Abstract
The aim of this study was comparative evaluation of loop-mediated isothermal amplification (LAMP) and polymerase chain reaction (PCR) assay for rapid and inexpensive detection of shiga toxin-producing E. coli in animal origin foods by targeting stx1 and stx2 genes. The LAMP assay was performed using a water bath. The standardized LAMP assay was evaluated on 122 E. coli field isolates obtained from various animal origin food samples to ensure its reliability and usefulness. The result showed that conventional PCR could detect 68 (55.73%) and 75 (61.47%) positive E. coli isolates for stx1 and stx2 genes. Whereas, LAMP showed higher sensitivity by detecting 79 (64.75%) and 87 (71.31%) positive isolates of E. coli for stx1 and stx2 genes, respectively. LAMP assay was found to be highly specific and 10 times more sensitive as it could detect 1.11 × 102 cfu/ml for both stx1 and stx2 genes of E. coli isolates, whereas conventional PCR could detect 1.85 x 103 cfu/ml for both stx1 and stx2 genes of E. coli isolates. The rapidness, sensitivity, specificity, easiness and cost-effectiveness of LAMP assays will be very useful for the detection of foodborne pathogens for improving food sanitation and maintaining food safety.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Escherichia coli is among the first bacterial species to colonise in intestine during infancy [14]. On the basis of their virulence and disease manifestation, there are five distinct groups of E. coli, which include toxin-producing strains like enterotoxigenic (ETEC), enterohaemorrhagic (EHEC) or verocytotoxigenic E. coli (VTEC), enteroaggregative (EAggEC), non-toxic strains like enteropathogenic (EPEC) and enteroinvasive (EIEC) E. coli [3]. These groups are associated with diarrhoea, haemorrhagic colitis (HC), dysentery, bladder and kidney infections, surgical wound infection, septicaemia, haemolytic uraemic syndrome (HUS), pneumonia and meningitis, and some of these conditions result in death. Pathogenic types of E. coli also occur in animal origin foods, and in particular, verocytotoxigenic E. coli (VTEC) are zoonotic agents that cause severe diseases and are responsible for many foodborne outbreaks worldwide [17].
According to the World Health Organization (WHO) report, approximately 11 million children under the age of 5 years died because of E. coli-mediated gastroenteritis [23]. Shiga toxin-producing E. coli (STEC), also known as Vero toxin-producing E. coli (VTEC), comprises a serologically diverse group of pathogens that cause disease in humans and animals characterized by the production of cytotoxins that disrupt protein synthesis within host cells. These toxins are synonymously either called verocytotoxins (VT), because of their activity on Vero cells, or Shiga toxins (Stx) because of their similarity with the toxin produced by S. dysenteriae. The two main groups consist of Stx1, which is nearly identical to the toxin of S. dysenteriae type 1 and Stx2, which shares less than 60 percentage amino acid sequence with Stx1 [1]. There are at least 100 serotypes of E. coli that produce Shiga toxins [9] The cattle are considered the primary reservoir of both O157 and non-O157 STEC bacteria [2].
Most of the infections are caused due to the ingestion of contaminated foods, particularly undercooked ground beef. Other foods of bovine origin including roast beef, raw unpasteurized milk and other dairy products like yogurt, curd, cheese and foods are derived from other species, including pork, chevon, mutton, fish, shellfish meat of wild or exotic mammals. In India, there is little information available on the prevalence of Shiga toxin-producing E.coli across the country. The STEC from non-diarrhoeic animal sources in India was first isolated in 1999 [18].
In the past few decades, several molecular methods have been developed to overcome the shortcomings of the classical diagnostics methods, especially the in vitro amplification of a pathogen-specific nucleic acid sequence. Loop-mediated isothermal amplification technology developed by Notomi et al. [10] is a novel DNA amplification method which can amplify target gene under isothermal conditions with high efficiency and sensitivity [24]. Developing countries like India require low-cost detection techniques for detection of these pathogens at district, block as well as at field level. Loop-mediated isothermal amplification (LAMP) has attracted a lot of attention as a potentially rapid, accurate and cost-effective novel nucleic acid amplification method.
Materials and Methods
Isolation of E. coli from Animal Origin Foods
A total of 298 animal origin food samples comprising 139 chicken, 52 buffalo meat, 32 mutton, 39 pork, 16 milk and 10 each of fish and eggs were collected from retail shops located in and around Mumbai city over a period of 6 months during 2015–16. These samples were further processed for isolation of E. coli spp. following standard technique as per IS 5887(Part 1): 1976.
