Abstract
Background and Objectives
The genetic polymorphism of cytochrome P450 (CYP) 2D6 is characterized by an excessive impact on positive and adverse drug reactions to antipsychotics, such as risperidone. Consequently, the pharmacokinetics of the drug and metabolite can be substantially altered and exhibit a high variability between the different phenotypes. The goal of this study was to develop a physiologically based pharmacokinetic (PBPK) model considering the CYP2D6 genetic polymorphism for risperidone and 9-hydroxyrisperidone (9-OH-RIS) taking CYP3A4 into account. Additionally, risperidone dose adjustments, which would compensate for genetically caused differences in the plasma concentrations of the active moiety (sum of risperidone and 9-OH-RIS) were calculated.
Methods
Based on available knowledge about risperidone, 9-OH-RIS, and relevant physiological changes according to different CYP2D6 phenotypes, several PBPK models were built. In addition, an initial model was further evaluated based on the plasma concentrations of risperidone and 9-OH-RIS from a single-dose study including 71 genotyped healthy volunteers treated with 1 mg of oral risperidone.
Results
PBPK models were able to accurately describe risperidone exposure after single-dose administration, especially in the concentration range ≥ 1 µg/L, illustrated by a minimal bias and a good precision. About 90.3% of all weighted residuals versus observed plasma concentrations ≥ 1 µg/L were in the ± 30% range. The risperidone/9-OH-RIS ratio increased progressively according to reduced CYP2D6 activity, resulting in a mean ratio of 4.96 for poor metabolizers. Simulations demonstrate that dose adjustment of the drug by − 25% for poor metabolizers and by – 10% for intermediate metabolizers results in a similar exposure to that of extensive metabolizers. Conversely, the risperidone/9-OH-RIS ratio can be used to determine the phenotype of individuals.
Conclusion
PBPK modelling can provide a valuable tool to predict the pharmacokinetics of risperidone and 9-OH-RIS in healthy volunteers, according to the different CYP2D6 phenotypes taking CYP3A4 into account. These models are able to ultimately support decision-making regarding dose-optimization strategies, especially for subjects showing lower CYP2D6 activity.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Schizophrenia is a chronic and severe mental disorder, affecting more than 23 million people worldwide in 2018 [1, 2]. Patients suffering from schizophrenia are two to three times more likely to die early than the general population due to cardiovascular or metabolic diseases and infections [1]. Therefore, an effective and individual treatment is important. One treatment option for schizophrenia is risperidone.
Risperidone is an antipsychotic agent chemically classified as a benzisoxazole derivative that has selective antagonistic properties at serotonin 5-HT2 and dopamine D2 receptors [3]. Depending on the dosage of risperidone, the incidence of antipsychotic-induced extrapyramidal symptoms (EPS) occurs more frequently than with other second-generation antipsychotics [4,5,6]. Besides the limiting EPS profile, risperidone therapy is associated with an increased prolactin level, which is plasma concentration dependent [7].
The antipsychotic agent risperidone is mainly metabolized in the liver via the cytochrome P450 (CYP) 2D6 enzymatic pathway [7,8,9]. Furthermore, in vitro studies [8, 9] as well as clinical dug–drug interaction studies [10, 11] have suggested that CYP3A4 is also involved in the enzymatic pathway of risperidone, but plays a minor role compared with CYP2D6 [9]. The contribution of both enzymes was evaluated with specific in vitro inhibitors using quinidine (CYP2D6 inhibitor) and ketoconazole (CYP3A4 inhibitor) [8, 9]. Consequently, CYP2D6 and CYP3A4 are the relevant enzymes for the metabolism of risperidone to its active metabolite 9-hydroxyrisperidone (9-OH-RIS), which has pharmacologic effects equivalent to that of risperidone [8, 12]. Other metabolites such as 7-hydroxyrisperidone, among others, do not exhibit significant pharmacologic activities [7]. The clinical effect of the drug is determined by the sum of the concentration of risperidone and its active metabolite 9-OH-RIS, called active moiety [13]. The therapeutic reference range of the active moiety is 20–60 ng/mL according to the Arbeitsgemeinschaft für Neuropsychopharmakologie und Pharmakopsychiatrie (AGNP) consensus guideline on therapeutic drug monitoring (TDM) published by Hiemke et al. [14].
There is evidence of several differences in risperidone treatment showing large inter-individual variability in risperidone and 9-OH-RIS concentrations [15, 16]. The wide pharmacokinetic variability of many drugs is largely explained by the CYP2D6 genetic polymorphism [17], but can also be influenced by age, renal and hepatic function, disease status, and co-medication [17, 18]. To date, > 100 allelic variants of the CYP2D6 genotype have been proposed in four different phenotypes [19, 20]: extensive metabolizer (EM), intermediate metabolizer (IM), poor metabolizer (PM), and ultra-rapid metabolizer (UM). The frequency of metabolizers is ethnically dependent and was estimated for a white population by Xie and Feng [21] to be 60–85% EM, 10–15% IM, and 1–10% UM. In the white population, approximately 7–10% are PM [21], completely lacking CYP2D6 activity because of two non-functional alleles [19]. Due to the non-functional alleles of the CYP2D6 gene, the PM phenotype appears to be associated with more adverse drug reactions (ADRs) and frequent discontinuation of therapy due to ADRs [22]. Especially for risperidone, Ereshefsky [23] proposed that a risperidone/9-OH-RIS ratio > 1 is an index indicating a CYP2D6 PM in the absence of a potent CYP2D6 inhibitor. CYP2D6 PMs do not have CYP2D6 activity but are able to produce 9-OH-RIS. Wu [24] supposed that in an individual who produces an inactive variant for an enzyme in the major metabolic pathway, the drug can be metabolized through an alternate or minor pathway, assuming that these enzymes are not affected genetically. Therefore, it is likely that the messenger RNA (mRNA) of those genes are up-regulated to compensate for the presence of the deficient enzyme [24]. One possible alternative metabolic pathway in CYP2D6 PMs may be the alicyclic dehydroxylation and oxidative N-dealkylation via CYP3A [25]. All in all, the relative contribution of CYP3A4 in PMs is not yet fully understood.
