it could be a good traceable molecular marker for prediction of lithium responsiveness in BD [82]. Nevertheless, a combined set of miRNAs is more beneficial than a single miRNA for clinical applications [83]. Studies showed that most of the known lithium-responsive miRNAs are enriched in the hippocampus [84], suggesting the hippocampal neural cell cultures as a good strategy to investigate the mechanism of action of lithium in vitro. These findings are also consistent with our data of brain regions linked to lithium responsiveness (Fig. 3c).
Our study is limited by the lack of information about the newly discovered genes whose genomic loci were associated with lithium responsiveness (e.g., AL157359.3 and AL157359.4 [2]). Since the function and interaction properties of such genes are not known yet, they could not be detected and enriched in a particular biological process or signaling pathway by the available databases. Through functional characterization and identification of their interacting molecules (e.g., RNAs and proteins), they could be included in futures studies to complement our findings. Furthermore, the findings of this study need to be further investigated through experimental validations in order to translate them into clinical practice.
5 Conclusion
In this study, we analyzed well documented LRAGs to provide an integrated mechanism for lithium therapeutic effects in BD. Understanding these mechanisms could translate into tests to discover which patients are most likely to respond to lithium, which could enable tailored treatment [85]. Moreover, gaining insight into the mechanism of action of lithium will be crucial to develop new safer treatments for BD [86].
References
Sher L. Manic-depressive illness: bipolar disorders and recurrent depression. Am J Psychiatry. 2008;165(4):541–2.
Hou L, Heilbronner U, Degenhardt F, Adli M, Akiyama K, Akula N, et al. Genetic variants associated with response to lithium treatment in bipolar disorder: a genome-wide association study. Lancet. 2016;387(10023):1085–93.
Alda M. Lithium in the treatment of bipolar disorder: pharmacology and pharmacogenetics. Mol Psychiatry. 2015;20(6):661.
Rybakowski JK, Chlopocka-Wozniak M, Suwalska A. The prophylactic effect of long-term lithium administration in bipolar patients entering treatment in the 1970s and 1980s. Bipolar Disord. 1980s;3(2):63–7.
Tighe SK, Mahon PB, Potash JB. Predictors of lithium response in bipolar disorder. Ther Adv Chronic Dis. 2011;2(3):209–26.
Wei Shan G, Makmor-Bakry M, Salihah OM. Long term use of lithium and factors associated with treatment response among patients with bipolar disorder. Psychiatria Danubina. 2016;28(2):146–53.
Kapur V, Nadella RK, Raghuraman BS, Saraf G, Mishra S, Srinivasmurthy N, et al. Clinical factors associated with lithium treatment response in bipolar disorder patients from India. Asian J Psychiatry. 2018;39(38):165–8.
Sportiche S, Geoffroy PA, Brichant-Petitjean C, Gard S, Khan J-P, Azorin J-M, et al. Clinical factors associated with lithium response in bipolar disorders. Aust N Z J Psychiatry. 2017;51(5):524–30.
Aronoff MS, Epstein RS. Factors associated with poor response to lithium carbonate: a clinical study. Am J Psychiatry. 1970;127(4):472–80.
Eugene AR, Masiak J, Eugene B. Predicting lithium treatment response in bipolar patients using gender-specific gene expression biomarkers and machine learning. F1000Research. 2018;7:474–99.
Mertens J, Wang Q-W, Kim Y, Diana XY, Pham S, Yang B, et al. Differential responses to lithium in hyperexcitable neurons from patients with bipolar disorder. Nature. 2015;527(7576):95.
Stern S, Santos R, Marchetto M, Mendes A, Rouleau G, Biesmans S, et al. Neurons derived from patients with bipolar disorder divide into intrinsically different sub-populations of neurons, predicting the patients’ responsiveness to lithium. Mol Psychiatry. 2018;23(6):1453.