All of these positive isolates were further characterized by biochemical tests and the results were interpreted and validated as per bacteriological analytical manual for E. coli (2007). Further, 122 positive E. coli isolates were subjected for detection by standardized conventional polymerase chain reaction (PCR) and loop-mediated isothermal amplification (LAMP) methods.
Bacterial Strains and DNA Extraction
The reference strain E. coli (MTCC 443) was procured from Institute of Microbial Technology (MTCC), Chandigarh, India. Additionally, 122 field isolates of E. coli isolated were also included in the study.
Genomic DNA of E. coli was extracted as per the protocol [12] with slight modifications. A colony of E. coli isolate on nutrient agar was picked and mixed with 1000 µl of NSS in centrifuge tube. It was then centrifuged at 10,000 rpm for 10 min. After centrifugation, the pellet formed was dissolved in 100 µl of nuclease-free water (NFW), vortexed and further boiled at 100 °C for 10 min. The centrifuge tube was subjected to rapid cooling in ice which was followed by centrifugation at 10,000 rpm for 10 min. Then, the upper aqueous phase which contained DNA was transferred to sterile micro-centrifuge tube. These extracted DNAs were further used for amplification. Until use, these were stored at freezing temperature (− 20 °C to − 80 °C) in sterile micro-centrifuge tube.
Primer Used for LAMP and PCR Reactions
Each LAMP primer set used in this study consisted of two outer (F3, B3), two inner (FIP, BIP) and two loop primers (Loop F, Loop B), which recognized eight different regions of the gene target and were commercially synthesized by Integrated DNA Technologies (IDT) obtained from Sigma Aldrich, Bangalore, India. The LAMP primer sets for each of the VTEC gene targets (stx1 and stx2) were selected from previous study [4]. The primers used in PCR for the specific detection of E. coli were previously described [18] for stx1 gene and [5] stx2 genes. The sequences of the primers are summarized in Table 1.
Optimization of LAMP Assay
The optimization of LAMP assay was carried out by conducting the trials at different temperatures 58 °C, 60 °C, 62 °C, 63 °C, 65 °C and 66 °C and 58 °C, 60 °C, 62 °C, 63 °C, 65 °C, 65.2 °C and 66 °C for both stx1 and stx2 genes, respectively, and also at different time periods 50 min, 60 min and 70 min for both the genes. The LAMP reaction mixture was optimized using different concentrations of inner primers, outer primers, MgSO4 and dNTPs. However, 65 °C was chosen as the optimal temperature since there was presence of significant visual turbidity due to formation of large amount of by-product, pyrophosphate ion, being produced, yielding an insoluble white precipitate of magnesium pyrophosphate in reaction mixture and fluorescence on addition of SYBR green dye under ultraviolet illumination. After completion of LAMP, amplified DNA was analysed by electrophoresis on 1.5% agarose gel at 98 V for 45 min. A 100 bp DNA ladder was run along with LAMP products.
Optimization of PCR
The PCR assay for the detection of E. coli was standardized as per the method of [5, 19] with slight modifications. PCR was performed using a Thermocycler PCR machine (Eppendorf Mastercycler gradient, Germany). The cycle times are standardized for both stx1 and stx2 genes of E. coli positive isolate (Table 2). Following the last cycle, there was 7-min incubation at 72 °C for the final elongation. The tubes were then held at 4 °C for both the genes. Amplified PCR products were analysed by agarose gel electrophoresis on 1.5% agarose gel.
Sensitivity of the LAMP Assay
Sensitivity (detection limit) of LAMP assay was evaluated using 18-h-old E. coli culture on trypton soya agar, incubated for overnight at 37 °C. Tenfold serial dilution was carried out in PBS up to 10−7 dilutions. 10−4–10−7 dilutions were used for both LAMP and PCR assays. To determine the total viable count (TVC) of each dilution, the culture was plated onto nutrient agar. After incubation at 37 °C for 18 h, the numbers of colonies were counted.
Specificity of LAMP Assay
The specificity of LAMP assay was tested using standard E. coli DNA template and four other templates from non-E. coli strain. The DNA templates were prepared as described previously. The specificity of E. coli-specific LAMP was performed by testing it with four other bacterial species viz. Pseudomonas aeruginosa, Salmonella spp., Proteus vulgaris and Klebsiella pneumoniae. The reaction was performed at 65 °C for 60 min, and the results of this assay were compared with conventional PCR assay.
