Abstract
Anti‐CD20 antibodies such as ofatumumab has demonstrated efficacy in relapsed/refractory chronic lymphocytic leukemia, are among the most successful therapies to date. In this study, we have designed an immunotoxin composed of Granzyme B and the high affinity variant of Ofatumumab. Different simulation software applied to explore the structure of Granzyme B, a serine protease in cytotoxic lymphocytes granules as an apoptosis mediator was attached to its specific antibody structure (Ofatumumab) via an adaptor sequence. The accuracy, energy minimization and characterization of biological properties of the final structure were evaluated. Our computational outcomes indicated that the employed method for structure prediction has been successfully managed to design the immunotoxin structure. The precise and accurate design of the immune-therapeutic agents against cancer cells can be confirmed by employment of in-silico approaches. Consequently, based on this approach we could introduce a capable immunotoxin which specifically targeting CD20 in an accurate orientation and initiates cancer cell destruction by its toxin domain.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Chronic lymphocytic leukemia (CLL) is one of the most common leukemia in adults. It accounts for 11% of all blood cancers and more that 25% of the various leukemia types (Garg et al. 2011). Different therapeutic approaches are already used to treat cancer such as chemotherapy, radiation therapy, and surgery. However, they failed to reduce the cancer related mortalities. Tumor relapse is commonly encountered in surgical treatments. The radiotherapy and chemotherapy approaches harbor the risk for damaging the normal cells of the body, due to low specificity or lack of specificity for cancer cells. Chemotherapy could also lead to systemic toxicity and adverse effects for patients (Mohan et al. 2019).
CD20 is one of the cell surface markers found in 90% of adult B-cell leukemia and lymphomas. This glycoprotein (279 amino acids) is located in the cell membranes. It acts as a calcium channel and plays a key role in regulation of the cell cycle and cell differentiation. This marker is normally found on the surface of B cells and is expressed when B cells change from the pre-B stage to the activated B stage. However, it is not expressed in stem cells, plasma cells, and other cell lines (Huff and Matsui 2008; Thomas et al. 2009; Tsai et al. 2019). Given these circumstances, targeting this marker using immune chemotherapy agents could be deemed as a viable treatment strategy for inhibition of CLL progress. Prior studies have showed that the expression of CD20 is significantly high in CLL cells. Based on this feature of cancer cells, monoclonal antibodies (mAbs) such as Rituximab and Ofatumumab were designed to treat this type of cancer (Lin 2010; Payandeh et al. 2019a). Ofatumumab, under the brand name Arzerra, is a second-generation antibody against the CD20 transmembrane protein. The in vitro studies of Ofatumumab have showed that compared to Rituximab, this antibody is highly effective in CLL treatment. The high efficacy of Ofatumumab is thought to be rooted in the fact that it’s binding site on CD20 (a small binding site on a large loop on the structure of CD20) which is close to the cell membrane and is completely different from the Rituximab binding site. This feature has made the Ofatumumab to be effective even for B-cells that have a low expression rate of CD20. So, Ofatumumab is more efficient in killing of the target cells (Pawluczkowycz et al. 2009).
The cytotoxic activity of mAbs could be intensified by their conjugation to drugs, radioisotopes, toxins, and enzymes. Immunotoxins are among the most effective anticancer cytotoxic agents which are designed based on an antibody part and a toxic moiety (Lin 2010; Czuczman and Gregory 2010; Danilov 2013; Klein et al. 2013; Xu et al. 2019).
The Granzyme B (GrB), as an important molecule, could trigger apoptotic pathways in the tumor or virus-infected cells. Regarding its enzymatic cytotoxicity, the GrB could be amended to a toxic candidate for immunotoxin design (Hehmann-Titt et al. 2013). Moreover, it is an important molecule due to its potential for activation of apoptosis via Caspase activation. The GrB carrying immunotoxins could initiate its entrance into the target cells via perforin releasing and cleaving Glu and Asp residues; finally, DNA is fragmented (Ganji et al. 2020; Rousalova and Krepela 2010).
Development of novel therapeutic strategies for B‐CLL is of particular importance (Herishanu et al. 2013). Anti-CD20 monoclonal antibodies have demonstrated high efficiency in the elimination of B-cell cancers. In our previous study, we have designed a high affinity variant of Ofatumumab against CD20 antigen (Payandeh et al. 2018, 2019b).