Cruceanu C, Alda M, Turecki G. Lithium: a key to the genetics of bipolar disorder. Genome Med. 2009;1(8):79.
Manchia M, Adli M, Akula N, Ardau R, Aubry J-M, Backlund L, et al. Assessment of response to lithium maintenance treatment in bipolar disorder: a Consortium on Lithium Genetics (ConLiGen) report. PLoS One. 2013;8(6):e65636.
Schulze TG, Alda M, Adli M, Akula N, Ardau R, Bui ET, et al. The International Consortium on Lithium Genetics (ConLiGen): an initiative by the NIMH and IGSLI to study the genetic basis of response to lithium treatment. Neuropsychobiology. 2010;62(1):72–8.
Berridge MJ. Inositol 1, 4, 5-trisphosphate-induced calcium mobilization is localized in Xenopus oocytes. Proc R Soc Lond B Biol Sci. 1989;238(1292):235–43.
Berridge MJ, Downes CP, Hanley MR. Neural and developmental actions of lithium: a unifying hypothesis. Cell. 1989;59(3):411–9.
Klein PS, Melton DA. A molecular mechanism for the effect of lithium on development. Proc Natl Acad Sci. 1996;93(16):8455–9.
Benedetti F, Bernasconi A, Lorenzi C, Pontiggia A, Serretti A, Colombo C, et al. A single nucleotide polymorphism in glycogen synthase kinase 3-β promoter gene influences onset of illness in patients affected by bipolar disorder. Neurosci Lett. 2004;355(1–2):37–40.
Benedetti F, Serretti A, Pontiggia A, Bernasconi A, Lorenzi C, Colombo C, et al. Long-term response to lithium salts in bipolar illness is influenced by the glycogen synthase kinase 3-β-50 T/C SNP. Neurosci Lett. 2005;376(1):51–5.
Song J, Bergen S, Di Florio A, Karlsson R, Charney A, Ruderfer D, et al. Genome-wide association study identifies SESTD1 as a novel risk gene for lithium-responsive bipolar disorder. Mol Psychiatry. 2016;21(9):1290.
McCarthy MJ, Leckband SG, Kelsoe JR. Pharmacogenetics of lithium response in bipolar disorder. Pharmacogenomics. 2010;11(10):1439–65.
Papiol S, Schulze TG, Alda M. Genetics of lithium response in bipolar disorder. Pharmacopsychiatry. 2018;51(5):206–11.
Moher D, Liberati A, Tetzlaff J, Altman DG. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. Ann Intern Med. 2009;151(4):264–9.
Buyske S, Bates ME, Gharani N, Matise TC, Tischfield JA, Manowitz P. Cognitive traits link to human chromosomal regions. Behav Genet. 2006;36(1):65.
Franchini LF, Pollard KS. Genomic approaches to studying human-specific developmental traits. Development. 2015;142(18):3100–12.
Li H, Chen H, Liu F, Ren C, Wang S, Bo X, et al. Functional annotation of HOT regions in the human genome: implications for human disease and cancer. Sci Rep. 2015;5:11633.
Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci. 2005;102(43):15545–50.
Chen EY, Tan CM, Kou Y, Duan Q, Wang Z, Meirelles GV, et al. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinform. 2013;14(1):128.
Kuleshov MV, Jones MR, Rouillard AD, Fernandez NF, Duan Q, Wang Z, et al. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016;44(W1):W90–W9797.
Palasca O, Santos A, Stolte C, Gorodkin J, Jensen LJ. TISSUES 2.0: an integrative web resource on mammalian tissue expression. Database. 2018;2018:1–12.
Sunkin SM, Ng L, Lau C, Dolbeare T, Gilbert TL, Thompson CL, et al. Allen Brain Atlas: an integrated spatio-temporal portal for exploring the central nervous system. Nucleic Acids Res. 2012;41(D1):D996–D1008.