Although risperidone metabolism is regulated by the genetic polymorphism of CYP2D6, several authors detect only little differences between the pharmacokinetics and effects of the active moiety between the different types of metabolizer [26,27,28,29,30]. However, physiological changes according to the different CYP2D6 phenotypes may influence the pharmacokinetics of the administrated drug. So, the aim of the current investigation was to develop a whole-body physiologically based pharmacokinetic (PBPK) model to analyze the relationship between the pharmacokinetics of risperidone and 9-OH-RIS and the CYP2D6 phenotype. Furthermore, the influence of the several types of CYP2D6 metabolizer on the dosage should be investigated to be able to treat patients individually according to their phenotype. In this context, it should also be investigated whether it is possible to draw conclusions on the different phenotypes using the risperidone/9-OH-RIS ratio, to avoid expensive genotyping. To our knowledge, this is the first study examining the effects of CYP2D6 phenotype-related physiological alterations on the pharmacokinetics of risperidone using a PBPK approach.
2 Materials and Methods
2.1 Physiologically Based Pharmacokinetic (PBPK) Modelling
The modelling was performed using PK-Sim® version 7.1.0 build 63 [31] as part of the Open Systems Pharmacology Suite (Bayer Technology Services, Leverkusen, Germany), a freely available ‘whole-body’ PBPK simulation software that allows description of the fate of xenobiotics, especially drugs in humans, as well as in several mammalian organisms [32, 33]. The software implements a generic structure consisting of 17 different compartments, each representing a single tissue or organ [34, 35]. All compartments are connected via organ-specific blood flow to the arterial and venous blood pool to enable inter-compartmental mass transport [32, 34, 35]. Each organ is subdivided into subcompartments, such as vasuclar space containing erythrocytes and plasma, intercellular and tissue interstitial [35]. Based on physicochemical properties (e.g., plasma protein binding or fraction unbound in plasma, lipophilicity and molecular weight), compound-specific clearance information, as well as organ/tissue composition data (e.g., lipid, protein, water), PK-Sim® estimates drug-related model parameters to describe the rate and extent of drug disposition in each compartment [34]. For a detailed description of the generic model structure of PK-Sim® and the input/output parameters, refer to the user software manual [33], Willmann et al. [32, 34], or Kuepfer et al. [36].
2.2 General Workflow
The workflow for the development, evaluation, and transmission of the risperidone and 9-OH-RIS PBPK model is described in Fig. 1. First, an initial single-dose model was created based on a systematic literature survey to detect relevant anatomical, physicochemical, and drug-specific parameters of oral risperidone and its active metabolite. Additionally, properties relevant to absorption, distribution, metabolism, and elimination for risperidone and 9-OH-RIS were integrated. No difference was made between different CYP2D6 phenotypes. Based on the initial PBPK model, the model was evaluated using study data, supplied by two bioequivalence trials [37]. Plasma concentrations of risperidone and 9-OH-RIS were used to develop different PBPK models, according to each CYP2D6 phenotype: EM, IM, PM, and UM. Finally, simulations were performed in four different populations during steady-state. All PBPK simulations for risperidone and 9-OH-RIS were completed using PK-Sim® software [31].
Schematic workflow for the development of a physiologically based pharmacokinetic model for healthy individuals. The initial model includes absorption, distribution, metabolism, and elimination (ADME), physicochemical, and physiological data on literature values only. For the evaluated model the literature-based adaptions were modified by 71 genotyped healthy individuals from the study. Based on the evaluated single-dose model, the model was first scaled from risperidone 1–6 mg and subsequently simulated for steady-state. EM extensive metabolizer, IM intermediate metabolizer, PM poor metabolizer, RIS risperidone, UM ultra-rapid metabolizer
2.3 Development of a Single-Dose PBPK Model
Risperidone- and 9-OH-RIS-relevant anatomical, physicochemical, and drug-specific parameters were taken from the literature and integrated into the initial PBPK model. After oral administration, the absolute bioavailability of risperidone amounts to 70% [38] after rapid absorption. Risperidone is excreted in the urine as metabolites (65%) and as unchanged drug (5%) [39]. Additionally, 14% of risperidone is excreted as metabolites in the feces [39]. For the initial PBPK model, risperidone clearance were determined to be 5 ± 1 mL/min/kg [40] regarding all types of CYP2D6 metabolizer. Besides the metabolizing enzymes CYP2D6 and CYP3A4, the drug transporter P-glycoprotein (P-gp) was incorporated in the simulation, based on P-gp adenosine triphosphatase activity measurement [41]. The PK-Sim® Standard method [42, 43] was chosen for the estimation of partition coefficient and cellular permeabilities.
2.4 Evaluation of a Single-Dose PBPK Model
2.4.1 Study Data
For the PBPK model evaluation of risperidone and 9-OH-RIS, data were supplied by two bioequivalence trials [37] using Risperdal® 1 mg (Janssen-Cilag, Madrid, Spain) as reference formulation and one test formulation. As described previously [37], a single-dose, randomized, two-period, crossover study was conducted. Between May and July 2003, blood samples of 71 healthy volunteers (36 women and 35 men) were collected at the following times after dosing: 0, 0.25, 0.5, 1.0, 1.5, 2.0, 3.0, 4.0, 5.0, 6.0, 8.0, 10, 12, 24, 48, 72, and 96 h. All subjects were Caucasian, expect for three Hispanic persons (one from Mexico and two from Argentina). Inclusion criteria were 18–35 years of age, non-smoking, body mass index between 19 and 30 kg/m2, and results within the reference range of laboratory values for hematology, as well as other criteria [37]. Demographic data and patient characteristics are summarized in Table 1.
Genotyping of CYP2D6 was conducted by standard methods previously described in Cabaleiro et al. [44]. According to the activity score (AS) of the different CYP2D6 alleles, four phenotypic classes were determined: EM, IM, PM, and UM [20, 45]. A comparison of the genes and AS is provided in Table 2.