In this study, we have designed an immunotoxin composed of the GrB and the high affinity variant of Ofatumumab. In this regard, an integrated in silico approach and different computational tools were applied. Bioinformatics tools facilitate the understanding of the molecular mechanisms for different macromolecules and supports the rational design of the novel therapeutic agents. These predictions can reduce the failure rate in experimental methods and also decreases the expenses (Bahrami et al. 2019; Khalili et al. 2017). The designed immunotoxin could have an effective interaction with the CD20 that leads to GrB mediated toxic effects in the target cells.
Methods
Structure availability
According to our findings in the previous study, the variant 3 mutant have improved the characteristics of antibody binding compared to normal Ofatumumab antibodies (Payandeh et al. 2019b). Mutations for variant 3 include: (Y32L/H), (R91L/G), (S92L/W), (N93L/G), (D98H/I), (I99H/W), (Q100H/L), (Q100H/L), and (G102H/V).
Linker addition
In predicting the 3D structure, the light and heavy chains of the antibody are considered separately without any linking amino acids. As in recombinant antibody production, a linker was used to join two chains; The light and heavy chains were interconnected with three repeats of (Gly4Ser)3.
Immunotoxin design and modelling
The structure of GrB, the toxic domain of immunotoxin, was retrieved from Protein Data Bank (PDB). The acquired structure was in dimeric form with ligand interactions. The molecular operating environment (MOE) was employed to eliminate the ligands and one of the monomers. To increase the GrB toxicity, an adaptor sequence “RHRQPRGNRVRRSPLSSIFSRIGDPYVHDEVDRGP” was attached beforehand of the interpolation of antibody and GrB.
Thereafter, the final structure of immunotoxin combined with GrB and the antibody were predicted using Modeler software (homology modeling). Therefore, Modeler aligns the immunotoxin sequence with the pre-existing structures; the immunotoxin structure attained based on antibody (containing linker) and preparing GrB as templates.
To calculate the root-mean-square deviation (RMSD), a structural alignment between the whole immunotoxin structure and its determined separate parts (antibody, GrB, and linker) was performed using the MOE. This analysis checks any negative structural impacts that incorporation of several parts may have made on the final structure.
Energy minimization and structure refinement
To achieve a more stable and pseudo-native structure energy minimization and refinement was performed (Ganji et al. 2020).
To refine the conformation of the loops in energy minimized model we used the loop model class in MODELLER. The structure refinement depicts the initial models closer to their native state, in terms of hydrogen bonds, backbone topology and side-chain positioning. Also it makes considerable improvement in physical quality of local structures (Estébanez-Perpiñá et al. 2000).
MODELLER is used for homology or comparative modeling of protein three-dimensional structures. The user provides an alignment of a sequence to be modeled with known related structures. The MODELLER automatically calculates a model containing all non-hydrogen atoms. MODELLER implements comparative protein structure modeling by satisfaction of spatial restraints, and can perform many additional tasks, including de novo modeling of loops in protein structures, optimization of various models of protein structure with respect to a flexibly defined objective function, multiple alignment of protein sequences and/or structures, clustering, searching of sequence databases, comparison of protein structures, etc.
Model validation
In order to confirm the accuracy of the 3D structure, secondary structure of the immunotoxin was predicted applying PSIPRED (McGuffin et al. 2000), PRABI (Li et al. 2017), and Jpred (Drozdetskiy et al. 2015).
PSI-blast based secondary structure PREDiction (PSIPRED) is a method used to investigate protein structure. It uses artificial neural network machine-learning methods in its algorithm. It is a server-side program, featuring a website serving as a front-end interface, which can predict a protein's secondary structure (beta sheets, alpha helixes and coils) from the primary sequence. PRABI SOPMA ((Rhone-Alpes Bioinformatics Center Self-Optimized Prediction Method with Alignment) is based on the homologue method. JPred is a Protein Secondary Structure Prediction server. JPred incorporates the Jnet algorithm in order to make predictions that are more accurate.
Thereafter, the secondary structure of the modeled molecules was compared with the predicted secondary structure of Discovery Studio Visualizer to assert if they are in concordance with each other.
