Abstract
Purpose
To clarify the clinical and microbial characteristics of polymicrobial bacteremia (PMB) to contribute to improvements in clinical diagnosis and effective early treatment.
Methods
This retrospective multicenter study used data from three acute-care hospitals in Okayama Prefecture, Japan, collected between January 2014 and March 2019. We reviewed the demographics, comorbidities, organisms isolated, infectious focus, and 30-day mortality of patients with PMB.
Results
Of the 7233 positive blood cultures, 808 (11.2%) were positive for more than one organism. Of the patients with bacteremia, 507 (7.0%) had PMB, of whom 65.3% were male. Infectious foci were identified in 78.3% of the cases, of which intra-abdominal infections accounted for 47.1%. A combination of Gram-positive cocci (GPC) (chain form) and Gram-negative rods (GNR) accounted for 32.9% of the cases, and GPC/GNR and GNR/GNR patterns were significantly associated with intra-abdominal infections. The 30-day mortality rate of patients with PMB was 18.1%, with a median of 7.5 days from diagnosis to death. The mortality in patients with an infectious focus identified was significantly lower than that in patients with an unknown focus (16.3% vs. 24.5%; p = 0.031).
Conclusions
Intra-abdominal infections were the most common source of PMB, and were strongly associated with a Gram-staining combination pattern of GPC (chain form)/GNR. PMB cases with an unknown focus had a poorer prognosis, highlighting the importance of early diagnosis and appropriate treatment.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Bloodstream infections (BSI), which are diagnosed by blood culture, may result in serious conditions, such as sepsis and septic shock [1, 2]. Risk factors for BSI include male sex, aging, and underlying comorbidities such as alcoholism, immunosuppression, liver cirrhosis, diabetes mellitus, congestive cardiac failure, cancer, and chronic renal failure requiring hemodialysis [3,4,5]. Without appropriate treatment, BSIs have a high mortality rate [6, 7]; thus, clinicians should focus on early diagnosis and appropriate treatment.
The incidence of polymicrobial bacteremia (PMB), defined as the presence of at least two distinct microorganisms detected in blood culture, has been increasingly reported, with rates ranging from 2 to 20% of all BSI episodes [8,9,10,11,12]. Patients with PMB often require a longer hospital stay [11], and have a mortality rate approximately double that of patients with monomicrobial BSI; ranging from 30 to 48% [8, 9, 12]. The identification of an infectious focus is especially important for a favorable prognosis. Although the infectious foci vary according to the clinical situation, catheter-related infections [8], intra-abdominal infections [9], and respiratory tract infections [11] are the most commonly reported sources of PMB. Previous studies have found that Gram-negative bacteria are detected in more than 70% of cases of PMB [8, 9], while Gram-positive bacteria are detected in 42–52% of cases [8, 9, 11]. Staphylococcus spp. [13], Acinetobacter spp. [14], Klebsiella spp. [15], and Candida spp. [10] are common pathogens detected in PMB. Determining the clinical and microbiological characteristics of PMB may help to identify the focus of infection and contribute to appropriate antimicrobial treatment.
Despite past studies, information on the clinical characteristics of PMB is limited. The purpose of this study was to investigate the clinical characteristics of PMB to contribute to reducing the mortality rate.
Methods
Study design and setting
We performed a multicenter retrospective observational study at three acute-care medical institutions in Okayama Prefecture, Japan: Okayama University Hospital (865 beds), Okayama City Hospital (400 beds), and Tsuyama Chuo Hospital (515 beds) between January 2014 and March 2019. These hospitals were equipped with in-house microbiology laboratories containing automated blood culture systems: the BD BACTEC™ FX system (Becton, Dickinson, and Co., NJ, USA) at Okayama University Hospital and Okayama City Hospital, and the BACT/ALERT VIRTUO® R3.0 System (bioMérieux Japan Ltd.; Tokyo, Japan) at Tsuyama Chuo Hospital. Two sets of blood culture samples per patient were routinely submitted to the laboratory, and the cultures were incubated for 7 days at Okayama University Hospital, 6 days at Okayama City Hospital, and 5 days at Tsuyama Chuo Hospital.
