Abstract
Background
Type 2 diabetes mellitus (T2DM) is recognized as a complex metabolic which has affected the lives of millions of people around the world. Vitamin D receptor (VDR) gene polymorphisms have been suggested to be a vital contributor to the development of T2DM. However, the association between VDR gene polymorphisms and T2DM remains controversial. We have investigated the association between two VDR gene polymorphisms (rs731236 and rs1544410) and T2DM in an Iranian population.
Methods
A total of 148 T2DM patients and 100 normal controls were recruited in this study. We used polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) analysis to perform genotyping.
Results
The results of the present research revealed that the frequency of the rs731236 C allele was significantly higher in T2DM patients than in normal controls (p = 0.044). The CC genotype of rs731236 was connected with an increased risk of T2DM (OR = 2.85, 95% CI = 1.06–7.69, p = 0.039). However, no significant difference in the frequency of the rs1544410 C allele between T2DM patients and normal controls was observed (p = 0.918).
Conclusion
Our findings were suggestive of the rs731236 polymorphism of the VDR as a risk factor for developing T2DM in the Iranian population, while rs1544410 polymorphism may not be associated with T2DM susceptibility. Further research is needed to approve these findings in other populations and to clarify the underlying mechanisms involved in such an association.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The most common form of diabetes is type 2 diabetes mellitus (T2DM) which includes approximately 90% of all cases of diabetes. The International Diabetes Federation (IDF) reports that there were 424.9 million people worldwide with diabetes in 2017, and this number is projected to increase to 628.6 million by 2045 [1]. Diabetes accounts for nearly 4% of all causes of death and is the third leading cause of death due to non-communicable diseases globally [2]. In Iran, the prevalence of diabetes has risen from 11% to 2016 to 14% in 2021, representing a significant increase of 30% over five years that this trend is expected to continue [3].
The vitamin D endocrine system is characterized by the secosteroid hormone vitamin D, its receptor (VDR), and the metabolizing enzymes involved in the synthesis of the physiologically active form of the hormone. This system is critical in skeletal metabolism, particularly intestinal calcium absorption, but it has also been found to be essential in other metabolic pathways, such as those implicated in immune response and cancer [4].
Vitamin D has a vital role in the regulation of insulin secretion maintained by β-cells which tightly control calcium concentration and flux [5]. Low vitamin D status has been recognized as a risk factor for obesity, diabetes, and cardiovascular disease. Recent studies have revealed that high vitamin D levels offer protection against type 2 diabetes [6]. Vitamin D deficiency can lead to reduced insulin secretion in humans, while supplementation improves glucose tolerance. Vitamin D exerts its biological effects by interacting with its receptor, which belongs to the steroid receptor superfamily and possesses a DNA binding domain. Any alteration in the structure of the vitamin D receptor or changes in the vitamin D production gene can enhance the risk of developing type 2 diabetes [7].
The VDR gene, consisting of 11 exons, is located on chromosome 12q13.1 [8]. Single nucleotide polymorphisms (SNPs) in the VDR gene, such as Apal (C/A, rs795232, intron 8), Bms1 (G/A, rs1544410, intron 8), Fok1 (C/T, rs10735810, Exon 2), and Taq1 (T/C, rs731236, exon 9), may restrict the formation of the vitamin D-VDR complex and reduce vitamin D absorption even in the presence of normal serum levels [9]. These variations cause mRNA instability and elicit different isoforms or conformational changes in the protein’s structure (10).
Although some studies have examined the association between vitamin D levels and VDR polymorphisms, the results are indecisive [10]. Therefore, the present research aimed to evaluate the association between diabetes risk and VDR gene polymorphisms (Taq1 and Bsm1) in a group of Iranian patients with type 2 diabetes [11].