Results and Discussion
Standardization of LAMP
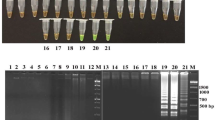
LAMP was standardized for the detection of stx1 and stx2 genes of E. coli from foods of animal origin. The LAMP conditions optimized for the amplification after standardization were 65 °C for 60 min followed by 80 °C for 2 min for termination of the reaction for both stx1 and stx2 genes. The presence of significant visual turbidity and fluorescence on addition of SYBR green dye was observed at 65 °C (Fig. 1). LAMP products observed under UV transilluminator of gel documentation system exhibited specific ladder-like pattern in case of DNA amplification (Fig. 2a, b). The PCR was standardized for stx1 and stx2 gene (348 and 478 bp, respectively) using reference strain (Fig. 3a, b).
a Ladder-like pattern of LAMP products on 1.5% agarose gel (stx1 gene). Lane 1–4: Ladder-like pattern of LAMP products of stx1 gene of E. coli, Lane 5: Negative control showing no ladder-like pattern, Lane M: TrackItTM 100bp DNA ladder (Invitrogen, Cat. No. 10488-058). b Ladder-like pattern of LAMP products on 1.5% agarose gel (stx2 gene). Lane 1–6: Ladder-like pattern of LAMP products of stx2 gene of E. coli, Lane 7: Negative control showing no ladder-like pattern, Lane M: TrackItTM 100bp DNA ladder (Invitrogen, Cat. No. 10488-058)
a Standardization of PCR for stx1 gene of E. coli. Lane 1–7: 348 bp PCR products of stx1 gene of E. coli isolates. Lane M: TrackItTM 100bp DNA ladder (Invitrogen, Cat. No. 10488-058). b Standardization of PCR for stx2 gene of E. coli. Lane 1 and 5: 478 bp PCR products of stx2 gene of E. coli, Lane N: Negative control for E. coli, Lane M: TrackItTM 100bp DNA ladder (Invitrogen, Cat. No.10488-058)
Analysis of Animal Origin Food Samples
In the present study, 122 (40.93%) positive isolates of E. coli were recovered from 298 animal origin food samples analysed (Table 3). Out of 122 E. coli analysed for virulence gene characterization using conventional PCR and LAMP. It was observed that conventional PCR could detect 68 and 75 (55.73% and 61.47%) positive stx1 and stx2 genes of E. coli isolates, whereas LAMP showed higher sensitivity by detecting 79 (64.75%) and 87 (71.31%) positive isolates of E. coli for stx1 and stx2 genes, respectively.
After successful standardization of LAMP, all the positive E. coli isolates (122) were subjected to LAMP technique. After subjecting all the 122 positive E. coli isolates to LAMP, it was observed that all of the isolates were found 79/122 (64.75%) and 87/122 (71.31%) positive for stx1 and stx2 genes of E. coli, respectively, using LAMP technique. The results of the present study are in agreement with the previous findings who could detect all the 24 strains (100%) of stx-producing E. coli. However, six strains of non-stx-producing E. coli were not detected by LAMP technique [4]. Similarly, LAMP technique developed for iapH gene of Shigella and enteroinvasive E. coli detected 38 out of 38 enteric pathogens [13]. This may be attributed to difference in the target gene of E. coli and primers used changing the sensitivity of detection.
The PCR technique could detect 68/122 (55.73%) and 75/122 (61.47%) of stx1 and stx2 genes of E. coli isolates, respectively. LAMP technique could detect 79/122 (64.75%) and 87/122 (71.31%) isolates positive for stx1 and stx2 genes of E. coli. This may be attributed to the presence of four specific primers targeting six distinct sites on the stx1 and stx2 genes of E. coli.
However, 71% positive isolates by PCR for stx gene was compared to 100% by LAMP method [8]. Moreover, in case of Salmonella 90% and 72.72% detection of positive Salmonella isolates by PCR, LAMP technique successfully identified all the Salmonella spp. analysed (100%) [11, 15].
Determination of Detection Limits (Sensitivity) and Specificity of LAMP
Sensitivity of LAMP
The sensitivity (detection limit) of LAMP was evaluated by using tenfold serial dilution method. The total viable count (TVC) of undiluted culture was 1.11 x 109 by calculation using plate-counting method.
Similar protocol of DNA dilution was adopted for evaluating sensitivity (detection limit) of conventional PCR assay. The sensitivity (detection limit) of the LAMP assay was noted to be tenfold greater than that of conventional PCR as LAMP could detect 1.11 x 102 cfu/ml for both stx1 and stx2 genes of E. coli isolates, whereas conventional PCR could able to detect 1.85 x 103 cfu/ml for both stx1 and stx2 genes of E. coli isolates. The sensitivity (detection limit) of the LAMP assay was noted to be tenfold greater than that of conventional PCR as LAMP could detect 1.11 x 102 cfu/ml for both stx1 and stx2 genes of E. coli isolates, whereas conventional PCR could able to detect 1.85 x 103 cfu/ml for both stx1 and stx2 genes of E. coli isolates. The results are in accordance with a study conducted using LAMP assay for detection of E. coli from diarrhoeal stool, who reported that the LAMP assay could detect 102 cfu/ml, whereas the PCR could detect 103 cfu/ml of E. coli indicating that LAMP was 10 times more sensitive than PCR [13].