2.4.2 PBPK Model Evaluation
For model evaluation, plasma concentrations of risperidone and 9-OH-RIS were used to predict the pharmacokinetics of a mean population of healthy volunteers. A differentiation between the various phenotypes was not performed. Only plasma concentrations relating to the Risperdal® formulation were used. Timepoints with very few measurements above the lower limit of quantification (LLOQ: 0.1 ng/mL) are not representative for the whole population, and taking these data into account would lead to a bias. Therefore, only those timepoints in which at least 25% of all subjects of the respective phenotype had plasma concentrations above the LLOQ were deemed eligible for analysis. A detailed overview of data selection is provided in Electronic Supplementary Material Figure S1. Measured data were used as observed values. In comparison to the initial model, lipophilicity (logP) for risperidone and 9-OH-RIS was modified according to predicted values from the literature [46,47,48]. Risperidone itself undergoes massive hepatic metabolism (65 % urine changed and 14% feces changed) mediated by the integration of CYP enzymes [39]. Additionally, the specific hepatic and renal clearances of 9-OH-RIS were fitted to the plasma concentration–time profile. Specific clearance represents the intrinsic clearance normalized to the volume where the process occurs [33] and amounts to 0.04/min (hepatic) and 0.67/min (renal). The model parameter partition coefficient and cellular permeability were adjusted according to the observed data. All other model-building parameters were kept from the initial model. Final PBPK parameters are listed in Table 3.
2.5 Cytochrome P450 2D6 Phenotype-Related Scaling of PBPK Model Parameters
Next, all 71 healthy volunteers were divided into different subgroups according to their CYP2D6 phenotype: EM (n = 33), IM (n = 26), PM (n = 6), and UM (n = 6). Following that, four separate populations were created for each type of metabolizer to allow a comparison among the different CYP2D6 phenotypes. All chemical and physicochemical parameters of risperidone and 9-OH-RIS, as well as anatomical and physiological information on the evaluated model, were transferred unchanged to all four models, as they did not vary between the several types of metabolizers. In addition to the compound properties, the administration protocol and the formulation were also adopted as they do not differ. Due to the genetic polymorphism of CYP2D6, the catalytic rate constant (kcat) (/min), also known as the turnover frequency, was changed during model adaption because of the different enzyme activities. kcat is defined as the catalytic efficiency of enzymes, or rather the number of substrate molecules, each enzyme site converts to product per unit of time, and in which the enzyme is working at maximum efficiency [31, 33, 36]. The extent of the model adjustments was based on the views of Gaedigk et al. [45] using the AS, describing the respective enzyme activity of the different metabolizers. Subjects with an AS of 0 were considered as PMs, an AS value of 0.5–1.0 as IMs, a value of 1.5–2.0 as EMs, and an AS > 2 as UMs [45]. Based on the different AS ratios in the literature, all kcat values were first estimated through parameter optimization by PK-Sim® and subsequently set into ratio according to their status (Table 4). kcat values for CYP2D6 9-hydroxylation, 7-hydroxylation, and N-dealkylation were set to 0/min for PMs, because this type of metabolizer has two non-functional alleles, which prevents the enzyme from functioning [19, 49]. In contrast, the CYP3A4 kcat value for N-dealkylation and 7-hydroxylation was increased from 0.15/min (EM, IM, and UM) to 2.5/min (PM), as this is more active in PMs [25]. UMs, who have at least three copies of functional alleles, CYP2D6 kcat values were set at 4.0/min (9-hydroxylation) and 2.0/min (7-hydroxylation and N-dealkylation).
2.6 Steady-State Simulation
Based on the single-dose PBPK models for EMs, IMs, PMs, and UMs, the simulations were conducted for steady-state conditions using PK-Sim®. This is possible because the concentrations of risperidone and its active metabolite 9-OH-RIS increase dose proportionally over the dosing range of 1–16 mg daily (0.5–8 mg bi-daily) [38], indicating that risperidone exhibits linear elimination pharmacokinetics [50, 51]. As risperidone has a half-life of 3.2 h and 9-OH-RIS of 24.7 h [52], steady-state is reached within 1 day for risperidone and within 5 days for 9-OH-RIS [50]. In addition, CYP2D6 PMs show a much longer risperidone half-life than EMs [7]. The dose of risperidone was fixed to 6 mg/day (3 mg twice daily), which is the recommended dose for the management of schizophrenia in adults [53]. All simulations were conducted for a treatment period of 420 h. According to the product information [51], the dose was escalated starting with 1 mg twice a day followed by 2 mg twice daily and 3 mg twice a day from the third treatment day [51]. The daily target dose of the active moiety corresponds to the therapeutic reference range, which was set to a range of 20–60 ng/mL, as published by Hiemke et al. [14]. For plasma concentrations outside the therapeutic range, the daily dose was adjusted until it was within the range. Risperidone/9-OH-RIS ratios for EMs, IMs, PMs, and UMs were calculated and used to compare each metabolizer type.
2.7 Model Evaluation and Statistical Analysis
The evaluation of the PBPK models was carried out in accordance with the Guideline on the Qualification and Reporting of Physiologically Based Pharmacokinetic Modelling and Simulation [54]. Assessment of the plasma concentration–time profiles of the PBPK models was initially based on visual inspections of the observed and predicted plasma concentration. Simulations were carried out with the aid of the computer-assisted Monte-Carlo study on populations with n = 1000. PBPK model quality was analyzed using goodness-of-fit (GOF) plots and weighted residuals versus observed concentration. To describe model accuracy and precision, prediction error (PE), mean prediction error (MPE) and mean absolute prediction error (MAPE) were calculated according to Eqs. 1–3:
Pharmacokinetic parameters, such as the median area under the systemic drug concentration–time curve from time zero to infinity (AUC∞), as well as the maximum concentration of a drug (Cmax), and the time it takes until the drug reach the Cmax (tmax), was calculated by PK-Sim®. All statistical and graphical pre-analytics were performed in R (version 3.4.2; R Foundation for Statistical Computing, Vienna, Austria) [55].
