Abstract
Molecular identity of dominant cultivars of soybean needs to complement the morphological descriptors, which are insufficient and influenced by environment, for protecting the interest of breeders and maintaining the genetic purity. In the present investigation, genomic DNA of six commercial Indian soybean varieties viz. JS95-60, JS93-05, NRC7, JS97-52, NRC37, JS335 was amplified using ten simple sequence repeat (SSR) markers, which were selected based upon the relatively higher polymorphic information content (PIC) value observed after polymorphism survey of 46 soybean genotypes through 58 SSR markers. PCR analysis showed nine polymorphic SSR primers which could differentiate unambiguously all the six varieties. Maximum similarity was observed between NRC7, JS93-05 and JS95-60. Two unique alleles amplified in each of the two varieties namely JS335 and NRC37 and four unique alleles in JS97-52 unambiguously differentiated them from the remaining varieties. Morphologically similar varieties JS93-05 and JS95-60 could be differentiated distinctly by primer pair Satt181, while NRC7 could be distinguished from both JS93-05 and JS95-60 through Sct_199. Results also showed that these nine polymorphic markers could distinguish the dominant cultivars from six non-trading varieties, which further confirmed the utility of nine SSR primers for protecting the breeders’ rights and maintaining the purity of dominant cultivars during seed production programme.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
In the backdrop of Protection of Plant Varieties and Farmers’ Rights Act, it is important to maintain the identity of commercial cultivars of the crops during production and seed processing. Morphological traits/descriptors like growth-type, days-to-flowering, flower colour, leaf shape, leaf colour, plant height, pod pubescence, stem pubescence, pod colour, days-to-maturity, seed size, seed coat colour, seed hilum colour, seed luster traditionally used for identification of varieties are limited in number and have very low variability. Further, some of these morphological traits are influenced by the environment [1]. The electrophoresis patterns of isozymes or contents of biochemical components of the seed are also not the perfect tools due to the influence of environmental factors on their de novo synthesis [2]. DNA fingerprinting technique is very handy to distinguish morphologically similar cultivars precisely, distinctly and rapidly. The International Union for the protection of new varieties of plants (UPOV) has already declared the results based upon DNA markers as the legal basis for variety protection and the guidelines for molecular profiling for identification of varieties adopted by UPOV have also been documented [3]. As per the UPOV guidelines, various DNA fingerprinting techniques viz. random amplified polymorphic DNA (RAPD), amplified fragment length polymorphism (AFLP), simple sequence repeat (SSR), inter simple sequence repeat (ISSR) and sequence tagged sites (STS) can be employed in cultivar identification. Of these molecular tools, simple sequence repeat markers (SSR) have been widely preferred for identification of varieties, owing to their abundance, co-dominance, polymorphism and reproducibility; and more importantly, they are easy to be handled through polymerase chain reaction. SSR fingerprinting technique for the identification of cultivars has been employed in several crops viz. potato [4], wheat [5], rice [6], cotton [7], chick pea [8], maize [9], onion [10], common bean [11], grapes [12], and chrysanthemum [13].
Reports on application of SSR markers in varietal identification in soybean, which is the leading oilseed crop and emerging functional food, are limited [14, 15]. Globally, India ranks fifth in soybean production. Ever since the commencement of commercial cultivation in 1970, 102 soybean varieties have been released for farmers. These varieties have played a pivotal role in raising the soybean production from 0.014 mt in 1970 to 14.6 mt in 2012, thereby contributing 47 % of 30.8 mt of total oilseed production in the country [16]. Six soybean cultivars namely JS95-60, JS93-05, NRC7, JS97-52, NRC37, JS335 dominate the commercial trading of soybean in India; popularity of which can be gauged by their share in the total breeder’s seed indent which is 36, 21, 2, 1, 4 and 29 %, respectively [17]. Morphological descriptors alone cannot distinguish these varieties. It is important to generate the molecular identity of these varieties, in order to effectively address the future intellectual property rights issues. The present investigation was undertaken to finger-print these dominant varieties using SSR markers.