Ethical approval
Ethical approval was obtained from the institutional review board of Okayama University Hospital (no. 1908-056). The requirement for informed consent was waived because the study was a retrospective analysis of anonymized routinely collected data.
Blood culture inclusion criteria
PMB was defined as the presence of two or more distinct species isolated from a single blood culture episode. Examples are given in Supplemental Fig. 1. Different organisms identified from blood cultures drawn on days within a one-week period were not included, as in a previous study [12]. Positive blood cultures from samples drawn a week or more apart were defined as different episodes of blood culture testing. Blood cultures were excluded from our study if they contained microorganisms associated with the skin, including coagulase-negative Staphylococci, Bacillus spp., Corynebacterium spp., Lactobacillus spp., Propionibacterium spp., or viridans group streptococci in only one of the two blood samples as these were regarded as contaminants [16,17,18]. When these skin-associated microorganisms grew in two or more blood culture samples, they were considered as pathogens [8, 10]. Hospital-acquired infections were defined as those occurring > 48 h after hospital admission and were detected from the first positive blood culture [19]. The organisms identified were divided into four groups: Gram-positive, Gram-negative, anaerobic, and yeast. Anaerobic bacteremia was defined as the isolation of obligate anaerobes, such as Bacteroides spp., Bifidobacterium spp., Clostridium spp., Eubacterium spp., Lactobacillus spp., Peptostreptococcus spp., Fusobacterium spp., Prevotella spp., Actinomyces spp., Treponema spp., Eggerthella spp., Veillonella spp., and Propionibacterium spp. [3].
Data collection
Clinical and microbiological records of all patients with positive blood cultures between January 2014 and March 2019 were investigated. First, patients with monomicrobial BSI were excluded, then patients with potential contaminants, multiple BSI cultures within one week in an individual patient, and patients aged < 16 years were excluded. We excluded patients aged < 16 years because in our hospitals, patients aged ≤ 15 years are usually treated by pediatricians and usually only have one sample submitted for blood culture. Data on age, sex, comorbidities, isolated organisms, infectious focus, and patient outcomes were extracted from the medical records. According to the diagnosis provided in the medical records, the sources of bacteremia were classified into urinary tract infection, lower respiratory tract infection, intra-abdominal infection, skin and soft tissue infection, catheter-related bloodstream infection (CRBSI), and others. A case was defined as a mixed infection if it contained two or more different foci of infection. Cases without an identified infectious focus were regarded as primary bacteremia. Data were recorded on the comorbidities of patients with PMB, including diabetes mellitus, chronic obstructive pulmonary disease, end-stage renal disease, cirrhosis, and immunosuppression, which have been reported as risk factors for BSI [3,4,5]. Patients with active hematologic malignancy, neutropenia (neutrophil count in peripheral blood < 1500/μL), post-splenectomy, or receiving therapy with immunosuppressive agents, such as anti-cancer chemotherapy, corticosteroids (≥ 1 mg per day), and any biologic drugs, were defined as immunosuppressed.
Primary outcome and statistical analysis
Continuous variables were described as medians and interquartile ranges (IQR) and groups were compared using the Mann–Whitney U test. Categorical variables were reported as frequencies and percentages and groups were compared using Chi-square tests or Fisher’s exact test, as appropriate. The primary outcome was the overall 30-day mortality rate from diagnosis of PMB. The survival data were analyzed by Kaplan–Meier plots and compared using log-rank tests. The data were analyzed using EZR software, a graphic user interface for the R 3.5.2 software (The R Foundation for Statistical Computing, Vienna, Austria) [20]. p values < 0.05 were considered statistically significant.