3 Results
3.1 Single-Dose PBPK Model
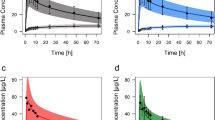
After incorporation of the known physicochemical properties of risperidone in the initial PBPK model, risperidone exposure after a single oral dose accurately described the plasma concentrations for a population of healthy volunteers (Fig. 2). Risperidone is rapidly absorbed and shows a Cmax between 4.2 and 8.8 µg/L within 1–2 h after dosing, according to the CYP2D6 phenotype (Table 5). In PMs the Cmax of risperidone tended to be higher and the elimination much slower than in EMs, IMs, and UMs. Due to the lack of CYP2D6 activity, PMs convert less risperidone into its active metabolite, so that the Cmax of the metabolite is significantly lower than in the other metabolizers (Fig. 2, Table 5). All PBPK models can accurately predict the influence of the CYP2D6 metabolizer status, as can be seen by the respective MPE and MAPE values shown in Table 6. Especially in the concentration range ≥ 1 µg/L, the models show a high accuracy illustrated by a minimal bias (MPE range − 13.8% to + 9.07%) and a good precision (MAPE range 9.70–20.9%) in their prediction. In accordance with this, 78.9% (EMs), 81.8% (IMs), 53.8% (PMs), and 82.4% (UMs) of the plasma concentration values in the concentration range ≥ 1 µg/L were in the 1.25-fold error range in the GOF plot (Fig. 3a). About 90.3% of all weighted residuals versus observed plasma concentrations ≥ 1 µg/L were in the ± 30% range (Fig. 3c), indicating that the distribution of risperidone and 9-OH-RIS in the early phase of administration is predicted more accurately. Especially for plasma concentrations lower than 1 µg/L, a poorer prediction can be seen for all types of metabolizer, suggesting that the clearance of risperidone is under-predicted. This in turn leads to an overestimation of risperidone and an underestimation of 9-OH-RIS plasma concentrations in the lower, subtherapeutic concentration range. The respective values for MPE and MAPE (Table 6) as well as the pharmacokinetic parameters (Table 5) support these observations.
Plasma concentration–time curve profiles (mean and standard deviation) of risperidone and 9-hydroxyrisperidone after the administration of a single 1 mg dose of risperidone in healthy adults according to their cytochrome P450 (CYP) 2D6 phenotype. Solid lines indicate the mean and the colored area indicates the standard deviation of the physiologically based pharmacokinetic model. Circles indicate observed plasma concentrations. a Extensive metabolizers (n = 33); b intermediate metabolizers (n = 26); c poor metabolizers (n = 6); and d ultra-rapid metabolizers (n = 6)
Goodness-of-fit plot for model prediction of risperidone and 9-hydroxyrisperidone plasma concentrations in healthy adults. b Weighted residuals vs. observed risperidone and 9-hydroxyrisperidone concentration for healthy adult volunteers over the entire concentration range. The solid black line indicates the line of identity, filled (risperidone) and open (9-hydroxyrisperidone) circles indicate observed data. c Weighted residuals vs. observed risperidone and 9-hydroxyrisperidone concentration for healthy adult volunteers for plasma concentrations ≥ 1 µ/L. The solid black line indicates the line of identity, dashed lines indicate ± 30% range, dotted lines indicate ± 50%, filled (risperidone) and open (9-hydroxyrisperidone) circles indicate observed data. a, b, c Black circles indicate extensive metbaolizer, blue intermediate metabolizer, red poor metaboizer, and green ultra-rapid metabolizer
3.2 Steady-State PBPK Model
Using the evaluated PBPK models, steady-state conditions with administration of risperidone 3 mg twice a day were simulated (Fig. 4). All predicted trough plasma concentrations for EMs, IMs, PMs, and UMs during steady-state were in the therapeutic reference range of 20–60 µg/L determined by Hiemke et al. [14]. However, the predicted Cmax values of the active moiety for PMs and IMs were outside this range. Especially in PMs, 57.1% of all predicted plasma concentrations were higher than the therapeutic reference range during steady-state. In the case of IMs, only 26.5% of all plasma concentrations were above the therapeutic reference range.
Mean predicted concentration–time profiles of total active moiety (sum of risperidone and 9-hydroxyrisperidone) after administration of multiple 6 mg daily doses (3 mg twice a day) of risperidone according to their cytochrome P450 (CYP) 2D6 phenotype. The shaded boxes represent the therapeutic reference range between 20 and 60 µg/L. a Extensive metabolizers; b intermediate metabolizers; c poor metabolizers; and d ultra-rapid metabolizers
3.2.1 Risperidone Dose Adjustment for Poor Metabolizers (PMs) and Intermediate Metabolizers (IMs)
Due to the high plasma concentrations of total active moiety in PMs and IMs during steady-state, the dose was adjusted (Fig. 5). The steady-state trough plasma concentrations of PMs were + 106% higher than those of EMs (25.3 µg/L in EMs and 52.4 µg/L in PMs) at the same dose. Even the Cmax of PM was significantly higher (+ 53.1%) in PMs than in EMs (50.5 µg/L in EMs and 77.4 µg/L in PMs). This was also evident in the IMs; the trough plasma concentration was + 62.3% higher than that of EMs (25.3 µg/L in EMs and 41.1 µg/L in IMs), and the Cmax was + 38.5% higher in IMs than in EMs (50.5 µg/L in EMs and 70.0 µg/L in IMs). Consequently, the original dose of risperidone 6 mg (3 mg twice a day) was reduced by − 10% to 5.4 mg for IMs and by − 25% to 4.5 mg for PMs. After dose adjustment trough plasma concentrations and Cmax values of the active moiety were within the therapeutic reference range and showed similar plasma concentrations to those of EMs and UMs with risperidone 6 mg (3 mg twice a day).