Material and Methods
Genomic DNA was extracted from the 10–15 days old young and tender leaves of six dominant soybean varieties viz. JS95-60, JS93-05, NRC7, JS97-52, NRC37, and JS335 and six non-trading varieties viz. Pusa16, Co Soya-2, Improved Pelican, Punjab1, PK262, and Bragg. The pedigree, year of release and the breeding centre of the respective variety is given in Table 1. The crude DNA was purified through standard protocol. Fifty eight SSR markers were screened for randomly selected 46 soybean genotypes. The sequences of these SSR markers were taken from the list of soybean SSR loci mapped by Agricultural Research Services, United States Department of Agriculture and are available at http://bldg6.arsusda.gov/cregan/soymap.htm. The synthesis of the oligonucleotide sequences of 58 SSR primer pairs was outsourced to Sigma Aldrich, Bangalore. Ten distinct and stutter free SSR primers viz. Satt538 (LGp A2), Satt577 (LGp B2), Satt267 (LGp D1a), Satt146 (LGP F), Satt352 (LGp G), Sct_199 (LGp G), Satt541 (LGp H), Satt181 (LGp H), Satt229 (LGp L), Satt009 (LGp N) (Table 2) exhibiting relatively high PIC value (0.5 or more) were finally used for PCR-based amplification of the genomic DNA of 6 soybean varieties. The PCR reactions were performed in a Thermocycler model PTC100 and the reaction mixture (10 µl) contained 2 µl DNA (20 ng/µl), 1 µl PCR (10×) buffer, 1.1 µl MgCl2 (25 mM), 0.1 µl dNTPs (25 mM), 0.4 µl each forward and reverse SSR primers (30 ng/µl), 0.068 µl Taq DNA polymerase (3 U/µl), and 4.932 µl distilled water. Initially, DNA was denatured at 94 °C for 1 min. followed by 30 cycles, each cycle comprising of denaturation at 94 °C for 2 min, primer annealing at 50 °C for 2 min., primer elongation at 72 °C for 3 min. Finally, elongation was carried out at 72 °C for 10 min. PCR products were resolved on 3 % metaphore gel. DNA ladder (50 bp) was also loaded to assess the size of the amplicons generated through each SSR primer in six popular varieties. Oligonucleotide sequences of the selected 10 SSR markers are given in Table 2. Polymorphic information content (PIC) of SSR markers were calculated as under.
where i denotes the SSR marker while pij is frequency of jth allele.
Amplicons were scored as 1 and 0 for the presence and absence of alleles, respectively. Similarity coefficients between varieties were computed using Jaccard’s similarity coefficient using SIMQUAL module. Unweighted Pair Group Method with Arithmetic Average (UPGMA) cluster analysis was carried out using NTSYS 2.02 [18].
Results and Discussion
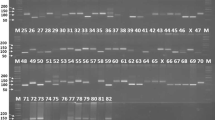
PCR amplification of the genomic DNA of six soybean varieties obtained through 10 SSR markers is depicted in Fig. 1 (Satt577, Satt267, and Satt009), Fig. 2 (Satt352, Sct_199, and Satt181) and Fig. 3 (Satt538, Satt229, Satt146, and Satt541). Figure 4 summarizes the details of PCR fragments generated through 10 SSR markers in six soybean varieties. Out of 10 primer pairs, one primer pair Satt229 was monomorphic, generating a single allele of size 200 bp in all the six varieties. For the remaining nine primer pairs, polymorphic information content ranged from 0.277 to 0.612, with five primer pairs showing PIC value ≥0.5 (Table 2). Of the total number of 24 alleles, five SSR primer pairs viz. Satt577, Satt352, Sct_199, Satt541 and Satt009 generated 3 alleles each, while 4 SSR primer pairs viz. Satt538, Satt267, Satt146 and Satt181 produced two alleles each. In total, eight unique alleles were generated, two each in three varieties namely JS335, JS97-52 and NRC37. Two unique alleles viz. 125 and 150 bp amplicon from Satt538 and Satt541, respectively, were amplified in JS335. Four unique alleles viz. 100, 170, 160 and 210 bp amplicon by Satt577, Satt352, Satt541 and Satt009, respectively, were generated in variety JS97-52. NRC37 could be differentiated with 110 and 220 bp size amplicon generated by Satt577 and Satt267, respectively.
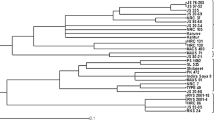
To assess the genetic relatedness among these 6 soybean varieties, UPGMA cluster analysis based upon the Jaccard’s similarity coefficient classified the six soybean varieties into two major groups viz. class I and II, separating at 21 % similarity (Fig. 5). Class I comprised of two varieties namely JS97-52 and NRC37 with 33.33 % genetic similarity. JS335 of class IIa separated from three varieties namely JS95-60, JS93-05 and NRC7 of Class IIb by a similarity level of 0.33. The pair- wise genetic similarity values ranged from 0.176 to 0.666. Maximum similarity (66.66 %) was observed between JS95-60, JS93-05 and NRC7. As evident from the pedigree data given in Table 1, JS93-05 and JS95-60 have been developed through selection from the same genotype i.e. PS73-22. Further, both these varieties are early maturing and similar in major morphological characters like growth habit (determinate), leaf shape (lanceolate), height (short-statured) and seed coat colour (yellow). Primer pairs Satt538, Satt577, Satt267, Satt146, Sct_199, Satt541, Satt229 and Satt009 generated amplicon of same size in both the varieties, which confirms the similarity between the two. These two varieties could be distinguished unambiguously by SSR marker Satt181 which generated a fragment of size 210 bp in variety JS93-05 while PCR fragment of 180 bp size in variety JS95-60. A 30 bp difference in the size of the PCR fragments generated distinct bands for the molecular identification of these two varieties. Satt352 also generated PCR fragments of 180 and 185 bp in JS93-05 and JS95-60, respectively. A 5 bp differing amplicon may be little difficult to distinguish in metaphore agarose gel; however, it can be resolved quite distinctly in polyacrylamide gel. NRC7 could be distinguished from JS93-05 and JS97-52 through Sct_199, which generated 190 bp amplicon in NRC7 but not in JS93-05 and JS97-52.