Results
Clinical characteristics of the study patients
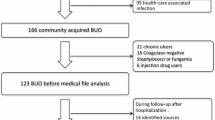
The enrolment flowchart is shown in Fig. 1. During the study period, the total number of positive blood cultures was 7233, with 6425 (88.8%) monomicrobial BSI cultures and 808 (11.2%) PMB cultures. A total of 301 cultures were excluded due to contamination, multiple cultures from the same patient within a week, and patients aged < 16 year. A total of 507 (7.0%) episodes of PMB were included in the analysis, of which 382 (5.3%) and 125 (1.7%) episodes were caused by two pathogens and three or more pathogens, respectively (referred to hereafter as “PMB 2 pathogens” and “PMB ≥ 3 pathogens”). The annual number of PMB episodes at each facility is shown in Supplementary Table 1.
The clinical characteristics of the eligible patients are shown in Table 1. The median age of patients was 78 years (IQR: 67–85 years) and 65.3% were male. The majority of patients with PMB (n = 437, 86.2%) were aged ≥ 60 years (Supplemental Fig. 2). Overall, 55.0% of the patients with PMB had no comorbidities, while 25.4% had diabetes mellitus and 17.4% were immunosuppressed. There were no statistically significant differences in the underlying comorbidities between cases of PMB 2 pathogens and PMB ≥ 3 pathogens. Of the total cases, 58.6% were community acquired, with no statistically significant difference between the two groups.
Kaplan–Meier curves of 30-days survival rates for polymicrobial bacteremia (PMB) patients, by a the number of causative pathogens and b infectious focus detected or unknown. Survival rates at 30 days between groups of PMB cases with a focus of infection detected and unknown were statistically different (16.3% and 24.5%; p = 0.031)
Pathogens detected in polymicrobial bacteremia
Collectively, 1,182 pathogens were isolated from patients with PMB (Table 2). Gram-positive and Gram-negative bacteria accounted for 38.6% and 46.2% of all bacterial species, respectively. More specifically, Enterococcus spp. was the most frequently isolated genus (16.4%), followed by Klebsiella spp. (13.9%) and E. coli (13.8%).
Infectious focus of polymicrobial bacteremia according to the Gram-staining pattern
The infectious focus was identified in 397 (78.3%) patients with PMB (Table 3). The most common source of PMB was intra-abdominal infection (n = 239, 47.1%), followed by primary bacteremia (n = 110, 21.7%) and urinary tract infection (n = 57, 11.2%). Cholangitis (n = 156) accounted for 65.3% of the intra-abdominal infection. More details in the clinical diagnoses of the patients with PMB are provided in Supplementary Table 2. There was no significant difference in the frequency of infectious focus between PMB 2 pathogens and PMB ≥ 3 pathogens.
The five most common foci of PMB among the study patients according to the Gram-staining pattern are shown in Table 4. The most common Gram-staining combination pattern was Gram-positive cocci (GPC)/Gram-negative rods (GNR) (204 cases, 40.2%). In cases of GPC (chain form)/GNR, 65.3% were derived from intra-abdominal infections. The GNR/GNR and GPC/GPC combinations accounted for 17.8% and 10.5% of the cases of PMB, respectively. These combination patterns may not be identified as PMB on Gram staining. The GNR/GNR pattern was frequently caused by intra-abdominal infections (64.4%) and UTIs (14.4%), and the most common source of the GPC/GPC combination was primary bacteremia (37.7%). When compared among various Gram-staining patterns, a combination of GPC (chain form)/GNR was significantly associated with a diagnosis of intra-abdominal infection, excepting GNR/GNR pattern (Table 5).
Mode of onset and infectious focus in patients with polymicrobial bacteremia
We first stratified the PMB cases as either community acquired or hospital acquired (Supplementary Table 3). Compared to patients with hospital-acquired PMB, patients with community-acquired PMB were older (median age, 81 years vs. 73 years), were less likely to have comorbidities (61.3% vs. 46.2%), and were less likely to be immunosuppressed (10.4% vs. 27.1%). As the infectious focus was identified in 93.9% and 56.2% of cases of community-acquired and hospital-acquired PMB, respectively (p < 0.001). Intra-abdominal infections were common in patients with community-acquired PMB (62.6%), while primary bacteremia (43.8%) and catheter-related infection (11.9%) were common in patients with hospital-acquired PMB.