Dose adjustment by – 10% for intermediate metabolizers and − 25% for poor metabolizers. Mean predicted plasma concentration–time profiles of total active moiety (sum of risperidone and 9-hydroxyrisperidone) after administration of multiple doses. The shaded boxes represent the therapeutic reference range between 20 and 60 µg/L. a Multiple daily doses of risperidone 5.4 mg for intermediate metabolizers; and b multiple daily doses of risperidone 4.5 mg for poor metabolizers
3.2.2 Risperidone/9-Hydroxyrisperidone (9-OH-RIS) Ratio
Table 7 describes how the risperidone/9-OH-RIS ratio increases progressively according to reduced CYP2D6 activity. For this purpose, steady-state schedules with 4 mg (2 mg twice a day) and 6 mg (3 mg twice a day) were simulated. The smallest risperidone/9-OH-RIS ratio was observed for UMs and ranged between 0.10 and 0.40 for risperidone 6 mg/day and 0.1 and 0.39 for 4 mg/day depending on the time of the measurement. In contrast, PMs showed greater ratios, ranging between 4.38 and 6.29 for risperidone 4 and 6 mg/day during steady-state. Thus, the mean risperidone/9-OH-RIS ratio increases progressively according to the number of functional CYP2D6 alleles. This resulted in the following gradation of the mean risperidone/9-OH-RIS ratio: 0.16 (UMs), 0.27 (EMs), 0.72 (IMs), and 5.00 (PMs).
4 Discussion
In the present study, we analyzed the influence of CYP2D6- and CYP3A4-related physiological changes on the pharmacokinetics of healthy subjects treated with risperidone as a single-dose and in a steady-state condition using a PBPK approach. The transmission of single-dose models to steady-state was possible due to linear pharmacokinetics of the active moiety [38, 50, 51]. The two phenotypes IM and PM were the focus of the investigations as they metabolize risperidone significantly slower (Fig. 2) due to a reduced number (IM) or lack (PM) of CYP2D6 active alleles [19, 49].
4.1 PBPK Model Development
For the first time, various PBPK models for the prediction of the plasma concentrations of risperidone and 9-OH-RIS were successfully developed in different CYP2D6 phenotypes (Fig. 2). Overall, the absorption, distribution, and elimination phases of the single-dose concentration–time courses were accurately predicted by the models. Especially in the plasma concentration range ≥ 1 µg/L, the models showed a high predictive power supported by minimal MPEs and good MAPE values (Table 6). Even the pharmacokinetic parameters Cmax and tmax were predicted very well. In contrast, plasma concentrations below 1 µg/L showed larger deviations between the observed and predicted values. However, this concentration range was considered less important during model development, which was for multiple reasons. First, we developed single-dose models instead of multiple doses as used in practice. During steady-state, risperidone and 9-OH-RIS accumulate in the body, which leads to higher plasma concentrations, recognizable by the therapeutic reference range of 20–60 µg/L [14]. Secondly, the clinical and pharmacological relevance of such low concentrations is questionable, as the therapeutic reference range of the active moiety is 20–60 times higher. For this purpose, plasma concentrations below 1 µg/L were neglected during model development.
4.2 Limitations
Potential limitations should be taken into account when interpreting the results of PBPK model development. First, data from healthy volunteers was used rather than patients, which may be limiting when using the results for clinical applications. The applicability of the model needs to be evaluated further with clinical pharmacokinetic data including elderly patients. Furthermore, it is apparent that not only genetic but also pharmacological (e.g., co-medications such as inhibitors or inducers) and other variables such as age or co-morbidity should be considered to personalize dosing in the clinical environment [56].
4.3 Dose Adjustment for IMs and PMs
As reported by de Leon et al. [22], the CYP2D6 PM phenotype appears to be associated with risperidone’s ADRs and discontinuation due to ADRs. According to the authors’ experience, CYP2D6 PMs show a three to six times higher risk of experiencing ADRs related to risperidone or discontinuations because of ADRs [22, 57]. PMs and IMs consistently showed significantly higher plasma concentrations in our PBPK models than EMs and UMs at a dose of risperidone 3 mg twice a day (Fig. 4), which leads to an active moiety within the supratherapeutic range (> 60 µg/L). As a consequence, drug plasma concentrations within the supratherapeutic range can lead to an increased occurrence of ADRs [14]. So, an optimal dose setting for PMs is very important to reduce the rate of ADRs and discontinuation due to ADRs.
In order to reduce the proportion of the plasma concentrations of the active moiety in the supratherapeutic area, a dose adjustment, especially for PMs and IMs, should be considered. The PBPK analysis results in a recommendation for a dose adjustment (Fig. 5) in subjects with a reduction in the number (IMs) or lack (PMs) of CYP2D6 active alleles of − 10% for IMs and − 25% for PMs. This observation is consistent with the results in the literature. Mas et al. [58] and Kirchheiner et al. [59] postulated a strong correlation between CYP2D6 status and risperidone dosage. Both authors recommended a dose adjustment for PMs of − 20% [58] and − 13% [59] and for IMs of − 13% [58] and – 5% [59]. Dose adjustment in PMs is also suggested in the clinical guideline for using CYP2D6 genotypes in patients taking antipsychotics published by de Leon et al. [57]. Williams [60] recommends that CYP2D6 PMs take half or less of the upper range in normal circumstances. However, no adjustment is necessary in UMs.
4.4 Risperidone/9-OH-RIS Ratio
Steady-state plasma concentration of risperidone and 9-OH-RIS can be used to calculate the risperidone/9-OH-RIS ratio, which is a marker of CYP2D6 activity. In patients taking oral risperidone, a risperidone/9-OH-RIS ratio > 1 indicates the patient is a genetic CYP2D6 PM or is taking a potent CYYP2D6 inhibitor that completely inhibits CYP2D6 activity [61]. Our calculated risperidone/9-OH-RIS ratio for PM ranged from 4.38 to 6.29 (Table 7) and was therefore above the ratio of 1. In comparison, the remaining phenotypes showed lower ratios, which ranged from 0.19 to 0.59 (EMs), 0.59 to 1.15 (IMs), and 0.10 to 0.40 (UMs). Thereby, the risperidone/9-OH-RIS ratio seems to be an inverse measurement of CYP2D6 activity. The increase in the number of active CYP2D6 alleles indicates a decreased ratio due to the metabolism of risperidone. As a result, UMs, who have at least three copies of functional alleles [45], show lower ratios because UMs metabolize risperidone faster and the proportion of the active metabolite 9-OH-RIS is higher. In contrast, PMs, who have two non-functional alleles [45], show high risperidone/9-OH-RIS ratios because only a small proportion of risperidone is converted into the active metabolite due to the lack of CYP2D6 enzymes. CYP2D6 PMs do not have CYP2D6 activity but are able to produce some 9-OH-RIS using CYP3A4 [9, 24].