JS335 is one of the most resilient soybean variety and adaptable to diverse agro-climatic zones in the country. Satt538 was the most robust polymorphic SSR marker for distinguishing this variety from the other five soybean varieties. In variety JS335, this SSR marker generated an amplicon of 125 bp in size, which was 15 bp longer than the amplicon generated (110 bp) in all the remaining five soybean varieties. Similarly, an amplicon of 150 bp size generated by Satt541 in variety JS335 was 10 bp shorter than the fragment amplified in variety JS97-52 (160 bp) while 20 bp shorter than the fragment (170 bp) produced in four varieties viz. NRC7, JS93-05, NRC37, JS95-60. SSR analysis has been applied in the identification of 15 certified soybean varieties of Thailand [15]. Recently, RAPD markers have been employed in diversity analysis of Serbian soybean varieties [19].
Six non-trading soybean varieties viz. Pusa16, Co Soya-2, Improved Pelican, Punjab1, PK262, Bragg, which are not in the commercial cultivation, were also surveyed by the nine SSR markers found to be polymorphic with six dominant cultivars. The details of PCR fragments generated are presented in Fig. 6. Pusa16 was distinguished from the other 5 non-trading varieties and six dominant varieties through 115 bp amplicon generated by Satt538 in this variety. In dominant varieties, alleles of either 110 or 125 bp appeared at Satt538. Co Soya-2 was quite distinct from other non-trading varieties and all the dominant varieties except JS97-52 in generating an amplicon of 160 bp through Satt541. Further, JS97-52 and Co Soya-2 could be distinguished from each other through SSRs Satt577 and Satt538, which generated amplicon of 100 and 110 bp in JS97-52 but amplified 120 and 125 bp alleles in Co Soya-2, respectively. Improved pelican could be distinguished from other non-trading varieties and all dominant varieties except JS97-52 through amplicon of 170 bp generated by Satt352. Identification between JS97-52 and Improved pelican can be made by allele of 170 bp at Satt541, which appeared in Improved Pelican but not in JS97-52. Punjab 1 could be distinguished from other non-trading varieties by allele of 290 bp at Satt146. The same allele was helpful in distinctly identifying this variety from six dominant varieties, wherein amplicon of different size (310 or 320 bp) appeared at this locus. PK262 and Bragg could be distinguished from all the six dominant varieties using SSR Sct_199, which generated an amplicon of 210 bp in these two varieties but alleles of 190, 200 or 220 bp size appeared in six dominant varieties. Among a set of six non-trading varieties, PK262 and Bragg were distinct in generating amplicon of 310 and 240 bp at locus Satt146 and Satt267, respectively.
Future Perspective
Further, molecular analysis along with the morphological data of proposed varieties has become mandatory while submitting the proposal for their release as varieties in All India Co-ordinated trial of several field crops including soybean. In this context, the use of SSR markers in DUS testing for released soybean varieties is being deliberated. Already, in countries like Brazil [20] and Argentina [21] SSR marker generated amplicons are being employed in the DUS testing of soybean crop. In Brazil, the dispute over soybean variety Conquista grown in 3 million ha and newly released variety Pioneira—which was suspected to be very similar to Conquista—could be resolved using molecular markers. Nine polymorphic SSR primers which could differentiate the six dominant varieties as well as six non-trading varieties can be used for DNA bar-coding of all the 102 released varieties of soybean.
Conclusion
Amplicon profile obtained with nine SSR markers could differentiate all the six dominant soybean varieties of India, which can be employed to address any intellectual property rights’ issue related to these varieties. Besides, the application of these SSR primer pairs would be very handy in establishing the genetic purity of these six soybean varieties during seed production and maintenance under DUS (Distinctness-Uniformity-Stability) trial.