Jones AR, Overly CC, Sunkin SM. The Allen brain atlas: 5 years and beyond. Nat Rev Neurosci. 2009;10(11):821.
Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2008;4(1):44.
Huang DW, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2008;37(1):1–13.
Mi H, Muruganujan A, Thomas PD. PANTHER in 2013: modeling the evolution of gene function, and other gene attributes, in the context of phylogenetic trees. Nucleic Acids Res. 2012;41(D1):D377–D386386.
Thomas PD, Campbell MJ, Kejariwal A, Mi H, Karlak B, Daverman R, et al. PANTHER: a library of protein families and subfamilies indexed by function. Genome Res. 2003;13(9):2129–41.
Szklarczyk D, Morris JH, Cook H, Kuhn M, Wyder S, Simonovic M, et al. The STRING database in 2017: quality-controlled protein–protein association networks, made broadly accessible. Nucleic Acids Res. 2016;45(1):362–8.
Kerner B, Brugman DL, Freimer NB. Evidence of linkage to psychosis on chromosome 5q33-34 in pedigrees ascertained for bipolar disorder. Am J Med Genet Part B Neuropsychiatr Genet. 2007;144(1):74–8.
Herzberg I, Jasinska A, García J, Jawaheer D, Service S, Kremeyer B, et al. Convergent linkage evidence from two Latin-American population isolates supports the presence of a susceptibility locus for bipolar disorder in 5q31–34. Hum Mol Genet. 2006;15(21):3146–53.
Hamshere ML, Schulze TG, Schumacher J, Corvin A, Owen MJ, Jamra RA, et al. Mood-incongruent psychosis in bipolar disorder: conditional linkage analysis shows genome-wide suggestive linkage at 1q3.23, 7p13 and 20q13.31. Bipolar Disord. 2009;11(6):610–20.
Liu J, Juo S, Dewan A, Grunn A, Tong X, Brito M, et al. Evidence for a putative bipolar disorder locus on 2p13–16 and other potential loci on 4q31, 7q34, 8q13, 9q31, 10q21–24, 13q32, 14q21 and 17q11–12. Mol Psychiatry. 2003;8(3):333.
Pergadia ML, Glowinski AL, Wray NR, Agrawal A, Saccone SF, Loukola A, et al. A 3p26-3p25 genetic linkage finding for DSM-IV major depression in heavy smoking families. Am J Psychiatry. 2011;168(8):848–52.
Lewis CM, Levinson DF, Wise LH, DeLisi LE, Straub RE, Hovatta I, et al. Genome scan meta-analysis of schizophrenia and bipolar disorder, part II: Schizophrenia. Am J Hum Genet. 2003;73(1):34–48.
Kerner B, Lambert CG, Muthen BO. Genome-wide association study in bipolar patients stratified by co-morbidity. PLoS One. 2011;6(12):e28477.
Chen C-H, Lee C-S, Lee M-TM, Ouyang W-C, Chen C-C, Chong M-Y, et al. Variant GADL1 and response to lithium therapy in bipolar I disorder. N Engl J Med. 2014;370(2):119–28.
Zanni G, Michno W, Di Martino E, Tjärnlund-Wolf A, Pettersson J, Mason CE, et al. Lithium accumulates in neurogenic brain regions as revealed by high resolution ion imaging. Sci Rep. 2017;7:40726.
Thellier M, Wissocq J, Heurteaux C. Quantitative microlocation of lithium in the brain by a (n, α) nuclear reaction. Nature. 1980;283(5744):299.
Thellier M, Heurteaux C, Wissocq J-C. Quantitative study of the distribution of lithium in the mouse brain for various doses of lithium given to the animal. Brain Res. 1980;199(1):175–96.
Smith FE, Thelwall PE, Necus J, Flowers CJ, Blamire AM, Cousins DA. 3D 7 Li magnetic resonance imaging of brain lithium distribution in bipolar disorder. Mol Psychiatry. 2018;23(11):2184.