Sensitivity of LAMP assay is ten times higher than the PCR-based method, and these findings are also in agreement with previous study reports [16], [20], [21] and [22]. They further stated that LAMP was more sensitive technique than PCR. However, some of the authors reported that the LAMP test developed for E. coli was 100 times more sensitive than conventional PCR [6]. This variation may be attributed to the difference in target gene and LAMP primers used in their study.
Specificity of LAMP
In the present study, the specificity of LAMP assay was tested using standard E. coli DNA template and four other templates from non-E. coli strains viz. P. aeruginosa, Salmonella spp., P. vulgaris and K. pneumoniae. The LAMP was carried out as per the standard protocol at 65 °C for 60 min in water bath. It was found that the LAMP assay successfully amplified E. coli DNA only, while it did not amplify any non-E. coli organisms. Similarly, the PCR detected E. coli successfully and did not give any positive result with non-E. coli strains. Thus, the specificity of both LAMP and conventional PCR was found to be 100%.
The present study indicated that LAMP could differentiate and specifically detect the E. coli from other non-E. coli strains. However, both LAMP and PCR assays were successfully able to identify only E. coli without giving any false-positive results for non-E. coli strains showing 100% specificity for both the assays. The specificity results (100%) observed in present study are also in accordance with [7] who reported that LAMP technique could amplify all the 35 enteric bacteria successfully but none of non-E. coli standard strains used under study viz. P. aeruginosa, Salmonella spp., P. vulgaris and K. pneumoniae. amplified using LAMP technique.
Conclusions
Shiga toxin-producing E. coli (STEC) strains are zoonotic foodborne pathogen of significant public health concern due to its frequent involvement in outbreak of haemorrhagic colitis (HC) and ability to cause life-threatening complications such as haemorrhagic uraemic syndrome (HUS) and thrombotic thrombocytopenic purpura. The LAMP method gives results similar to that of gold standard microbiological culture method. To ease the odds faced by PCR, LAMP stands out to be good and effective diagnostic test for empowering in developing countries as it does not require sophisticated equipment like thermocycler for DNA amplifications and well-trained personnel. Thus, this LAMP assay can help in improving food sanitation, maintaining food safety as well as developing international trade.
References
Barman NN, Deb R, Ramamurthy T, Sharma RK, Borah P, Wani SA, Kalita D (2007) Molecular characterization of Shiga toxin producing E. coli isolates from pigs oedema. Indian J Med Res 127:602–606
Bettelheim KA (2000) Role of non-O157 VTEC. J Appl Microbiol 88:38–50
Burns AJ, Herbert TM, Ward SM, Sanders KM (1997) Interstitial cells of Cajal in the guinea-pig gastrointestinal tract as revealed by C-Kit immunohistochemistry. Cell Tissue Res 290:11–20
Hara-Kudo Y, Nemoto J, Ohtsuka K, Segawa Y, Takatori K, Kojima T, Ikedo M (2007) Sensitive and rapid detection of verotoxin producing E. coli using loop-mediated isothermal amplification, World health organization (WHO) Fact Sheet. J Med Microbiol 56:398–406
Hazarika RA, Singh DK, Kapoor KN, Agrawal RK, Pandey AB, Purosottam (2007) Verotoxigenic E.coli (STEC) from beef and its products. Indian J Expt Biol 45:207–211
Hill J, Beriwal S, Chandra I, Paul VK, Kapil A, Singh AT, Wadowsky RM, Singh V, Goyal A, Jahnukainen T, Johnson JR, Tarr PI, Vats A (2008) Loop-mediated isothermal amplification assay for rapid detection of common strains of E. coli. J Clin Microbiol 46:2800–2804
Mahony J, Chong S, Stone C, Chui L (2016) Evaluation of four loop-mediated isothermal amplification (LAMP) assays for identification of Shiga toxin producing E.coli O157 (STEC) and non-O157 Strains. Adv Mol Diag 1(1):3–6
Matise I, Shelton M, Phillips G and Will LA (1998). Sensitive PCR method for detection of E.coli 0157:H7 and other Shiga toxin-producing bacteria in ground meat. Department of Microbiol, Immunol and Preventive Medicine, Iowa State University. paper- 33