Materials and methods
Patient’s characteristics
The study included 148 individuals with T2DM and a control group of 100 healthy individuals aged between 20 and 70 years old with a body mass index (BMI) ranging from 25 to 35. Participants with fasting blood sugar (FBS) levels ≥ 126 mg/dL and hemoglobin A1C (HbA1C) levels ≥ 6.5% were defined as the patient group while the control group was defined as those with fasting blood sugar levels < 100 mg/dL and anti-diabetic agent consumption and past history of diabetes were excluded. Patients with malignancies, endocrine disorders, chronic renal failure, severe liver disease, systemic inflammatory disease or corticosteroid treatment were excluded from the study. Demographic information including age, height, weight, diabetes duration and treatment duration with hypoglycemic drugs were collected from all subjects at baseline. Patients were also interviewed regarding their family history of diabetes, smoking habits, alcohol consumption patterns, exercise routines, hypertension, cardiovascular disease, nephropathy, retinopathy and neuropathy.
Genotyping
Ten milliliters of blood samples were collected from all patients and stored in tubes containing 0.5 M EDTA. DNA was extracted from peripheral blood leukocytes using a standard salting-out method [12].
Genotyping was performed using the polymerase chain reaction-restriction (PCR-RFLP) with the Bsm1 (G/A, rs1544410) and Taq1 (T/C, rs731236) variants in the VDR gene. The primer sequences used for amplifying DNA were listed in Table 1. PCR amplification was carried out in a final reaction volume of 25 µl, which contained 12 µl dH2O, 10 µl Red Master Mix, 1 µl each of forward and reverse primers (10 pmol/uL), and 1 µl DNA template. The PCR conditions were as follows: initial denaturation at 95ºC for 5 min, followed by 35 cycles with denaturation at 95ºC for 50s, annealing at 59.3ºC for 50s, and extension at 72ºC for 50s. The final extension was at 72ºC for 10 min. The PCR products were separated on a SYBR green-stained agarose gel to confirm their quality. Additionally, ten microliters of PCR product were digested with BsmI restriction enzyme for rs1544410 and Taq1 restriction enzyme for rs731236.
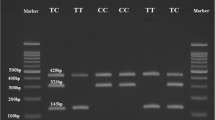
After restriction digestion, the homozygous AA genotype in rs1544410 was identified by an uncut fragment of 825 bp, while the homozygous mutant GG genotype was represented by two bands at 650 and 175 bps. The heterozygous AG genotype was observed at 825, 650, and 175 bps. In rs731236, the homozygous mutant TT genotype was identified by two bands at 495 and 245 bps, while the homozygous mutant CC genotype was represented by three bands at 290,245, and 205 bps. The heterozygous TC genotype was observed at 495, 290, 245 and 205bps (Table 2).
Statistical analysis
We tested our hypothesis using the Chi square test to compare diabetic and non-diabetic subjects regarding different genotypes of the polymorphism. Univariate and multivariate logistic regression models were applied to assess the association between genotypes and diabetes mellitus. All statistical analyses were performed using STATA software version 14. A P value less than 0.05 revealed strong evidence of statistical significance.
Results
During the study period, a total of 248 patients were genotyped, comprising 148 diabetic patients (59.68%) and 100 (40.32%) healthy controls. Of these, 165 (66.53%) were female and 140 (56.68%) had ethnicities other than Fars. The mean age of participants was 49.53 (SD = 14.72) years, ages ranging between 19 and 76 years. We observed no significant difference in gender distribution between diabetic and non-diabetic subjects (p = 0.333). However, Fars ethnicity had a significantly higher population among non-diabetics than diabetics in the present study (p < 0.001). Furthermore, patients with diabetes mellitus were significantly older (p = 0.0001) and had higher body mass indices (p = 0.001) compared to those without diabetes mellitus (Table 2). The frequencies of GG, AG, and AA genotypes of rs1544410 polymorphism among the study subjects were 22.18%, 60.08%, and 17.74%, respectively, while genotyping in rs731236 showed that 32.66%, 36.69%, and 30.65% had TT, TC, and CC genotypes, respectively.