Maddu N, Raghavendra PB. Review of lithium effects on immune cells. Immunopharmacol Immunotoxicol. 2015;37(2):111–25.
Takaesu Y. Circadian rhythm in bipolar disorder: a review of the literature. Psychiatry Clin Neurosci. 2018;72(9):673–82.
Melo MC, Abreu RL, Neto VBL, de Bruin PF, de Bruin VM. Chronotype and circadian rhythm in bipolar disorder: a systematic review. Sleep Med Rev. 2017;34:46–58.
Gallicchio VS. Effects of lithium on cell growth. Lithium and cell physiology. Berlin: Springer; 1990. p. 121–124.
Smits VA, Essers MA, Loomans DS, Klompmaker R, Rijksen G, Medema RH. Inhibition of cell proliferation by lithium is associated with interference in cdc2 activation. FEBS Lett. 1999;457(1):23–7.
de Groot T, Alsady M, Jaklofsky M, Otte-Höller I, Baumgarten R, Giles RH, et al. Lithium causes G2 arrest of renal principal cells. J Am Soc Nephrol. 2014;25(3):501–10.
Zhang W, Jüllig M, Connolly A, Stott N. Early gene response in lithium chloride induced apoptosis. Apoptosis. 2005;10(1):75–90.
Pietruczuk K, Jóźwik A, Ruckemann-Dziurdzińska K, Bryl E, Witkowski J. Cytoprotective effect of lithium against spontaneous and induced apoptosis of lymphoid cell line MOLT-4. Folia Histochem Cytobiol. 2009;47(4):639–46.
Uribe E, Wix R. Neuronal migration, apoptosis and bipolar disorder. Revista de Psiquiatría y Salud Mental (English Edition). 2012;5(2):127–33.
Fries GR, Vasconcelos-Moreno MP, Gubert C, Dos Santos BTMQ, Da Rosa ALST, Eisele B, et al. Early apoptosis in peripheral blood mononuclear cells from patients with bipolar disorder. J Affect Disord. 2014;152:474–7.
Coyle JT, Duman RS. Finding the intracellular signaling pathways affected by mood disorder treatments. Neuron. 2003;38(2):157–60.
Brown KM, Tracy DK. Lithium: the pharmacodynamic actions of the amazing ion. Ther Adv Psychopharmacol. 2013;3(3):163–76.
Feng H-L, Leng Y, Ma C-H, Zhang J, Ren M, Chuang D-M. Combined lithium and valproate treatment delays disease onset, reduces neurological deficits and prolongs survival in an amyotrophic lateral sclerosis mouse model. Neuroscience. 2008;155(3):567–72.
Spiliotaki M, Salpeas V, Malitas P, Alevizos V, Moutsatsou P. Altered glucocorticoid receptor signaling cascade in lymphocytes of bipolar disorder patients. Psychoneuroendocrinology. 2006;31(6):748–60.
Watson S, Thompson JM, Ritchie JC, Nicol Ferrier I, Young AH. Neuropsychological impairment in bipolar disorder: the relationship with glucocorticoid receptor function. Bipolar Disord. 2006;8(1):85–90.
Spijker A, Van Rossum E. Glucocorticoid sensitivity in mood disorders. Neuroendocrinology. 2012;95(3):179–86.
Zhou R, Gray NA, Yuan P, Li X, Chen J, Chen G, et al. The anti-apoptotic, glucocorticoid receptor cochaperone protein BAG-1 is a long-term target for the actions of mood stabilizers. J Neurosci. 2005;25(18):4493–502.
Miyazaki I, Nagamachi T, Shinomiya K, Matsunaga H, Sendo T, Kawasaki H, et al. Effects of imipramine and lithium on the suppression of cell proliferation in the dentate gyrus of the hippocampus in adrenocorticotropic hormone-treated rats. Acta Med Okayama. 2010;64(4):219–23.