We then stratified the PMB cases according to whether the infectious focus was identified (Supplementary Table 4). Patients with an identified infectious focus were older than those in whom no focus was identified (median age: 80 years vs. 71 years) and caused by community-onset disease (70.3% vs. 16.4%). The proportions of patients without any comorbidities were higher in those identified the infectious focus (59.2% vs. 40.0%).
Mortality rates in patients with polymicrobial bacteremia
The overall 30-day mortality rate of patients with PMB was 18.1% (Fig. 2). The 30-day mortality rates of patients with PMB 2 pathogens and PMB ≥ 3 pathogens did not differ significantly (16.2% and 24.0%, respectively; p = 0.073). However, the 30-day mortality rate was significantly higher in patients with an unknown focus (primary bacteremia) than in those with a focus detected (24.5% and 16.3%, respectively; p = 0.031).
Discussion
We determined the clinical and microbiological characteristics of patients with PMB in a multicenter study. PMB accounted for 7.0% of all bacteremia episodes, which is similar to previous studies [9, 10]. An infectious focus of PMB was identified in approximately 75% of cases, of which half occurred as a result of intra-abdominal infection. Based on the Gram staining findings, a combination of GPC (chain form)/GNR was indicative of intra-abdominal infection. The number of pathogens detected in blood culture was not significantly associated with prognosis; however, the prognosis of patients with primary PMB was significantly poorer than that of patients with an identified focus of infection.
According to previous reports, the proportion of PMB in all BSI episodes varies according to the clinical setting, ranging from 2% in a tertiary care hospital in Israel [8], to 6.7% and 10.9% in emergency departments in Taiwan and Spain [9, 10], and 20.2% in an intensive care unit in Spain [11]. In this study, the proportion of PMB 2 and PMB ≥ 3 pathogens were 75.3% and 24.7%, respectively. This is consistent with previous literature reporting that 77–85% of PMB cases were caused by two pathogens and 15–23% of cases were caused by three or more pathogens [9, 10]. The clinical backgrounds of patients in the present study were similar to those of previous studies, with a male predominance and diabetes mellitus being the most common comorbidity [8,9,10,11,12]. Notably, 55% of the patients with PMB in this study had no reported comorbidities. Other risk factors for PMB include recent invasive procedures, presence of foreign bodies, biliary tract infection, hospital-acquired infection, nursing home residence, stroke, afebrile presentation, neutropenia, presence of a biliary tract catheter, intra-abdominal infection, and abscess formation [8, 12, 13, 21]. In this study, there was no significant difference in risk factors between patients with PMB 2 and PMB ≥ 3 pathogens.
In this study, the most common causative bacteria of PMB were Enterococcus spp. (16.4%), followed by Klebsiella spp. and E. coli. Previous studies have also reported that Enterococcus spp. (13–20%) are frequently isolated in patients with PMB [8,9,10], despite E. coli and Staphylococcus spp. being the most common isolates in blood culture samples in general [3, 22]. These data suggest that it is important to treat patients with PMB with empiric antimicrobial therapy targeting Enterococcus spp. However, a recent, larger prospective study reported that E. coli was the most common pathogen (25.4%), followed by Staphylococcus aureus (15.2%) [23]. Another study conducted in two tertiary emergency departments in Taiwan found that Pseudomonas aeruginosa, Acinetobacter baumannii, Candida spp., and anaerobes were more commonly isolated in patients with PMB than in those with monomicrobial BSI [12]. The causative pathogens of PMB may thus vary according to the clinical situation; whether the infections are community-acquired or hospital-acquired. Patients with risk factors such as immunosuppression and intravascular devices may develop more complicated infections, including CRBSI and febrile neutropenia.