To determine the quality of the immunotoxin 3D model before and after refinement the QMEAN was used (Bahrami et al. 2020). QMEAN, the Qualitative Model Energy ANalysis, is a composite scoring function assessing the major geometrical aspects of protein structures (Benkert et al. 2008).
Immunotoxin characterization
The physicochemical features of the immunotoxin was characterized using ProtParam tool (Gasteiger et al. 2005). ProtParam is a tool, which allows the computation of various physical and chemical parameters. The computed parameters include the molecular weight, theoretical pI, amino acid composition, atomic composition, extinction coefficient, estimated half-life, instability index, aliphatic index, and grand average of hydropathicity (GRAVY).
In the following, the antigenicity of the designed immunotoxin was surveyed applying VaxiJen (Doytchinova and Flower 2007) and ToxiPred (Gupta et al. 2015).
VaxiJen is the first server for alignment-independent prediction of protective antigens. It was developed to allow antigen classification solely based on the physicochemical properties of proteins without recourse to sequence alignment. ToxiPred is an in silico method, which is developed to predict and design toxic/non-toxic peptides. The main dataset used in this method consists of 1805 toxic peptides (≤ 35 residues).
AlgPred (Saha and Raghava 2006) was used to determine the allergenicity of the designed molecule. Allergenic protein prediction (Algpred) allows prediction of allergens based on similarity of known epitope with any region of protein. Propred (Singh and Raghava 2001) and Propred-1 were utilized to specify T-cell epitopes 1 and 2, respectively. ProPred-1 is an online web tool for the prediction of peptide binding to MHC class-I alleles. The aim of Propred server is to predict MHC Class-II binding regions in an antigen sequence, using quantitative matrices.
Prediction of B-cell epitopes was performed using Ellipro (Ponomarenko et al. 2008), Bepipred (Jespersen et al. 2017), and SVMtrip (Yao et al. 2012). ElliPro predicts linear and discontinuous antibody epitopes based on a protein 3D structure. The BepiPred-2.0 server predicts B-cell epitopes from a protein sequence, using a Random Forest algorithm trained on epitopes and non-epitope amino acids determined from crystal structures. SVMtrip developed a new method to predict antigenic epitope with sequence input (Sefid et al. 2019).
Immunotoxin binding efficiency
The Possibility of immunotoxin binding to its receptor can be analyzed through molecular docking study. First, the 3D structure of CD20 was obtained from PDB and edited using MOE. As this structure was in complex form with Nivolumab-Fab, the extra domains and ligands so were removed by MOE.
The possible interaction of CD20-immunotoxin was predicted employing Zdock, ClusPro (Kozakov et al. 2017), HADDOCK (Dominguez et al. 2003) and Hex8 servers.
ZDOCK, an automatic protein docking serve, searches all possible binding modes in the translational and rotational space between the two proteins and evaluates each pose using an energy-based scoring function.
The ClusPro server is a widely used tool for protein–protein docking. The server provides a simple home page for basic use, requiring only two files in Protein Data Bank format. Six different energy functions can be used depending on the type of proteins. Hex Server is the First Fourier transform (FFT)-based protein docking server to be powered by graphics processors. Using two graphics processors simultaneously which is up to two orders of magnitude faster than conventional FFT-based docking approaches using comparable resolution and scoring functions. High Ambiguity Driven protein–protein DOCKing (HADDOCK) is an integrative platform for the modeling of biomolecular complexes that supports a large variety of input data and can deal multi-component assembles of proteins, peptide, small molecules and nucleic acids.
Moreover, PRODIGY server (Xue et al. 2016) (https://wenmr.science.uu.nl/prodigy/) was used to calculate the Gibbs free energy. The less the energy, the reaction between the two is more stable.
Results
Sequence retrieval
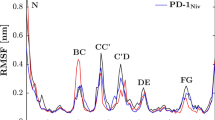
The UniProtKB P10144 was retrieved as the GrB protein sequence. The 6Y92 PDB record was detected for the structure of full-length CD20 in complex with Ofatumumab Fab. The acquired structure had 6 chains. Chain C, D, H, and L belongs to the Ofatumumab (Heavy and light chains) in dimer form, and chains A and B belong to the CD20 in dimer form. All non-protein atoms were eliminated to achieve the CD20 monomer structure using the MOE. Figure 1 indicates the CD20 structure before and after monomerization.