Rs1544410 variant of the VDR
Table 3 reveals the absence of statistically significant difference between diabetic patients and non-diabetic ones regarding AG and AA genotypes (p = 0.956). Crude and adjusted odds ratios for the heterozygote genotype were found to be non-significant at 1.1 (p = 0.77) and 0.93 (p = 0.88), respectively; corresponding figures for the homozygote genotype were also non-significant at p = 0.93 and p = 0.67 respectively. However, further studies with larger sample sizes are needed to confirm these results.
The frequency of the A allele was found to be similar in both diabetic and non-diabetic subjects (p = 0.918), with no statistically significant odds ratio for its presence (p = 0.918) (Table 4).
While the TC and CC genotypes of rs731236 polymorphism were more common in diabetic patients than non-diabetics, it didn’t show a statistically significant difference (p = 0.185). The crude odds ratios for TC and CC were also not statistically significant (p = 0.201 for TC and p = 0.078 for CC). However, after controlling for potential confounding demographic factors, the adjusted odds ratios for TC and CC were found to be 2.05 (p = 0.116) and 2.85 (p = 0.039), respectively (Table 5). Furthermore, analysis of the alleles revealed a higher frequency of the C allele in diabetic patients compared to non-diabetics (p = 0.044), resulting in a statistically significant odds ratio for its presence (p = 0.044) (Table 7).
Discussion
The vitamin D receptor (VDR) gene is a main regulator of vitamin D metabolism and signaling. Vitamin D is a hormone with a vital role in maintaining blood sugar levels. Low levels of vitamin D have been associated with an increased risk of T2D [13].
The VDR gene has several polymorphisms, which are variations in the DNA sequence. Two of these polymorphisms, rs731236 and rs1544410, have been investigated for their association with T2D [14]. The rs731236 polymorphism is commonly known as TaqI and is located in exon 9 (allele T is often referred to as allele T, and allele C is allele t). The rs1544410 polymorphism is a restriction fragment length polymorphism (RFLP), which means that a different length of DNA is present depending on the genotype. This variant is universally known as BsmI and is located in intron 8 (allele C is commonly referred to as allele b and allele T is referred as allele B) [15].
In our study, we observed that diabetic patients had higher frequencies of the C allele and its genotypes for rs731236 polymorphism. Although heterozygote genotype for rs731236 polymorphism had no significant association with developing diabetes mellitus, presence of homozygote genotype caused approximately a three-fold surge in the risk of developing diabetes mellitus. The same was true for carriers of the C allele, who had about a 45% higher risk of developing diabetes mellitus, indicating that this allele can be considered a risk factor for diabetes mellitus.
The presence of genotypes and alleles of the rs1544410 polymorphism of the VDR gene was found to be similar in diabetic patients compared to the general population. Although, the associations were not statistically significant but the statistical models demonstrated that the heterozygote genotype of this polymorphism reduced the chance of developing diabetes mellitus by 7%, while the homozygote genotype increased the risk by 29%. Therefore, this nonsignificant association demonstrates the importance of the necessity of more investigations with larger sample size.
Additionally, although the presence of the A allele increased the chance of diabetes by 2%, this association was also non-significant. Therefore, this polymorphism cannot be considered a risk factor for developing diabetes mellitus.
These findings suggest that rs731236 polymorphism seems to be linked with an increased risk of developing diabetes. Our study adds to a growing body of literature on this subject. Several other studies have also explored the association between rs731236 polymorphism and diabetes risk, with mixed results.
Several studies have been conducted to explore the association between rs731236 and rs1544410 T2D suggesting inconsistent results, with some studies showing an association and others showing no association.