4.5 Phenotyping Using the Risperidone/9-OH-RIS Ratio
Due to the significant differences in the ratio, only the risperidone/9-OH-RIS ratio needs to be calculated to determine whether the patient is a PM or not. For assessment of the PM, the risperidone/9-OH-RIS ratio is an equivalent alternative to genotyping. Therefore, determination of the plasma concentrations of risperidone and 9-OH-RIS would be sufficient to exclude a PM. If the observed plasma concentration value exceeds 4.38 (Table 7), the patient can be classified as a PM. Such a classification into the several types of metabolizers would make sense during TDM, which is recommended for risperidone by many experts [14]. With respect to the genetic polymorphism in CYP2D6, TDM of risperidone and its main metabolite is an important tool to improve patient safety. So, when TDM is available, phenotyping could be used to classify a patient as a PM.
5 Conclusion
The present study generated and evaluated a PBPK model using PK-Sim® for risperidone and its active metabolite 9-OH-RIS for CYP2D6 EMs, IMs, PMs, and UMs taking CYP3A4 into account. Especially in the concentration range ≥ 1 µg/L, the models show high accuracy which is illustrated by a minimal bias and good precision. PMs and IMs showed plasma concentrations of the active moiety above the therapeutic reference range. To reduce the proportion of plasma concentrations in the supratherapeutic range, the dose was adjusted by – 10% for IMs and – 25% for PMs when using risperidone 3 mg twice a day [53]. As a marker of CYP2D6, the risperidone/9-OH-RIS ratio during steady-state was calculated showing that PMs have a mean ratio of 4.96 whereas EMs show a mean ratio of 0.27.
In the future, calculation of the risperidone/9-OH-RIS ratio from TDM measurements could be used to phenotype each patient. To ensure the best therapy for each subject, dose adjustment, especially for PM and IMs, must be considered to reduce ADRs and discontinuation due to ADRs in different CYP2D6 phenotypes.
References
World Health Organization. Schizophrenia—fact sheet. 2018. http://www.who.int/news-room/fact-sheets/detail/schizophrenia. Accessed 14 Aug 2018.
World Health Organization. The world health report 2001: mental health: new understanding, new hope. 2018. http://www.who.int/whr/2001/en/whr01_en.pdf. Accessed 14 Aug 2018.
Janssen PA, Niemegeers CJ, Awouters F, Schellekens KH, Megens AA, Meert TF. Pharmacology of risperidone (R 64 766), a new antipsychotic with serotonin-S2 and dopamine-D2 antagonistic properties. J Pharmacol Exp Ther. 1988;244(2):685–93.
Leucht S, Cipriani A, Spineli L, et al. Comparative efficacy and tolerability of 15 antipsychotic drugs in schizophrenia: a multiple-treatments meta-analysis. Lancet. 2013;382(9896):951–62.
Bo Q-J, Li X-B, Wang Z-M, Li A-N, Ma X, Wang C-Y. Extrapyramidal symptoms during risperidone maintenance treatment in schizophrenia: a prospective, multicenter study. J Clin Psychopharmacol. 2016;36(2):125–9.
Kasper S, Tauscher J, Küfferle B, Barnas C, Pezawas L, Quiner S. Dopamine- and serotonin-receptors in schizophrenia: results of imaging-studies and implications for pharmacotherapy in schizophrenia. Eur Arch Psychiatry Clin Neurosci. 1999;249(Suppl 4):83–9.
Huang ML, van Peer A, Woestenborghs R, et al. Pharmacokinetics of the novel antipsychotic agent risperidone and the prolactin response in healthy subjects. Clin Pharmacol Ther. 1993;54(3):257–68.
Fang J, Bourin M, Baker GB. Metabolism of risperidone to 9-hydroxyrisperidone by human cytochromes P450 2D6 and 3A4. Naunyn Schmiedebergs Arch Pharmacol. 1999;359(2):147–51.
Yasui-Furukori N, Hidestrand M, Spina E, Facciolá G, Scordo MG, Tybring G. Different enantioselective 9-hydroxylation of risperidone by the two human CYP2D6 and CYP3A4 enzymes. Drug Metab Dispos. 2001;29(10):1263–8.
Bork JA, Rogers T, Wedlund PJ, Leon Jd. A pilot study on risperidone metabolism: the role of cytochromes P450 2D6 and 3A. J Clin Psychiatry 1999;60(7):469–76.
Spina E, Avenoso A, Facciolà G, et al. Plasma concentrations of risperidone and 9-hydroxyrisperidone: effect of comedication with carbamazepine or valproate. Ther Drug Monit. 2000;22(4):481–5.
Nasrallah HA. Atypical antipsychotic-induced metabolic side effects: insights from receptor-binding profiles. Mol Psychiatry. 2008;13(1):27–35.
Schoretsanitis G, de Leon J, Haen E, et al. Pharmacokinetics of risperidone in different application forms—comparing long-acting injectable and oral formulations. Eur Neuropsychopharmacol. 2018;28(1):130–7.
Hiemke C, Bergemann N, Clement HW, et al. Consensus guidelines for therapeutic drug monitoring in neuropsychopharmacology: update 2017. Pharmacopsychiatry. 2018;51(1–02):9–62.
Balant-Gorgia AE, Gex-Fabry M, Genet C, Balant LP. Therapeutic drug monitoring of risperidone using a new, rapid HPLC method: reappraisal of interindividual variability factors. Ther Drug Monit. 1999;21(1):105–15.
Aravagiri M, Marder SR, Wirshing D, Wirshing WC. Plasma concentrations of risperidone and its 9-hydroxy metabolite and their relationship to dose in schizophrenic patients: simultaneous determination by a high performance liquid chromatography with electrochemical detection. Pharmacopsychiatry. 1998;31(3):102–9.