Linker addition
The antibody structure in join with the linker was successfully modeled via Modeler 9.25. The obtained structure in which two chains of the antibody are linked as a single chain is shown in Fig. 2. The results of MOE confirmed that linker addition does not cause a notable change in the original antibody structure. The RMSD was calculated to be 0.83 Å. The superimposed structures of the antibody before and after linker addition is illustrated in Fig. 3.
Immunotoxin assembly
The PDB record of 1FQ3 was saved for the GrB structure. To monomerize the GrB, all the non-protein atoms and the extra structures of GrB were removed. Figure 4 indicates the GrB structure before/after monomerization. The final sequence of immunotoxin is represented in Fig. 5. In addition, the final structure of the immunotoxin after the homology modelling shows in Fig. 6. The calculated RMSD between the enzyme and immunotoxin was 0.21 Å, while this value was 0.20 Å between the antibody and the immunotoxin. These results indicate that there are no significant changes in the native structure of each region. All immunotoxin domains can have their own performance without being compromised. Figure 7 illustrates the immunotoxin structure after alignment.
Energy minimization, refinement and Model validation
Energy minimization run have improved the structure and reduced the total energy to a more stable state. The energy minimized structure used as an input for refinement. The comparison of refined structure to the initial model showed slight difference between these two. The calculated RMSD of 1.447 Å implies that energy minimization and refinement did not make significant changes on the structure. Figure 8 shows the structural alignment of the immunotoxin before and after refinement.
The QMEAN score of immunotoxin structure before and after refinement calculated − 3.68 and − 2.81, respectively (the higher the score, the more reliable is the structure). Figure 9 shows the validation of the immunotoxin structure before and after refinement.
Immunotoxin characterization
The isoelectric point (pI) and molecular weight of the immunotoxin were computed to be 9.53 and 55,712.16 Da, respectively. The protein half-life was within E. coli was estimated 10 h.
The ToxiPred predicts no toxic peptide along the immunotoxin sequence. Based on the AlgPred results, the designed immunotoxin has no significant allergen potency (Score = − 1.2397132, Threshold = − 0.4). The result of antigenicity demonstrated that the designed immunotoxin is a probable ANTIGEN (Score = 0.7615, Threshold = 0.4).
Based on the results of Propred and Propred-1, the immunotoxin contains 75 and 94 T-cell epitopes of type 1 and 2, respectively. The Ellipro clarified 12 linear and 6 structural B-cell epitopes. Bepipred and SVMtrip also identified 25 and 7 linear epitopes in the immunotoxin sequence, respectively.
Immunotoxin binding efficiency
The Zdock, Cluspro, Hex8 and HADDOCK results displayed the possibility of the immunotoxin interaction to the CD20. The residues 74–80 (AGIYAPI) and residues 145–161 (HFLKMESLNFIRAHTPY) from CD20, residues 32L, 91L, 92L, 93L, 98H, 99H, 100H and 102H from the antibody structure were specified as active residues (directly involved in the interaction).
Figure 10 indicates the docking results for the interaction of immunotoxin-CD20 in comparison to Ofatumumab-CD20. The results demonstrated that CD20 loops contribute to react with the immunotoxin. Also, the function of immunotoxin in binding with CD20 is shown in Fig. 11.
Table 1 shows the docking information for immunotoxin/antibody with CD20 antigen. The results of binding energy calculation indicated that immunotoxin binds to the CD20 with high affinity. The antibody-CD20 complex was checked in term of binding affinity as a positive control to compare with the immunotoxin-CD20 complex.
Discussion
Given the high specificity and affinity of antibodies against the target molecules, they are known as authentic drug candidates (Adams and Weiner 2005). Identification of novel tumor‐associated antigens leads to provide developed therapeutic cancer‐targeting antibodies.
Increasingly, antibodies are used to treat leukemia. Phage display technique plays an essential role in the development of human antibodies (Jaglowski et al. 2010). In last decades, anti‐CD20 mAbs have changed the landscape of CLL patient management (Jaglowski et al. 2010). Rituximab was the first mAb to be approved in 1998. Since then, two other anti‐CD20 mAbs were granted EMA approval for CLL (Payandeh et al. 2014). The mechanisms of action anti‐CD20 mAbs to induce tumor cell death include the antibody‐dependent cell‐mediated cytotoxicity (ADCC), complement‐dependent cytotoxicity (CDC), and direct cellular cytotoxicity (DCC). Single‐agent activity of Rituximab is limited in B‐cell malignancy. New treatment options are Rituximab plus Fludarabine and Cyclophosphamide (Robak et al. 2010).