Sigitova E, Fišar Z, Hroudová J, Cikánková T, Raboch J. Biological hypotheses and biomarkers of bipolar disorder. Psychiatry Clin Neurosci. 2017;71(2):77–103.
Hannestad JO, Cosgrove KP, DellaGioia NF, Perkins E, Bois F, Bhagwagar Z, et al. Changes in the cholinergic system between bipolar depression and euthymia as measured with [123I] 5IA single photon emission computed tomography. Biol Psychiatry. 2013;74(10):768–76.
Ashok AH, Marques TR, Jauhar S, Nour MM, Goodwin G, Young AH, et al. The dopamine hypothesis of bipolar affective disorder: the state of the art and implications for treatment. Mol Psychiatry. 2017;22(5):666.
Can A, Frost DO, Cachope R, Cheer JF, Gould TD. Chronic lithium treatment rectifies maladaptive dopamine release in the nucleus accumbens. J Neurochem. 2016;139(4):576–85.
Vizi E, Illes P, Ronai A, Knoll J. The effect of lithium on acetylcholine release and synthesis. Neuropharmacology. 1972;11(4):521–30.
Dehpour A, Farsam H, Azizabadi-Farahani M. Inhibition of the morphine withdrawal syndrome and the development of physical dependence by lithium in mice. Neuropharmacology. 1995;34(1):115–21.
Budde M, Degner D, Brockmöller J, Schulze T. Pharmacogenomic aspects of bipolar disorder: an update. Eur Neuropsychopharmacol. 2017;27(6):599–609.
Liu S, Zhang F, Wang X, Shugart YY, Zhao Y, Li X, et al. Diagnostic value of blood-derived microRNAs for schizophrenia: results of a meta-analysis and validation. Sci Rep. 2017;7(1):15328.
Reinbold CS, Forstner AJ, Hecker J, Fullerton JM, Hoffmann P, Hou L, et al. Analysis of the influence of microRNAs in lithium response in bipolar disorder. Front Psychiatry. 2018;9:207.
Chen H, Wang N, Burmeister M, McInnis MG. MicroRNA expression changes in lymphoblastoid cell lines in response to lithium treatment. Int J Neuropsychopharmacol. 2009;12(7):975–81.
Hunsberger J, Chibane F, Elkahloun A, Henderson R, Singh R, Lawson J, et al. Novel integrative genomic tool for interrogating lithium response in bipolar disorder. Transl Psychiatry. 2015;5(2):e504.
Chao Y-L, Chen C-H. An introduction to microRNAs and their dysregulation in psychiatric disorders. Tzu Chi Medical Journal. 2013;25(1):1–7.
Kim AH, Reimers M, Maher B, Williamson V, McMichael O, McClay JL, et al. MicroRNA expression profiling in the prefrontal cortex of individuals affected with schizophrenia and bipolar disorders. Schizophr Res. 2010;124(1–3):183–91.
Lawrie CH. MicroRNA expression in lymphoid malignancies: new hope for diagnosis and therapy? J Cell Mol Med. 2008;12(5a):1432–44.
Biswas S, Haleyurgirisetty M, Lee S, Hewlett I, Devadas K. Development and validation of plasma miRNA biomarker signature panel for the detection of early HIV-1 infection. EBioMedicine. 2019;43:307–16.
Zhou R, Yuan P, Wang Y, Hunsberger JG, Elkahloun A, Wei Y, et al. Evidence for selective microRNAs and their effectors as common long-term targets for the actions of mood stabilizers. Neuropsychopharmacology. 2009;34(6):1395.
Malhi GS, Tanious M, Das P, Berk M. The science and practice of lithium therapy. Aust N Z J Psychiatry. 2012;46(3):192–21111.
Malhi GS, Outhred T. Therapeutic mechanisms of lithium in bipolar disorder: recent advances and current understanding. CNS Drugs. 2016;30(10):931–49.