References
Khan AZ, Shah Khalil SK, Taj FH (2003) Influence of planting date on planting date and plant density on morphological traits of determinate and indeterminate soybean cultivars under temperate environments. J Agron 2:146–152
Carrera CS, Dardenellie JL, Soldini DO (2014) Genotypic and environmental variation in seed nutraceutical and industrial composition of non-transgenic soybean (Glycine max) genotypes. Crop Pasture Sci 65:1311–1322
UPOV/INF/17/1 (2010) Guidelines for DNA-profiling: Molecular marker selection and database construction. Adopted by the Council in 44th session on 21 Oct 2010, Geneva
Novakova A, Simackova Barta J, Curn V (2010) Utilizaion of DNA markers based upon micro-sattelite polymorphism for identification of potato varieties cultivated in Czech Republic. J Central Eur Agric 11(4):415–422
Zhu Y, Hu J, Han R, Wang Y, Zhu S (2011) Fingerprinting and identification of closely related wheat cultivars using ISSR and fluorescence-labeled TP-M13-SSR markers. Aust J Crop Sci 5(7):846–850
Zhu Y, Qin GC, Hu J, Wang JC, Zhu SJ (2012) Fingerprinting and variety identification of rice based upon simple sequence repeat markers. Plantomics J 5(4):421–426
Ahemed MM, Guo H, Huang C, Zhang X, Lin Z (2013) Selection of core SSR markers for fingerprinting upland cotton cultivars and hybrids. Aus J Crop Sci 7(12):1912–1920
Joshi N, Sharma S, Subramanian Rao KS (2013) Genetic DNA fingerprinting of chick pea germplasm using morphological and molecular markers. Asian J Expt Biol 4:398–405
Sudharani M, Rao PS, SubbaRao LV (2014) Identification of SSR markers for testing of hybridity and seed genetic purity in maize. Int J Sci Res 3(10):92–95
Anadhan S, Mote SR, Gopal J (2014) Evaluation of onion varietal identity using SSR markers. Seed Sci Tech 42:279–285
Cordoso PCB, Brondani C, Menezes IPP, Valdisser PAMR, Borba TCO, Del Peloso MJ, Vianello RP (2014) Discrimination of common bean cultivars using multiplexed microsatellite markers. Genet Mol Biol 13:1964–1978
Tessier C, David J, Boursiquot PJM, Charrier A (1999) Optimization of the choice of molecular markers for varietal identification in Vitis vinifera L. Theor Appl Genet 98:171–177
Zhang Y, Dai S, Hong Y, Song X (2014) Application of genomic SSR locus polymorphisms on the identification and classification of Chrysanthemum cultivars in China. PLoS ONE 9:e104856
Yun-lai GAO, Rong-Sheng Z, Chun-Yan L, Wen-Fu L, Can-Dong Hong-Wei, Li Bing-Che Y, Guo-Hua Hu, Qing-Shan C (2009) Constructing molecular identity for soybean varieties from Heilongjiang Province, China. Acta Agron Sinica 35(2):211–218
Tantasawat P, Trongchuen J, Prajongjai T, Jenweerawat S, Chaowiset W (2011) SSR analysis of soybean :genetic relationship and variety identification in Thailand. Aust J Crop Sci 5:283–290
All India Co-ordinated Research Project System on Soybean Report (2014) In: Proceeding and technical programme 44 th annual group meeting at Birsa Agricultural University, Ranchi, Jharkhand, May 25–27, p 10
Rohalf FJ (1998) NTSYS pc, Numerical taxonomy and multivariate analysis system, Version 2.02. Exter Software, New York
Peric V, Nicolic A, Babic V, Sudaric A, Srebric M, Dordevic V, Mladenovic Drinic S (2014) Genetic relatedness of soybean genotypes based upon agro-morphological and RAPD markers. Genetika 46:839–854
Pacheco LG (2011) Organization of DUS trials in Brazil based upon the use of molecular markers. A presentation in 13th session of Working Group on biochemical and molecular techniques and DNA profiling in particular, held at Brasilia from Nov 22 to 24
Vicario A, Giancola S, Echaide M, Hopp E (2001) Application of micro-satellite markers for the assessment of distinctness, uniformity and stability (DUS testing) of commercial soybean varieties. International Union for the Protection of New Varieties of Plants, Geneva Nov, 12. Working Group on biochemical and molecular techniques and DNA profiling in particular. Seventh session Hanover, Nov 21–23 BMT/7/13:2-10
Acknowledgments
Authors thankfully acknowledge Indian Council of Agricultural Research for the financial help.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Kumar, V., Rani, A., Jha, P. et al. Molecular Identification of Dominant Cultivars of Soybean Using Simple Sequence Repeat Markers. Proc. Natl. Acad. Sci., India, Sect. B Biol. Sci. 87, 647–653 (2017). https://doi.org/10.1007/s40011-015-0641-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40011-015-0641-7