To use mAbs as therapeutic agents, several properties should be optimized including binding affinity and specificity, folding stability, solubility, pharmacokinetics, effector functions, compatibility with the binding to additional antibody domains (bispecific antibodies), and cytotoxic drugs (antibody‐drug conjugates). Although current experimental approaches are costly and ponderous, antibody engineering can profoundly progress the therapeutic efficiency of the mAbs. Moreover, the potential of in vitro selection methods can be further enhanced through rational design (Payandeh et al. 2018).
In our previous study, protein-engineering strategies have been exploited to increase and optimize the affinity of Ofatumumab antibody against the CD20 marker. Finding the functionally pivotal amino acids of the Ofatumumab and replacing them with suitable amino acids have increased the antibody affinity for its marker. These alterations could increase the Mab activity compared to the original commercial type (Payandeh et al. 2019b).
In a study by Luca Bologna et al., the rituximab and Ofatumumab antibodies were compared. They have reported that the Ofatumumab antibody lysed the whole blood cells faster and more efficient than rituximab. The rate of cell lyses by Ofatumumab was about 56% in 24 h, while it was about 16% for rituximab (Bologna et al. 2013).
Based on reports single‐agent Ofatumumab has resulted in overall response rates of 42–51% in relapsed/refractory CLL (up to 80% when combined with chemotherapy). In the case of de novo CLL, overall response rates of 77–78% have been achieved (Payandeh et al. 2019a; Pawluczkowycz et al. 2009; Taylor et al. 2011).
GrB induces apoptosis by mimicking caspase activity and activating caspase-3 and other caspases. Studies showed that various fusion constructs targeting tumor cells and tumor endothelium and containing GrB have impressive proapoptotic and cytotoxic activity (Kanatani et al. 2011; Liu et al. 2003). Because endogenous GrB is present in plasma in both normal and pathologic states, it is unlikely that this molecule would engender an immune response. Studies by Wang and colleagues suggested that incorporation of a furin-sensitive linker into GrB-based fusion constructs may promote effective cytoplasmic delivery of an active GrB fragment into target cells (Wang et al. 2007).
The designed immunotoxin consists of an anti-CD20 antibody (engineered Ofatumumab) connected with GrB as its toxic part. A valid sequence of Ofatumumab was employed to predict the 3D structure of the binding region for the designed immunotoxin. Since each server performs based on specific algorithms and exclusive methods. Therefore, the usage of several tools and software to predict is more reliable. We used fully humanized sequence of the antibody to ensure its safety for in vivo applications. GrB is able to activate apoptosis in various ways, which this ability is considered one of its advantages. This feature could help to bypass resistant cells after frequent stimulations. The pivotal role of the linker length in the formation of complexes with different valences and the orientation of the scFv assembly is already shown. Complementary Vh and Vl pairs could be associate to form a bivalent dimer called a diabody by using flexible linkers. These linkers are mainly a combination of serine and glycine amino acids with a length of 12 (Kortt et al. 2001).
In antibody studies the (Gly4Ser)n is the most usual linker that utilized to join two chains (Ganji et al. 2020).
In order to design the recombinant antibody, we added a linker to join two chains. The selected linker could be effective in the reduction of interference amongst domains (Kreitman 2006; Akbari et al. 2017). In addition, the added linker sequence to our designed molecule did not change the features of the immunotoxin (Bahrami et al. 2020). The RMSD calculations indicated that the final molecule did not undergo a drastic structural change. Homology modeling was performed on the final structure of the immunotoxin and the results represented the stability of each segment, confirmed by RMSD calculation. The finally modeled immunotoxin represented no changes in the native structure of each part and can be employed as a valid complex for more analysis. The calculated RMSD between the granzyme/antibody and the immunotoxin showed which the final structure has the qualified functionality.
The directional binding of the immunotoxin reveals its ability to bind to the CD20 through integrin-binding region. This interaction enables the immunotoxin to hinder leukocytes from binding to PD-L1 expressing cells as well as directing the immunotoxin to its desired target. Thus, increases the cytotoxic activity.