The most common cause of PMB in this study was intra-abdominal infection (47.1%), which is consistent with the findings of a study conducted in Taiwan [9, 12]. However, other studies have found that catheter-related [8] or respiratory tract infections [11] were the most common causes of PMB. The differences in the infectious focus of PMB may be attributable to differences in the study setting. In this study, we found that intra-abdominal infections were most common in patients with community-acquired PMB, while CRBSI and primary bacteremia were more common in patients with hospital-acquired PMB. Usually, patients undergo catheter insertion in hospital settings; thus, it is understandable that CRBSI was mostly observed in patients with hospital-acquired PMB. In this study, the proportion of cases of PMB attributable to CRBSI was 5.1% which is lower than the 20–28% reported in previous studies [8, 11]. Notably, in this study 21.7% of cases of PMB were attributable to primary bacteremia, with primary bacteremia being more common in patients with hospital-acquired PMB than in patients with community-acquired PMB. A previous study found that there was less accuracy in diagnosing CRBSIs than in diagnosing BSIs from other sources (68% vs. 78%; p < 0.001) [24]. Accordingly, we presume that some of the patients in the study with hospital-acquired PMB, which was classified as primary bacteremia, may have had undiagnosed CRBSI. Moreover, hospitalized patients tend to have more complicated conditions due to factors such as the use of various invasive devices, surgery, use of antimicrobials, and immunosuppression. The combination of these background factors in hospitalized patients can make it difficult for clinicians to identify the infectious focus correctly in patients who develop hospital-acquired BSI. In another study, the infectious source could not be determined in 24% of patients with hospital-acquired PMB [11].
This study shows that the combination patterns found by Gram staining can help in diagnosing the infectious source of PMB. This can help to identify the source of PMB rapidly and enable appropriately treatment to be initiated. The most common combination pattern in this study was GPC/GNR, which accounted for 40.2% of PMB. Of these, 81.9% had GPC (chain form)/GNR pattern, and 65.3% of these cases had an intra-abdominal source. Thus, the GPC (chain form)/GNR pattern indicates that the gastrointestinal tract is the most likely source.
The mortality of patients with PMB has been reported to be higher than patients with monomicrobial BSI. For example, the 30-day mortality rates in PMB and monomicrobial BSI in a tertiary care hospital in Israel were 48% and 33%, respectively [8]. Compared to these previous studies, the overall 30-day mortality rate of patients with PMB in this study was relatively low at 18.1%, although we did not compare PMB with monomicrobial BSI. To date, few studies have reported differences in prognosis based on the number of isolated pathogens. Our study showed that the 30-day mortality of patients with PMB ≥ 3 pathogens was higher than that of patients with PMB 2 pathogens, although the difference was not statistically significant. When an increased number of pathogens are involved in bacteremia, the probability of treatment failure is higher. This could contribute to the increased mortality in cases of PMB ≥ 3 pathogens. Although, there was a significant difference in 30-day mortality between the patients with infectious focus detected and those with unknown focus. Bacteremia of unknown origin generally has a higher mortality than bacteremia with an identified source [25]. Thus, it is important to identify the source of bacteremia in cases of PMB to enable appropriate treatment and a better prognosis.
A key strength of this study was using clinical and microbiological data from multiple locations. In particular, a novel finding of this study is the usefulness of Gram-staining patterns in identifying the infectious focus of PMB. However, this study has several limitations. First, the data were retrospectively collected, and the infectious sources were determined according to medical records. Although systemic examinations with computed tomography were performed in many patients, the infectious focus may have been under ascertained. Thus, the reported clinical diagnosis may not comply with the standard diagnostic criteria. Second, some cases of the contamination may have been included because our protocol included all cases with positive blood cultures that detected two or more pathogens. Third, the blood culture environments (equipment, culture bottles, and culture periods) among the three facilities were different, which may have affected the results. Fourth, we did not investigate the relationship between antimicrobial treatment and prognosis in this study. This should be investigated in future studies. Finally, we did not compare the clinical backgrounds and prognoses of patients with PMB and monomicrobial BSI.
In summary, we have highlighted the clinical and microbiological features of PMB cases in a multicenter study. Half of the PMB cases were due to intra-abdominal infection and the Gram-staining combination pattern of GPC (chain form)/GNR was strongly associated with intra-abdominal infection. PMB cases with unknown etiology had a poorer prognosis. The study results could contribute to the early diagnosis, proper treatment, and improvement in the outcomes of patients with PMB.
Availability of data and material
Detailed data is available on request from the corresponding author.
Code availability
Not applicable.