One study conducted by Nosratabadi et al. (2011) investigated the association between rs731236 polymorphism and T2DM in a group of southeastern Iranian patients. The findings showed that individuals with the CC genotype had a significantly higher chance of developing type 2 diabetes compared to those with the TT or TC genotypes, even after adjusting for confounding factors such as age, family history of diabetes, and BMI [16]. A study conducted in a Chinese population by Zhang et al. (2012) investigated the association between the rs731236 polymorphism of VDR and the risk of developing T2DM. The study revealed a significantly higher risk of developing T2DM among individuals with the CC genotype compared to those with the TT or TC genotypes [17]. Another study conducted by Al-Daghri et al. (2012) examined the association between the Taq1 and Bsm-I SNPs and T2DM in a Saudi Arabian population. The study found that individuals with the CC genotype had a significantly greater risk of developing T2DM compared to those with the TT or TC genotypes, even after adjusting for confounding factors such as age, gender, and BMI [18]. In another study conducted by Al- Darraji et al. (2017), the association between rs731236 polymorphism and T2DM was evaluated in the obese diabetic Iraqi population. The study found that individuals with the TT genotype had a significantly higher risk of developing T2DM compared to controls [19]. Similarly, a study by Al-Hazmi (2019) found that individuals with the T2DM were more likely to have the TT genotype for Taq1 polymorphism. These results didn’t confirmed any significant differences between T2DM and control groups in both Bsm1 and Taq1 [20].
Aravindhan et al. (2020) in his meta-analysis of 47 studies to explore the association between VDR gene polymorphisms and susceptibility to type 2 diabetes (T2D) demonstrated that the FokI polymorphism was associated with an augmented possibility of T2D in the overall population, as well as in ethnic-specific analyses of Asian and European populations. The BsmI polymorphism was linked to an increased risk of T2D in the overall population, but only in the heterozygous model. This meta-analysis also revealed an association between rs731236 polymorphism and T2DM. The analysis included 23 studies composing of 4579 cases and 4815 controls. The study of overall population results reflected no significant association between rs731236 and rosk of developing T2DM risk. However, the meta-regression analysis showed that the association between rs731236 and T2DM risk was stronger in studies with higher quality and in studies conducted in Asian populations. These findings suggest a risk factor role of 731,236 for T2DM, particularly in Asian populations [21]. In another study conducted by Al-Kashwan et al. (2021), the association between BsmI rs1544410 SNP, TaqI rs731236 SNP and T2DM was evaluated in the Iraqi population. The study found that BsmI rs1544410 SNP has correlated with the risk of incident type 2 DM, while VDR TaqI polymorphism is no associated with the risk of type 2 diabetes mellitus in the Iraqi population [22].
However, in the majority of literature no significant association have been reported between rs731236 and rs1544410 polymorphism and diabetes risk. For example, Maleki et al. (2003) examined the association between VDR gene polymorphisms and type 2 diabetes (T2D) in a Polish population. The study included 548 individuals, 308 with T2D and 240 controls. The researchers genotyped the participants for four VDR polymorphisms: FokI, ApaI, BsmI, and TaqI. The results indicated no significant alteration in the allele frequency or genotype distribution between the T2DM patients and controls for any of the four VDR polymorphisms. They suggest that VDR gene polymorphisms are not associated with T2D in the Polish population [23] Dilmek et al.(2010) did not find a link between the VDR TaqI genotype and T2DM in the Turkish population [24]. However, one limitation of the study was its relatively small sample size. On the same basis, larger studies are needed to confirm these findings.
A meta-analysis of 14 studies was conducted by Li et al. (2013) to investigate the association between VDR genetic variants and susceptibility to type 2 diabetes (T2D). The researchers included a total of 5,424 T2D patients and 5,396 controls. The VDR gene polymorphisms studied were FokI, BsmI, ApaI, and TaqI. The results showed that the FokI polymorphism was associated with an increased risk of T2D in the overall population. However, the association was only significant in the Asian population. The BsmI, ApaI, and TaqI polymorphisms were not associated with T2D in any population [25].
The conflicting results raised by these studies may have their roots in differences in sample size, ethnicity, or other factors such as lifestyle or environmental factors that can enhance diabetes risk.
This preliminary study could be shed only some light on the genetic components of T2D in a group of Iranian patients, which needs more investigations with larger sample size. Further research is warranted to confirm our findings and seek the potential mechanisms underlying this association.
Conclusion
In summary, the current research found that similar frequency of A allele of rs1544410 was seen in both diabetic and non-diabetic subjects. However, diabetic patients had higher frequencies of C allele as well as its genotypes of rs731236 polymorphism, and after adjusting for demographic factors, the odds ratios for these genotypes were statistically significant. These findings suggest an association between rs731236 polymorphism with an increased risk of developing diabetes. But rs1544410 polymorphism of VDR gene was not associated with diabetes mellitus.