According to our results, the designed immunotoxin has a great chance of soluble expression in the periplasm of E. coli. In this study, the Granzyme B molecule is used as a part of the design. This enzyme consists of three disulfide bonds and the periplasm of the E. coli is the only oxidizing compartment in genetically unmodified E. coli cells that has the ability to create these disulfide bonds. Furthermore, this compartment has several different chaperones and foldases. These characteristics improve the proper protein folding and disulfide bond formation, which is common to restore the biological functions of expressed proteins. Soluble proteins have simpler downstream purification processes and they are more likely to obtain correct folding and biological activity (Sørensen and Mortensen 2005). Since the designed immunotoxin is predicted with significant soluble expression, it represents which the designed antigen could be suitable to expressed, purified, and used for therapeutic applications.
Conclusion
Anti‐CD20 mAbs have changed the landscape of CLL patient management. Designing an efficient immunotoxin against B-CLL could be an effective alternative strategy in fight against this disease. In this regard, we have designed an immunotoxin composed of Ofatumumab and GrB, which would bring about less immunogenicity. This bifunctional, immunotoxin-like protein employing human GrB as an effector domain that could be readily expressed with high yield in the bacterial expression systems. Upon specific binding to ofatumumab on the surface of tumor B-cells, the chimeric molecules are internalized that could be released into the cytosol by an endosomolytic reagent, resulting in rapid and efficient cell killing. According to the obtained results, the designed immunotoxin is endowed with desirable properties and the capability to interact with the CD20 molecule in correct orientation.
References
Adams GP, Weiner LM (2005) Monoclonal antibody therapy of cancer. Nat Biotechnol 23(9):1147–1157
Akbari B, Farajnia S, AhdiKhosroshahi S, Safari F, Yousefi M, Dariushnejad H et al (2017) Immunotoxins in cancer therapy: review and update. Int Rev Immunol 36(4):207–219
Bahrami AA, Payandeh Z, Khalili S, Zakeri A, Bandehpour M (2019) Immunoinformatics: in silico approaches and computational design of a multi-epitope, immunogenic protein. Int Rev Immunol 38(6):307–322
Bahrami AA, Bandehpour M, Khalesi B, Kazemi B (2020) Computational design and analysis of a poly-epitope fusion protein: a new vaccine candidate for hepatitis and poliovirus. Int J Pept Res Ther 26(1):389–403
Benkert P, Tosatto SC, Schomburg D (2008) QMEAN: A comprehensive scoring function for model quality assessment. Proteins Struct Funct Bioinform 71(1):261–277
Bologna L, Gotti E, Da Roit F, Intermesoli T, Rambaldi A, Introna M et al (2013) Ofatumumab is more efficient than rituximab in lysing B chronic lymphocytic leukemia cells in whole blood and in combination with chemotherapy. J Immunol 190(1):231–239
Czuczman MS, Gregory SA (2010) The future of CD20 monoclonal antibody therapy in B-cell malignancies. Leuk Lymphoma 51(6):983–994
Danilov AV (2013) Targeted therapy in chronic lymphocytic leukemia: past, present, and future. Clin Ther 35(9):1258–1270
Dominguez C, Boelens R, Bonvin AM (2003) HADDOCK: a protein− protein docking approach based on biochemical or biophysical information. J Am Chem Soc 125(7):1731–1737
Doytchinova IA, Flower DR (2007) VaxiJen: a server for prediction of protective antigens, tumour antigens and subunit vaccines. BMC Bioinform 8(1):4
Drozdetskiy A, Cole C, Procter J, Barton GJ (2015) JPred4: a protein secondary structure prediction server. Nucleic Acids Res 43(W1):W389–W394