References
Goto M, Al-Hasan MN. Overall burden of bloodstream infection and nosocomial bloodstream infection in North America and Europe. Clin Microbiol Infect. 2013;19:501–9. https://doi.org/10.1111/1469-0691.12195.
Timsit JF, Ruppé E, Barbier F, Tabah A, Bassetti M. Bloodstream infections in critically ill patients: an expert statement. Intensive Care Med. 2020;46:266–84. https://doi.org/10.1007/s00134-020-05950-6.
Ngo JT, Parkins MD, Gregson DB, Pitout JDD, Ross T, Church DL, et al. Population-based assessment of the incidence, risk factors, and outcomes of anaerobic bloodstream infections. Infection. 2013;41:41–8. https://doi.org/10.1007/s15010-012-0389-4.
van Hal SJ, Jensen SO, Vaska VL, Espedido BA, Paterson DL, Gosbell IB. Predictors of mortality in staphylococcus aureus bacteremia. Clin Microbiol Rev. 2012;25:362–86. https://doi.org/10.1128/CMR.05022-11.
Ballouz T, Aridi J, Afif C, Irani J, Lakis C, Nasreddine R, et al. Risk factors, clinical presentation, and outcome of Acinetobacter baumannii bacteremia. Front Cell Infect Microbiol. 2017;7:156. https://doi.org/10.3389/fcimb.2017.00156.
Shankar-Hari M, Harrison DA, Rubenfeld GD, Rowan K. Epidemiology of sepsis and septic shock in critical care units: comparison between sepsis-2 and sepsis-3 populations using a national critical care database. Br J Anaesth [Internet]. 2017;119:626–36. https://doi.org/10.1093/bja/aex234.
Bauer M, Gerlach H, Vogelmann T, Preissing F, Stiefel J, Adam D. Mortality in sepsis and septic shock in Europe, North America and Australia between 2009 and 2019-results from a systematic review and meta-analysis. Crit Care Crit Care. 2020;24:1–9.
Goldman S, Itshaki O, Shochat T, Gafter-Gvili A, Yahav D, Rubinovitch B, et al. Risk factors and outcome of polymicrobial bacteremia: a retrospective cohort study. Isr Med Assoc J. 2020;22:279–84.
Lin JN, Lai CH, Chen YH, Chang LL, Lu PL, Tsai SS, et al. Characteristics and outcomes of polymicrobial bloodstream infections in the emergency department: A matched case-control study. Acad Emerg Med. 2010;17:1072–9. https://doi.org/10.1111/j.1553-2712.2010.00871.x.
Bouza E, Burillo A, Muñoz P, Guinea J, Marín M, Rodríguez-Créixems M. Mixed bloodstream infections involving bacteria and Candida spp. J Antimicrob Chemother. 2013;68:1881–8. https://doi.org/10.1093/jac/dkt099.
Sancho S, Artero A, Zaragoza R, Camarena JJ, González R, Nogueira JM. Impact of nosocomial polymicrobial bloodstream infections on the outcome in critically ill patients. Eur J Clin Microbiol Infect Dis. 2012;31:1791–6. https://doi.org/10.1007/s10096-011-1503-8.
Yo CH, Hsein YC, Wu YL, Hsu WT, Ma MHM, Tsai CH, et al. Clinical predictors and outcome impact of community-onset polymicrobial bloodstream infection. Int J Antimicrob Agents. 2019;54:716–22. https://doi.org/10.1016/j.ijantimicag.2019.09.015.
Park SY, Park KH, Bang KM, Chong YP, Kim SH, Lee SO, et al. Clinical significance and outcome of polymicrobial Staphylococcus aureus bacteremia. J Infect. 2012;65:119–27. https://doi.org/10.1016/j.jinf.2012.02.015.
Wang YC, Ku WW, Yang YS, Kao CC, Kang FY, Kuo SC, et al. Is polymicrobial bacteremia an independent risk factor for mortality in Acinetobacter baumannii bacteremia? J Clin Med. 2020;9:153. https://doi.org/10.3390/jcm9010153.
