Abstract
We presented the synthesis of a new Schiff base and its complexes properties with some transition metal ions in this study. (1-amino-2-thioxo-4-p-tolyl-1,2-dihydropyrimidin-5-yl)(p-tolyl)methanone was synthesized as the starting material, and (1-((2-hydroxynaphthalen-1-yl)methyleneamino)-2-thioxo-4-p-tolyl-1,2-dihydropyrimidin-5-yl)(p-tolyl)methanone (Hnafmmp) (1) and its Ni(II) (2), Pd(II) (3), Pt(II) (4), Cu(II) (5), Co(II) (6) complexes were prepared using above-mentioned starting material. The structures of these new compounds (ligand and its complexes) were characterized with UV–Vis, FTIR, LC–MS, 1H NMR, 13C NMR, magnetic susceptibility, conductivity measurement, X-ray powder diffraction method, and thermal analyses techniques. The 1H NMR chemical shifts of all complexes were calculated by using the gauge-invariant atomic orbital HF (3.21G) method in DMSO phases. All measured results were compared with the experimental data. The ligand and its complexes were also evaluated as antimicrobial agents against representative strains bacteria. Furthermore, we studied the catalytic activity of these compounds in Suzuki–Miyaura cross-coupling reaction in aqueous media.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Research on metal organic complexes represents one of the most active areas of material science and chemical research. Major advances have been made in these materials due to their interesting properties. Schiff bases are some of the most widely used organic compounds in this area. They are used as catalysts, pigments, dyes, polymer stabilizers, and intermediates in organic synthesis [1]. In particular, pyrimidine derivatives have demonstrated a broad range of biological activities, including antimalarial, antifungal, antiproliferative, antibacterial, anti-inflammatory, antiviral, and antipyretic properties and sedative-hypnotic activity [1, 2]. In addition, these derivatives have attracted significant attention because of the application in drug development. Schiff base ligands with sulfur and nitrogen donor atoms in their structures act as good chelating agents for the transition and non-transition of metal ions [3,4,5,6]. There is a continuing interest in the metal complexes of Schiff bases. Because of the presence of both hard nitrogen or oxygen and soft sulfur donor atoms in the backbones of these ligands, they readily coordinate with a wide range of transition metal ions yielding stable and intensely colored metal complexes, some of which have been shown to exhibit interesting physical and chemical properties [7] and potentially useful biological activities [8,9,10].
In this work, we synthesized a new aromatic ligand (1). Subsequently, a series of new Schiff base complexes (1–5) were prepared and characterized by spectroscopic and analytic techniques (Figs. 1, 2). We studied their antibacterial activities against bacteria using the microdilution method. SAR and theoretical studies were performed. The catalytic activities of these compounds were tested in Suzuki–Miyaura cross-coupling reaction. The coupling products were obtained in good yields.
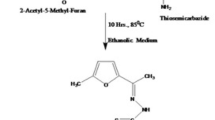
(1-amino-2-thioxo-4-p-tolyl-1,2-dihydropyrimidin-5-yl)(p-tolyl)methanone obtained from (Z)-2-(1-phenylethylidene)hydrazinecarbothioamide with 4-(4-methylbenzoyl)-5-(4-methylphenyl) furan-2,3-dione has been reported before [11]. Schematic representation of the synthesis route of the compound is shown in Scheme 1.
Experimental
Physical measurements
The following instrumental devices were used in this work: Mattson 1000 FTIR spectrometer, AGILENT 1100 (LC–MS), Bruker 400 MHz spectrometer, Bruker AXS D8 diffractometer, Unicam-UV 2-100 spectrometer, PerkinElmer Diamond TG/DTA thermal analyzer, Opti MELT 3 hot-stage apparatus, and Siemens WPA C 35 conductometer.