Estébanez-Perpiñá E, Fuentes-Prior P, Belorgey D, Braun M, Kiefersauer R, Maskos K et al (2000) Crystal structure of the caspase activator human granzyme B, a proteinase highly specific for an Asp-P1 residue. Biol Chem 381(12):1203–1214
Ganji M, Khalili S, Mard-Soltani M, Khalesi B, Karkhah A, Amani J (2020) A precisely designed immunotoxin against VCAM1 consisting of a humanized antibody variable domain fused to granzyme: an in silico approach. Int J Pept Res Ther 26(1):129–137
Greenberg EM, Probst A (2013) Chronic leukemia. Crit Care Nurs Clin 25(4):459–470
Gasteiger E, Hoogland C, Gattiker A, Wilkins MR, Appel RD, Bairoch A (2005) Protein identification and analysis tools on the ExPASy server. In: The proteomics protocols handbook. pp 571–607
Gupta S, Kapoor P, Chaudhary K, Gautam A, Kumar R, Raghava GP (2015) Peptide toxicity prediction. In: Computational peptidology, pp 143–157. Humana Press, New York, NY
Hehmann-Titt G, Schiffer S, Berges N, Melmer G, Barth S (2013) Improving the therapeutic potential of human granzyme B for targeted cancer therapy. Antibodies 2(1):19–49
Herishanu Y, Katz B-Z, Lipsky A, Wiestner A (2013) Biology of chronic lymphocytic leukemia in different microenvironments: clinical and therapeutic implications. Hematology/Oncol Clin 27(2):173–206
Huff CA, Matsui W (2008) Multiple myeloma cancer stem cells. J Clin Oncol 26(17):2895
Jaglowski SM, Alinari L, Lapalombella R, Muthusamy N, Byrd JC (2010) The clinical application of monoclonal antibodies in chronic lymphocytic leukemia. Blood 116(19):3705–3714
Jespersen MC, Peters B, Nielsen M, Marcatili P (2017) BepiPred-20: improving sequence-based B-cell epitope prediction using conformational epitopes. Nucleic Acids Res 45(W1):W24–W29
Kanatani I, Lin X, Yuan X, Manorek G, Shang X, Cheung LH et al (2011) Targeting granzyme B to tumor cells using a yoked human chorionic gonadotropin. Cancer Chemother Pharmacol 68(4):979–990
Khalili S, Rasaee MJ, Mousavi SL, Amani J, Jahangiri A, Borna H (2017) In silico prediction and in vitro verification of a novel multi-epitope antigen for HBV detection. Mol Genet Microbiol Virol 32(4):230–240
Klein C, Lammens A, Schäfer W, Georges G, Schwaiger M, Mössner E et al (eds) (2013) Epitope interactions of monoclonal antibodies targeting CD20 and their relationship to functional properties. MAbs, Taylor & Francis
Kortt AA, Dolezal O, Power BE, Hudson PJ (2001) Dimeric and trimeric antibodies: high avidity scFvs for cancer targeting. Biomol Eng 18(3):95–108
Kozakov D, Hall DR, Xia B, Porter KA, Padhorny D, Yueh C et al (2017) The ClusPro web server for protein–protein docking. Nat Protoc 12(2):255
Kreitman RJ (2006) Immunotoxins for targeted cancer therapy. AAPS J 8(3):E532–E551
Li Z, Wang J, Zhang S, Zhang Q, Wu W (2017) A new hybrid coding for protein secondary structure prediction based on primary structure similarity. Gene 618:8–13
Lin TS (2010) Ofatumumab: a novel monoclonal anti-CD20 antibody. Pharmacogenom Pers Med 3:51
Liu Y, Cheung LH, Hittelman WN, Rosenblum MG (2003) Targeted delivery of human pro-apoptotic enzymes to tumor cells: In vitro studies describing a novel class of recombinant highly cytotoxic agents. Mol Cancer Ther 2(12):1341–1350
McGuffin LJ, Bryson K, Jones DT (2000) The PSIPRED protein structure prediction server. Bioinformatics 16(4):404–405
Mohan G, TP AH, Jijo AJ, KM SD, Narayanasamy A, Vellingiri B (2019) Recent advances in radiotherapy and its associated side effects in cancer—a review. J Basic Appl Zool 80(1)
Pawluczkowycz AW, Beurskens FJ, Beum PV, Lindorfer MA, van de Winkel JG, Parren PW et al (2009) Binding of submaximal C1q promotes complement-dependent cytotoxicity (CDC) of B cells opsonized with anti-CD20 mAbs ofatumumab (OFA) or rituximab (RTX): considerably higher levels of CDC are induced by OFA than by RTX. J Immunol 183(1):749–758