Acknowledgements
The authors thank the Allen Institute for Brain Science (551 North 34th Street, Seattle, Washington 98103, USA) and the Broad Institute (415 Main Street, Cambridge, Massachusetts 02142, USA) for their valuable data, which are publicly available for analyses. The authors also thank Bahman Razi (PhD candidate in Hematology, Department of Hematology and Blood Transfusion, Faculty of Medicine, Tarbiat Modares University, Tehran, Iran) for his kind scientific comments on the manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Funding
No funding has been received for conducting this study and/or preparation of this manuscript.
Conflict of interest
The authors declare that they have no conflicts of interest.
Electronic supplementary material
Below is the link to the electronic supplementary material.
40263_2020_723_MOESM1_ESM.xlsx
Supplemental file S1. The list of the lithium responsiveness-associated genes (LRAGs), collected from previously published articles. These genes are listed by their official symbols and also by their full names (XLSX 17 kb)
40263_2020_723_MOESM2_ESM.xlsx
Supplemental file S2. A subset of LRAGs that were specifically enriched in four signaling pathways. These genes were used as the input data of PANTHER analysis of protein classes. LRAGs lithium responsiveness-associated genes, PANTHER Protein ANalysis THrough Evolutionary Relationships (XLSX 10 kb)
40263_2020_723_MOESM3_ESM.pdf
Supplemental figure S1. Circadian rhythm signaling pathway (KEGG number: map04710). The first-ranked enriched signaling pathway for the LRAGs was circadian rhythm pathway. It consists of cell-autonomous transcription-translation feedback loops that drives rhythmic expression patterns of core clock components. The first negative feedback loop is a rhythmic transcription of period (Per) genes and chryptochrome (Cry) genes. Per and Cry proteins form a heterodimer, which acts on the Clock/Bmal1 heterodimer to repress its own transcription. Per and Cry proteins are phosphorylated by casein kinase epsilon (CkIε), which leads to degradation and restarting of the cycle. The second loop is a positive feedback loop, driven by the Clock/Bmal1 heterodimer, which initiates transcription of target genes containing E-box cis-regulatory enhancer sequences. The components with the red stars denote LRAGs. KEGG Kyoto Encyclopedia of Genes and Genomes, LRAGs lithium responsiveness-associated genes (PDF 1056 kb)
40263_2020_723_MOESM4_ESM.pdf
Supplemental figure S2. Synaptic signaling pathways and their LRAGs. (a) Signaling by a dopaminergic synapse (KEGG number: map04728). (b) Signaling by a cholinergic synapse (KEGG number: map04725). In both pathways, signaling events are shared between neuronal (bottom left and bottom right cells) and glial cells (top). A significant number of LRAGs were enriched for these pathways (denoted as red asterisks). KEGG Kyoto Encyclopedia of Genes and Genomes, LRAGs lithium responsiveness-associated genes (PDF 1922 kb)
40263_2020_723_MOESM5_ESM.pdf
Supplemental figure S3. The signaling pathway of morphine addiction (KEGG number: map05032). The interactions between components of this pathway conditions are illustrated in five different conditions: (a) in the absence of morphine (control). (b) Acute use of morphine. (c) Chronic use of morphine. (d) Withdrawal from chronic use of morphine. (e) Increased adenosine tone, mediated by morphine withdrawal. Red asterisks denote the LRAGs of the pathway. KEGG Kyoto Encyclopedia of Genes and Genomes, LRAGs lithium responsiveness-associated genes (PDF 2827 kb)
Rights and permissions
About this article
Cite this article
Najafi, H., Totonchi, M. & Sadeghizadeh, M. Predicted Cellular and Molecular Actions of Lithium in the Treatment of Bipolar Disorder: An In Silico Study. CNS Drugs 34, 521–533 (2020). https://doi.org/10.1007/s40263-020-00723-7
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40263-020-00723-7