General procedure for the synthesis
Chemistry of 1
The synthesis of (1-((2-hydroxynaphthalen-1-yl)methyleneamino)-2-thioxo-4-p-tolyl-1,2-dihydropyrimidin-5-yl)(p-tolyl)methanone (1) was carried out according to general pathway illustrated in our previous study [12]. Its details are below: This compound was prepared using (1-amino-2-thioxo-4-p-tolyl-1,2-dihydropyrimidin-5-yl)(p-tolyl)methanone and 2-hydroxy-1-naphthaldehyde. It was obtained as a light-yellow solid, yield 58%; m.p. 203 °C (disintegration); UV (EtOH) λ max (log ε) 520 (3.84) nm; FTIR γ max 3616, 3080, 1639, 764 cm−1; 1H NMR (DMSO d6, 400 MHz,): δ: 10.76 (1 H, s), 9.08 (1 H, d, J: 8 Hz), 8.90 (1 H), 7.90–6.96 (14 H, m). 13C NMR (DMSO-d6, 400 MHz,): δ: 191.58, 165.72, 163.87, 160.87, 145.89, 145.14, 141.71, 136.82, 134.20, 133.70, 132.51, 130.61, 130.40, 129.84, 129.59, 129.42, 129.35, 129.31, 128.39, 128.30, 21.67, 21.41. LC/MZ (70 eV, APC1): 490.90 [M+ + H+].
Chemistry of 2, 3, 4, 5, 6 complexes
All complexes were prepared by the following standard method [12]. The following results were found:
Bis [(1-((2-hydroxynaphthalen-1-yl)methyleneamino)-2-thioxo-4-p-tolyl-1,2-dihydropyrimidin-5-yl)(p-tolyl)methanone] dichloro nickel(II) (2) was prepared using (1-amino-2-thioxo-4-p-tolyl-1,2-dihydropyrimidin-5-yl)(p-tolyl)methanone, 2-hydroxy-1-naphthaldehyde, and nickel(II)chloride. It was obtained as a light-brown solid, yield 58%; m.p. 220 °C (disintegration); UV (EtOH) λ max (log ε) 514.5 (5.63) nm; FTIR γ max 1618, 1280, 751 cm−1; 1H NMR (DMSO-d6, 400 MHz,): δ: 10.72 (1 H, s), 10.25 (1 H, s), 9.61 (2 H, d, J: 8 Hz), 8.30 (2 H), 7.99–5.38 (28 H, m), 2.66 (2 H, m, J: 12 Hz), 2.49 (2 H, m, J: 12 Hz), 2.35 (2 H, m, J: 12 Hz), 2.27 (2 H, m, J: 12 Hz), 2.16 (2 H, m, J: 12 Hz), 2.07 (2 H, m, J: 12 Hz). 13C NMR (DMSO-d6, 400 MHz,): δ: 193.22, 164.73, 145.17, 145.08, 141.70, 138.86, 134.02 (2C), 130.88 (4C), 130.62 (4C), 130.42, 130.23, 129.86, 129.78, 129.60, 129.40, 124.69, 124.56, 122.71, 119.19, 21.79, 21.66, 21.42, 21.30. LC/MZ(70 eV, APC1): 1039.70 [M+ + 2H+].
Bis [(1-((2-hydroxynaphthalen-1-yl)methyleneamino)-2-thioxo-4-p-tolyl-1,2-dihydropyrimidin-5-yl)(p-tolyl)methanone] palladium(II) chloride (3) was prepared using [(1-amino-2-thioxo-4-p-tolyl-1,2-dihydropyrimidin-5-yl)(p-tolyl)methanone], 2-hydroxy-1- naphthaldehyde, and palladium(II) chloride. It was obtained as a reddish-brown solid, yield 52%; mp 206 °C (disintegration); UV (EtOH) λ max (log ε) 514.5 (5.63) nm; FTIR γ max 751, 1279, 1600 cm−1; 1H NMR (DMSO-d6, 400 MHz,): δ: 10.76 (1 H, s), 10.75 (1 H, s), 9.08 (1 H, s), 9.07 (1 H, s), 8.91 (1 H, s), 8.90 (1 H, s), 7.89–7.25 (28 H, m), 2.53 (2 H, d, J: 8 Hz), 2.49 (2 H, d, J: 8 Hz), 2.41 (2 H, d, J: 12 Hz), 2.35 (2 H, d, J: 12 Hz), 2.32 (2 H, d, J: 12 Hz), 2.31 (2 H, d, J: 12 Hz), 2.27 (2 H, d, J: 12 Hz), 2.26 (2 H, d, J: 12 Hz), 2.24 (2 H, d, J: 12 Hz), 2.18 (2 H, d, J: 12 Hz), 2.16 (2 H, d, J: 12 Hz). 13C NMR (DMSO-d6, 400 MHz,): δ: 193.12, 191.59, 176.38, 164.53, 145.95, 144.16, 143.49, 141.69, 138.77, 134.19, 133.70, 132.47, 132.08, 130.96, 130.62, 130.22, 130.06, 129.95, 129.75, 129.59, 129.42, 129.26, 124.65, 122.81, 120.77, 119.24, 39.32, 21.75, 21.68, 21.41. LC/MZ (70 eV, APC1): 1086.59 [M+ + H+].