Payandeh Z, Rasooli I, MousaviGargari SL, RajabiBazl M, Ebrahimizadeh W (2014) Immunoreaction of a recombinant nanobody from camelid single domain antibody fragment with Acinetobacter baumannii. Trans R Soc Trop Med Hyg 108(2):92–98
Payandeh Z, Rajabibazl M, Mortazavi Y, Rahimpour A, Taromchi AH (2018) Ofatumumab monoclonal antibody affinity maturation through in silico modeling. Iran Biomed J 22(3):180
Payandeh Z, Bahrami AA, Hoseinpoor R, Mortazavi Y, Rajabibazl M, Rahimpour A et al (2019a) The applications of anti-CD20 antibodies to treat various B cells disorders. Biomed Pharmacother 109:2415–2426
Payandeh Z, Rajabibazl M, Mortazavi Y, Rahimpour A, Taromchi AH, Dastmalchi S (2019b) Affinity maturation and characterization of the ofatumumab monoclonal antibody. J Cell Biochem 120(1):940–950
Ponomarenko J, Bui H-H, Li W, Fusseder N, Bourne PE, Sette A et al (2008) ElliPro: a new structure-based tool for the prediction of antibody epitopes. BMC Bioinformatics 9(1):514
Robak T, Dmoszynska A, Solal-Céligny P, Warzocha K, Loscertales J, Catalano J et al (2010) Rituximab plus fludarabine and cyclophosphamide prolongs progression-free survival compared with fludarabine and cyclophosphamide alone in previously treated chronic lymphocytic leukemia. J Clin Oncol 28(10):1756–1765
Rousalova I, Krepela E (2010) Granzyme B-induced apoptosis in cancer cells and its regulation. Int J Oncol 37(6):1361–1378
Saha S, Raghava G (2006) AlgPred: prediction of allergenic proteins and mapping of IgE epitopes. Nucleic Acids Res 34(suppl_2):W202–W209
Sefid F, Bahrami AA, Darvish M, Nazarpour R, Payandeh Z (2019) In silico analysis for determination and validation of iron-regulated protein from Escherichia coli. Int J Pept Res Ther 25(4):1523–1537
Singh H, Raghava G (2001) ProPred: prediction of HLA-DR binding sites. Bioinformatics 17(12):1236–1237
Sørensen HP, Mortensen KK (2005) Soluble expression of recombinant proteins in the cytoplasm of Escherichia coli. Microb Cell Fact 4(1):1
Taylor PC, Quattrocchi E, Mallett S, Kurrasch R, Petersen J, Chang DJ (2011) Ofatumumab, a fully human anti-CD20 monoclonal antibody, in biological-naive, rheumatoid arthritis patients with an inadequate response to methotrexate: a randomised, double-blind, placebo-controlled clinical trial. Ann Rheum Dis 70(12):2119–2125
Thomas DA, O’Brien S, Jorgensen JL, Cortes J, Faderl S, Garcia-Manero G et al (2009) Prognostic significance of CD20 expression in adults with de novo precursor B-lineage acute lymphoblastic leukemia. Blood J Am Soc Hematol 113(25):6330–6337
Tsai D-Y, Hung K-H, Chang C-W, Lin K-I (2019) Regulatory mechanisms of B cell responses and the implication in B cell-related diseases. J Biomed Sci 26(1):64
Wang T, Zhao J, Ren J-L, Zhang L, Wen W-H, Zhang R et al (2007) Recombinant immunoproapoptotic proteins with furin site can translocate and kill HER2-positive cancer cells. Can Res 67(24):11830–11839
Xu Y, Li S, Wang Y, Liu J, Mao X, Xing H et al (2019) Induced CD20 expression on B-cell malignant cells heightened the cytotoxic activity of chimeric antigen receptor engineered T cells. Hum Gene Ther 30(4):497–510
Xue LC, Rodrigues JP, Kastritis PL, Bonvin AM, Vangone A (2016) PRODIGY: a web server for predicting the binding affinity of protein–protein complexes. Bioinformatics 32(23):3676–3678
Yao B, Zhang L, Liang S, Zhang C (2012) SVMTriP: a method to predict antigenic epitopes using support vector machine to integrate tri-peptide similarity and propensity. PLoS ONE 7(9):e45152
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
All authors declare no potential conflicts of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Sefid, F., Alagheband Bahrami, A., Payandeh, Z. et al. Ofatumumab and Granzyme B as immunotoxin against CD20 antigen. In Silico Pharmacol. 10, 6 (2022). https://doi.org/10.1007/s40203-022-00120-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s40203-022-00120-6