Zhou S-F. Polymorphism of human cytochrome P450 2D6 and its clinical significance: part I. Clin Pharmacokinet. 2009;48(11):689–723.
Snoeck E, van Peer A, Sack M, et al. Influence of age, renal and liver impairment on the pharmacokinetics of risperidone in man. Psychopharmacology. 1995;122(3):223–9.
Puangpetch A, Vanwong N, Nuntamool N, Hongkaew Y, Chamnanphon M, Sukasem C. CYP2D6 polymorphisms and their influence on risperidone treatment. Pharmgenomics Pers Med. 2016;9:131–47.
Gaedigk A, Simon SD, Pearce RE, Bradford LD, Kennedy MJ, Leeder JS. The CYP2D6 activity score: translating genotype information into a qualitative measure of phenotype. Clin Pharmacol Ther. 2008;83(2):234–42.
Xie H-G, Feng X, editors. Applying pharmacogenomics in therapeutics. Boca Raton: CRC Press; 2016.
de Leon J, Susce MT, Pan R-M, Fairchild M, Koch WH, Wedlund PJ. The CYP2D6 poor metabolizer phenotype may be associated with risperidone adverse drug reactions and discontinuation. J Clin Psychiatry. 2005;66(1):15–27.
Ereshefsky L. Pharmacokinetics and drug interactions: update for new antipsychotics. J Clin Psychiatry. 1996;57(Suppl 11):12–25.
Wu AH. Drug metabolizing enzyme activities versus genetic variances for drug of clinical pharmacogenomic relevance. Clin Proteomics. 2011;8(1):12.
Bozina N, Jovanović N, Lovrić M, Medved V. Clinical significance of a CYP2D6 poor metabolizer–a patient with schizophrenia on risperidone treatment. Ther Drug Monit. 2008;30(6):748–51.
Bertilsson L, Dahl M-L, Dalén P, Al-Shurbaji A. Molecular genetics of CYP2D6: Clinical relevance with focus on psychotropic drugs. Br J Clin Pharmacol. 2002;53(2):111–22.
Olesen OV, Licht RW, Thomsen E, Bruun T, Viftrup JE, Linnet K. Serum concentrations and side effects in psychiatric patients during risperidone therapy. Ther Drug Monit. 1998;20(4):380–4.
Scordo MG, Spina E, Facciolà G, Avenoso A, Johansson I, Dahl ML. Cytochrome P450 2D6 genotype and steady state plasma levels of risperidone and 9-hydroxyrisperidone. Psychopharmacology. 1999;147(3):300–5.
Roh HK, Kim CE, Chung WG, Park CS, Svensson JO, Bertilsson L. Risperidone metabolism in relation to CYP2D6*10 allele in Korean schizophrenic patients. Eur J Clin Pharmacol. 2001;57(9):671–5.
Mihara K, Kondo T, Yasui-Furukori N, et al. Effects of various CYP2D6 genotypes on the steady-state plasma concentrations of risperidone and its active metabolite, 9-hydroxyrisperidone, in Japanese patients with schizophrenia. Ther Drug Monit. 2003;25(3):287–93.
Open Systems Pharmacology. Software PK-Sim®: 7.3.0. 2018. https://github.com/Open-Systems-Pharmacology/Suite/releases/tag/v7.3.0. Accessed 7 Sep 2018.
Willmann S, Lippert J, Sevestre M, Solodenko J, Fois F, Schmitt W. PK-Sim®: a physiologically based pharmacokinetic ‘whole-body’ model. BIOSILICO. 2003;1(4):121–4.
Open Systems Pharmacology. PK-Sim® software manual. http://www.open-systems-pharmacology.org. Accessed 7 Sep 2018.
Willmann S, Höhn K, Edginton A, et al. Development of a physiology-based whole-body population model for assessing the influence of individual variability on the pharmacokinetics of drugs. J Pharmacokinet Pharmacodyn. 2007;34(3):401–31.
Eissing T, Kuepfer L, Becker C, et al. A computational systems biology software platform for multiscale modeling and simulation: integrating whole-body physiology, disease biology, and molecular reaction networks. Front Physiol. 2011;2:4.
Kuepfer L, Niederalt C, Wendl T, et al. Applied concepts in PBPK modeling: how to build a PBPK/PD model. CPT Pharmacometrics Syst Pharmacol. 2016;5(10):516–31.
Novalbos J, López-Rodríguez R, Román M, Gallego-Sandín S, Ochoa D, Abad-Santos F. Effects of CYP2D6 genotype on the pharmacokinetics, pharmacodynamics, and safety of risperidone in healthy volunteers. J Clin Psychopharmacol. 2010;30(5):504–11.
Janssen Pharmaceutical Ltd. Product information Risperdal® (risperidone) tablets. FDA Accessdata. 2018. https://www.accessdata.fda.gov/drugsatfda_docs/label/2005/020272s042,020588s030,021444s016,021346s010lbl.pdf. Accessed 7 Sep 2018.
Sheehan JJ, Sliwa JK, Amatniek JC, Grinspan A, Canuso CM. Atypical antipsychotic metabolism and excretion. Curr Drug Metab. 2010;11(6):516–25.
Mauri MC, Volonteri LS, Colasanti A, Fiorentini A, de Gaspari IF, Bareggi SR. Clinical pharmacokinetics of atypical antipsychotics: a critical review of the relationship between plasma concentrations and clinical response. Clin Pharmacokinet. 2007;46(5):359–88.
Ejsing TB, Pedersen AD, Linnet K. P-glycoprotein interaction with risperidone and 9-OH-risperidone studied in vitro, in knock-out mice and in drug-drug interaction experiments. Hum Psychopharmacol. 2005;20(7):493–500.
Rodgers T, Leahy D, Rowland M. Physiologically based pharmacokinetic modeling 1: predicting the tissue distribution of moderate-to-strong bases. J Pharm Sci. 2005;94(6):1259–76.