Abbreviations
- T2DM:
-
type 2 diabetes mellitus International Diabetes Federation (IDF)
- VDR:
-
Vitamin D receptor
- SNPs:
-
single nucleotide polymorphisms
- HbA1c:
-
hemoglobin A1c
- FBS:
-
fasting blood sugar
- BMI:
-
body mass index
- PCR:
-
polymerase chain reaction
- SD:
-
standard deviation
- OR:
-
odds ratios
- CI:
-
confidence intervals
References
Rodrigues KF, Pietrani NT, Bosco AA, de Sousa MCR, Silva IFO, Silveira JN, et al. Lower vitamin D levels, but not VDR polymorphisms, influence type 2 diabetes mellitus in brazilian population independently of obesity. Medicina. 2019;55(5):188.
Robert AA, Al Dawish MA. The worrying trend of diabetes mellitus in Saudi Arabia: an urgent call to action. Curr Diabetes Rev. 2020;16(3):204–10.
Khodakarami R, Abdi Z, Ahmadnezhad E, Sheidaei A, Asadi-Lari M. Prevalence, awareness, treatment and control of diabetes among iranian population: results of four national cross-sectional STEPwise approach to surveillance surveys. BMC Public Health. 2022;22(1):1–12.
Haussler MR, Whitfield GK, Haussler CA, Hsieh JC, Thompson PD, Selznick SH, et al. The nuclear vitamin D receptor: biological and molecular regulatory properties revealed. J Bone Miner Res. 1998;13(3):325–49.
Al Safar H, Chehadeh SEH, Abdel-Wareth L, Haq A, Jelinek HF, ElGhazali G, et al. Vitamin D receptor gene polymorphisms among emirati patients with type 2 diabetes mellitus. J Steroid Biochem Mol Biol. 2018;175:119–24.
Uitterlinden AG, Fang Y, Van Meurs JB, Pols HA, Van Leeuwen JP. Genetics and biology of vitamin D receptor polymorphisms. Gene. 2004;338(2):143–56.
Sarma D, Chauhan VS, Saikia KK, Sarma P, Nath S. Prevalence pattern of key polymorphisms in the vitamin D receptor gene among patients of type 2 diabetes mellitus in Northeast India. Indian J Endocrinol Metab. 2018;22(2):229–35.
Bid HK, Konwar R, Aggarwal C, Gautam S, Saxena M, Nayak VL, et al. Vitamin D receptor (FokI, BsmI and TaqI) gene polymorphisms and type 2 diabetes mellitus: a north indian study. Indian J Med Sci. 2009;63(5):187–94.
Yin X, Wang H, Guo J, Zhang L, Zhang Y, Li L et al. Association of vitamin D receptor BsmI rs1544410 and ApaI rs7975232 polymorphisms with susceptibility to adolescent idiopathic scoliosis: a systematic review and meta-analysis. Medicine. 2018;97(2).
Hong YJ, Kang ES, Ji MJ, Choi HJ, Oh T, Koong S-S, et al. Association between Bsm1 polymorphism in vitamin D receptor gene and diabetic retinopathy of type 2 diabetes in korean population. Endocrinol Metab. 2015;30(4):469–74.
Cyganek K, Mirkiewicz-Sieradzka B, Malecki M, Wolkow P, Skupien J, Bobrek J, et al. Clinical risk factors and the role of VDR gene polymorphisms in diabetic retinopathy in polish type 2 diabetes patients. Acta Diabetol. 2006;43(4):114.
MWer S, Dykes D, Polesky H. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res. 1988;16(3):1215.
Palomer X, González-Clemente J, Blanco‐Vaca F, Mauricio D. Role of vitamin D in the pathogenesis of type 2 diabetes mellitus. Diabetes Obes Metab. 2008;10(3):185–97.