Nataro JP, Kaper JB (1998) Diarrheagenic E. coli. Clin Microbiol Rev 11:142–201
Notomi T, Okayama H, Masubuchi H, Yonekawa T, Watanabe K, Amino N, Hase T (2000) Loop-mediated isothermal amplification of DNA. Nucleic Acids Res 28:63
Ohtsuka K, Yanagawa K, Takatori K, Hara-Kudo Y (2005) Detection of Salmonella enterica in naturally contaminated liquid eggs by Loop-mediated Isothermal Amplification, and characterization of Salmonella isolates. Appl Environ Microbiol 71(11):6730–6735
Rawool DB, Malik SVS, Barbuddhe SB, Shakuntala I, Aurora R (2007) A multiplex PCR for detection of virulence associated genes in Listeria monocytogenes. Int J Food Safety 9:56–62
Song T, Toma C, Nakasone N, Iwanaga M (2005) Sensitive and rapid detection of Shigella and enteroinvasive E.coli by a loop-mediated isothermal amplification method. FEMS Microbiol. 243:259–263
Stratakos AC, Linton M, Millington S, Grant IR (2016) A loop-mediated isothermal amplification method for rapid direct detection and differentiation of non-pathogenic and verocytotoxigenic E. coli in beef and bovine faeces. J Appl Microbiol 122:817–828
Tang T, Cheng A, Wang M, Li X, He Q, Jia R, Zhu D, Chen X (2012) Development and clinical verification of a loop-mediated isothermal amplification method for detection of Salmonella species in suspect infected ducks. Poult Sci 91:979–986
Teh CSJ, Chua KH, Lim YAL, Lee SC, Thong KL (2014) Loop-mediated isothermal amplification assay for detection of generic and vero cytotoxin producing E. coli among indigenous individuals in Malaysia. Hindawi Publishing Corporation Scientific World Journal 2014:6. https://doi.org/10.1155/2014/457839
Tenaillon O, Skurnik D, Picard B, Denamur E (2010) The population genetics of commensal Escherichia coli. Nat Rev Microbiol 8:207–217
Verma S, Kumar M, Kashyap S, Singh M, Venkatesh V (2013) Current scenario of E. coli and its serotype “O157:H7” in Indian subcontinent. Int J Innovative Res Sci Eng Technol 2(7):2642–2644
Vidal M, Kruger E, Duran C, Lagos R, Levine M, Prado V, Toro C, Vidal R (2005) Single multiplex PCR assay to identify simultaneously the six categories of diarrheagenic E.coli associated with enteric Infections. J Clin Microbiol 24:5362–5365
Wang D, Liu F, Huo G, Ren D, Li Y (2009) Development and evaluation of loop- mediated isothermal amplification method for detecting E. coli O157 in raw milk. J Rapid Methods Autom Microbiol 17:55–66
Wang F, Jiang L, Ge B (2011) Loop-mediated isothermal amplification assays for detecting shiga toxin-producing E. coli in ground beef and human stools. J Clin Microbiol 50:91–97
Wang F, Jiang L, Yang Q, Prinyawiwatkul W, Ge B (2012) Rapid and specific detection of E. coli serogroups O26, O45, O103, O111, O121, O145 and O157 in ground beef, beef trim produce by loop-mediated isothermal amplification. Appl Environ Microbiol 78(8):2727–2736. https://doi.org/10.1128/AEM.07975-11
World Health Organization (2007) Food safety–foodborne diseases and value chain management for food safety. (“Forging links between agriculture and health” CGIAR on agriculture and health meeting in WHO/HQ)
Zende R, Kshirsagar D, Vaidya V, Waghamare R, Todankar R, Shirke A (2017) Loop-mediated isothermal amplification assay (LAMP): a rapid tool for diagnosis of foodborne and zoonotic pathogens: a review. Int J Livest Res 7(5):23–35
Acknowledgements
This work was supported by grants from Indian Council of Agricultural Research, New Delhi, under the Project “All India Co-Ordinated Research Project on Post Harvest Engineering and Technology” implemented at Bombay Veterinary College, Parel, Mumbai.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Rights and permissions
About this article
Cite this article
Baraily, P., Zende, R.J., Kshirsagar, D.P. et al. Rapid Detection of Shiga toxin-Producing E. Coli in Animal Origin Foods Using Loop-Mediated Isothermal Amplification (LAMP) Assay. Agric Res 8, 490–496 (2019). https://doi.org/10.1007/s40003-018-0391-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40003-018-0391-x