Rodgers T, Rowland M. Physiologically based pharmacokinetic modelling 2: predicting the tissue distribution of acids, very weak bases, neutrals and zwitterions. J Pharm Sci. 2006;95(6):1238–57.
Cabaleiro T, Ochoa D, López-Rodríguez R, et al. Effect of polymorphisms on the pharmacokinetics, pharmacodynamics, and safety of risperidone in healthy volunteers. Hum Psychopharmacol. 2014;29(5):459–69.
Gaedigk A, Bradford LD, Marcucci KA, Leeder JS. Unique CYP2D6 activity distribution and genotype-phenotype discordance in black Americans. Clin Pharmacol Ther. 2002;72(1):76–89.
Wishart DS, Feunang YD, Guo AC, et al. DrugBank 5.0: a major update to the DrugBank database for 2018. Nucleic Acids Res 2017;46(D1):D1074–D1082.
Viswanadhan VN, Ghose AK, Revankar GR, Robins RK. Atomic physicochemical parameters for three dimensional structure directed quantitative structure-activity relationships. 4. Additional parameters for hydrophobic and dispersive interactions and their application for an automated superposition of certain naturally occurring nucleoside antibiotics. J Chem Inf Comput Sci. 1989;29(3):163–72.
Mannens G, Meuldermans W, Snoeck E, Heykants J. Plasma protein binding of risperidone and its distribution in blood. Psychopharmacology. 1994;114(4):566–72.
de Leon J, Susce MT, Johnson M, et al. DNA microarray technology in the clinical environment: the AmpliChip CYP450 test for CYP2D6 and CYP2C19 genotyping. CNS Spectr. 2009;14(1):19–34.
Heykants J, Huang ML, Mannens G, et al. The pharmacokinetics of risperidone in humans: a summary. J Clin Psychiatry. 1994;55(Suppl):13–7.
Janssen-Cilag GmbH. Fachinformation RISPERDAL® 0.5 mg, Filmtabletten. 2018. https://www.gelbe-liste.de/produkte/RISPERDAL-0-5-mg-Filmtabletten_355913. Accessed 10 Aug 2018.
Zhou Z-l, Li X, Peng H-y, et al. Multiple dose pharmacokinetics of risperidone and 9-hydroxyrisperidone in Chinese female patients with schizophrenia. Acta Pharmacol Sin 2006;27(3):381–6.
Chouinard G, Jones B, Remington G, et al. A Canadian multicenter placebo-controlled study of fixed doses of risperidone and haloperidol in the treatment of chronic schizophrenic patients. J Clin Psychopharmacol. 1993;13(1):25–40.
European Medicines Agency. Guideline on the qualification and reporting of physiologically based pharmacokinetic (PBPK) modelling and simulation: draft. London. 2016. http://www.ema.europa.eu/docs/en_GB/document_library/Scientific_guideline/2016/07/WC500211315.pdf. Accessed 24 Aug 2018.
R Development Core Team. R: a language and environment for statistical computing. Vienna: R Foundation for Statistical Computing; 2008. ISBN 3-900051-07-0. http://www.R-project.org.
Spina E, de Leon J. Clinical applications of CYP genotyping in psychiatry. J Neural Transm (Vienna). 2015;122(1):5–28.
de Leon J, Armstrong SC, Cozza KL. Clinical guidelines for psychiatrists for the use of pharmacogenetic testing for CYP450 2D6 and CYP450 2C19. Psychosomatics. 2006;47(1):75–85.
Mas S, Gassò P, Alvarez S, Parellada E, Bernardo M, Lafuente A. Intuitive pharmacogenetics: spontaneous risperidone dosage is related to CYP2D6, CYP3A5 and ABCB1 genotypes. Pharmacogenomics J. 2012;12(3):255–9.
Kirchheiner J, Nickchen K, Bauer M, et al. Pharmacogenetics of antidepressants and antipsychotics: the contribution of allelic variations to the phenotype of drug response. Mol Psychiatry. 2004;9(5):442–73.
Williams R. Optimal dosing with risperidone: updated recommendations. J Clin Psychiatry. 2001;62(4):282–9.
Schoretsanitis G, Spina E, Hiemke C, de Leon J. A systematic review and combined analysis of therapeutic drug monitoring studies for long-acting risperidone. Expert Rev Clin Pharmacol. 2017;10(9):965–81.
Poulin P, Schoenlein K, Theil FP. Prediction of adipose tissue: plasma partition coefficients for structurally unrelated drugs. J Pharm Sci. 2001;90(4):436–47.
Poulin P, Theil FP. A priori prediction of tissue:plasma partition coefficients of drugs to facilitate the use of physiologically-based pharmacokinetic models in drug discovery. J Pharm Sci. 2000;89(1):16–35.
Deutscher Apotheker Verlag. Monographien H - Z. 9. Ausgabe, Grundwerk 2017. Europäisches Arzneibuch, 9.2017, Band 3. Stuttgart: Deutscher Apotheker Verlag; Govi-Verlag—Pharmazeutischer Verlag. 2017.
Okubo M, Morita S, Murayama N, Akimoto Y, Goto A, Yamazaki H. Individual differences in in vitro and in vivo metabolic clearances of antipsychotic risperidone from Japanese subjects genotyped for cytochrome P450 2D6 and 3A5. Hum Psychopharmacol. 2016;31(2):93–102.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Funding
No sources of funding were used to conduct this study or prepare this manuscript.
Conflict of interest
Lisa Alina Kneller, Francisco Abad-Santos, and Georg Hempel have no potential conflicts of interest to declare.
Ethical approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki Declaration and its later amendments or comparable ethical standards.
Informed consent
Informed consent was obtained from all participants included in the study
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Kneller, L.A., Abad-Santos, F. & Hempel, G. Physiologically Based Pharmacokinetic Modelling to Describe the Pharmacokinetics of Risperidone and 9-Hydroxyrisperidone According to Cytochrome P450 2D6 Phenotypes. Clin Pharmacokinet 59, 51–65 (2020). https://doi.org/10.1007/s40262-019-00793-x
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40262-019-00793-x