Hitman GA, Mannan N, McDermott MF, Aganna E, Ogunkolade BW, Hales C, et al. Vitamin D receptor gene polymorphisms influence insulin secretion in bangladeshi Asians. Diabetes. 1998;47:688–90.
Poon AH, Gong L, Brasch-Andersen C, Litonjua AA, Raby BA, Hamid Q, et al. Very important pharmacogene summary for VDR. Pharmacogenetics Genom. 2012;22(10):758.
Nosratabadi R, Arababadi MK, Salehabad VA. Vitamin D receptor polymorphisms in type 2 diabetes in southeastern iranian patients. Lab Med. 2011;42(1):32–4.
Zhang J, Li W, Liu J, Wu W, Ouyang H, Zhang Q, et al. Polymorphisms in the vitamin D receptor gene and type 1 diabetes mellitus risk: an update by meta-analysis. Mol Cell Endocrinol. 2012;355(1):135–42.
Al-Daghri NM, Al-Attas O, Alokail MS, Alkharfy KM, Draz HM, Agliardi C, et al. Vitamin D receptor gene polymorphisms and HLA DRB1* 04 cosegregation in saudi type 2 diabetes patients. J Immunol. 2012;188(3):1325–32.
Al-Darraji S, Al-Azzawie H, Al-Kharsani A. Vitamin D status and its receptor genes bsmi, foki, apai, taqi polymorphism in relation to glucose metabolism in obese iraqi type 2 diabetes mellitus patients. J Mol Genet Med. 2017;11(1000260):1747–08621000260.
Al-Hazmi AS. Association of vitamin D deficiency and vitamin D receptor gene polymorphisms with type 2 diabetes mellitus saudi patients. Afr Health Sci. 2019;19(4):2812–8.
Aravindhan S, Almasoody MFM, Selman NA, Andreevna AN, Ravali S, Mohammadi P, et al. Vitamin D receptor gene polymorphisms and susceptibility to type 2 diabetes: evidence from a meta-regression and meta-analysis based on 47 studies. J Diabetes Metab Disord. 2021;20:845–67.
Al-Kashwan TA, Algenabi AHA, Omara AM, Kaftan AN. Association of vitamin D receptor gene polymorphisms BsmI (rs 1544410) and TaqI rs (731236) with the type 2 diabetes mellitus in iraqi patients from the middle Euphrates region. Meta Gene. 2021;28:100854.
Malecki M, Frey J, Moczulski D, Klupa T, Kozek E, Sieradzki J. Vitamin D receptor gene polymorphisms and association with type 2 diabetes mellitus in a polish population. Exp Clin Endocrinol Diabetes. 2003;111(08):505–9.
Dilmec F, Uzer E, Akkafa F, Kose E, van Kuilenburg AB. Detection of VDR gene ApaI and TaqI polymorphisms in patients with type 2 diabetes mellitus using PCR-RFLP method in a turkish population. J Diabetes Complicat. 2010;24(3):186–91.
Li L, Wu B, Liu J-Y, Yang L-B. Vitamin D receptor gene polymorphisms and type 2 diabetes: a meta-analysis. Arch Med Res. 2013;44(3):235–41.
Acknowledgements
The authors thank to T2DM patients who participated in this study.
Funding
No funding was obtained.
Author information
Authors and Affiliations
Contributions
SHS and SR: Investigation, Methodology; MA: Statistical Analysis; MA, FAM and SHJ; Resources; and MH Conceptualization, Validation. All authors contributed to and approved the final version of the manuscript.
Corresponding authors
Ethics declarations
Competing of interest
There are no conflicts of interest declared by the authors.
Consent for publication
The present study research is approved by endocrinology and metabolism clinical institute ethics committee (IR.IAU.PS.REC.1399.032; IR.IAU.PS.REC.1399.031) and all T2DM patients signed written consent forms.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Salehizadeh, S., Ramezani, S., Asadi, M. et al. Association analysis between the VDR gene variants and type 2 diabetes. J Diabetes Metab Disord 23, 633–638 (2024). https://doi.org/10.1007/s40200-023-01323-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40200-023-01323-0