Liu Q, Wu J, Wang Z, Wu X, Wang G, Ren J. Polymicrobial bacteremia involving Klebsiella pneumoniae in patients with complicated intra-abdominal infections: frequency, co-pathogens, risk factors, and clinical outcomes. Surg Infect (Larchmt). 2019;20:317–25. https://doi.org/10.1089/sur.2018.207.
Han XY, Kamana M, Rolston KVI. Viridans streptococci isolated by culture from blood of cancer patients: clinical and microbiologic analysis of 50 cases. J Clin Microbiol. 2006;44:160–5. https://doi.org/10.1128/JCM.44.1.160-165.2006.
Weinstein MP. Blood culture contamination: Persisting problems and partial progress. J Clin Microbiol. 2003;41:2275–8. https://doi.org/10.1128/JCM.41.6.2275-2278.2003.
Weinstein MP. Current blood culture methods and systems: clinical concepts, technology, and interpretation of results. Clin Infect Dis. 1996;23:40–6. https://doi.org/10.1093/clinids/23.1.40.
Rello J, Ricart M, Mirelis B, Quintana E, Gurgui M, Net A, et al. Nosocomial bacteremia in a medical-surgical intensive care unit: epidemiologic characteristics and factors influencing mortality in 111 episodes. Intensive Care Med. 1994;20:94–8. https://doi.org/10.1007/BF01707661.
Kanda Y. Investigation of the freely available easy-to-use software “EZR” for medical statistics. Bone Marrow Transplant. 2013;48:452–8. https://doi.org/10.1038/bmt.2012.244.
Al Majid F, Aldrees A, Barry M, Binkhamis K, Allam A, Almohaya A. Streptococcus anginosus group infections: management and outcome at a tertiary care hospital. J Infect Public Health. 2020;13:1749–54. https://doi.org/10.1016/j.jiph.2020.07.017.
Rhodes J, Jorakate P, Makprasert S, Sangwichian O, Kaewpan A, Akarachotpong T, et al. Population-based bloodstream infection surveillance in rural Thailand, 2007–2014. BMC Public Health. 2019;10:19–521. https://doi.org/10.1186/s12889-019-6775-4.
Schöneweck F, Schmitz RPH, Rißner F, Scherag A, Löffler B, Pletz MW, et al. The epidemiology of bloodstream infections and antimicrobial susceptibility patterns in Thuringia, Germany: a five-year prospective, state-wide surveillance study (AlertsNet). Antimicrob Resist Infect Control. 2021;10:132. https://doi.org/10.1186/s13756-021-00997-6.
Ruiz-Giardin JM, Ochoa CI, Velázquez RL, Jaqueti AJ, García Arat MI, SanMartín López JV, et al. Blood stream infections associated with central and peripheral venous catheters. BMC Infect Dis. 2019;19:1–841. https://doi.org/10.1186/s12879-019-4505-2.
Courjon J, Demonchy E, Degand N, Risso K, Ruimy R, Roger PM. Patients with community-acquired bacteremia of unknown origin: Clinical characteristics and usefulness of microbiological results for therapeutic issues: a single-center cohort study. Ann Clin Microbiol Antimicrob. 2017;16:40. https://doi.org/10.1186/s12941-017-0214-0.
Acknowledgements
We would like to thank Editage (http://www.editage.jp) for English language editing.
Funding
No funding was received for conducting this study.
Author information
Authors and Affiliations
Contributions
Study concept: HH. Data extraction: SF, KF, SK, HY, and MK. Statistical analysis: SF and HH. Drafting of the manuscript: SF. Revision of the manuscript: HH. Critical revision: FO.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Ethics approval
Ethics approval was obtained from the institutional review board of Okayama University Hospital (No. 1908–056).
Informed consent
The requirement for informed consent was waived because the study was a retrospective analysis of routinely collected data that were anonymized.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Fukushima, S., Hagiya, H., Fujita, K. et al. Clinical and microbiological characteristics of polymicrobial bacteremia: a retrospective, multicenter study. Infection 50, 1233–1242 (2022). https://doi.org/10.1007/s15010-022-01799-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s15010-022-01799-7