Bis [(1-((2-hydroxynaphthalen-1-yl)methyleneamino)-2-thioxo-4-p-tolyl-1,2-dihydropyrimidin-5-yl)(p-tolyl)methanone] platinum(II) chloride (4) was prepared using [(1-amino-2-thioxo-4-p-tolyl-1,2-dihydropyrimidin-5-yl)(p-tolyl)methanone], 2-hydroxy-1- naphthaldehyde, and platinum(II) chloride. It was obtained as a reddish-brown solid, yield 44%; m.p. 291 °C (disintegration); UV (EtOH) λmax (log ε) 344.6 (5.33) nm; FTIR γ max 750, 1280, 1602 cm−1; 1H NMR (DMSO-d6, 400 MHz,): δ: 10.00 (1 H, s), 10.01 (1 H, s), 8.90 (1 H, s), 8.80 (1 H, s), 8.70 (1 H, s), 8.60 (1 H, s), 8.30–6.80 (28 H, m) 2.49 (2 H, d, J: 8 Hz), 2.41 (2 H, d, J: 12 Hz), 2.37 (H, s), 2.33 (H, s), 2.31 (H, s), 2.29 (H, s), 2.26 (H, s), 2.24 (H, s), 2.18 (H, s), 2.16 (H, s). 13C NMR (DMSO-d6, 400 MHz,): δ: 193.21, 192.49, 175.28, 164.55, 146.95, 144.22, 143.54, 141.80, 139.22, 134.23, 133.81, 132.39, 132.05, 131.06, 130.80, 130.32, 130.04, 130.05, 129.88, 129.49, 129.40, 129.15, 124.45, 122.92, 121.07, 119.22, 39.32, 21.80, 21.88, 21.54.LC/MZ(70 eV, APC1): 1175.25 [M+ + H+].
Bis [(1-((2-hydroxynaphthalen-1-yl)methyleneamino)-2-thioxo-4-p-tolyl-1,2-dihydropyrimidin-5-yl)(p-tolyl)methanone] dichloro cupper(II) (5) was prepared using [(1-amino-2-thioxo-4-p-tolyl-1,2-dihydropyrimidin-5-yl)(p-tolyl)methanone], 2-hydroxy-1-naphthaldehyde, and cupper(II) chloride. It was obtained as a light-brown solid, yield 48%; m.p. 176 °C (disintegration); UV (EtOH) λ max (log ε) 475.8 (5.59) nm; FTIR γ max 1602, 1280, 759 cm−1. 1H NMR (DMSOd6, 400 MHz,): δ: 11.86 (1 H, s), 11.81 (1 H, s), 9.92 (1 H, s), 9.86 (1 H, s), 9.76 (1 H, s), 9.59 (1 H, s), 9.56–6.60 (28 H, m), 2.85 (2 H, d, J: 8 Hz), 2.80–2.29 (10 H, m). 13C NMR (DMSO-d6, 400 MHz,) δ: 191.58, 160.91, 145.93, 145.15, 141.73, 134.19, 130.65, 130.24, 129.96, 129.63, 129.44, 128.99, 128.28, 124.60, 119.12, 25.62, 21.82, 21.48, 20.80.LC/MZ(70 eV, APC1): 1044.70 [M+ + 2H+].
Bis [(1-((2-hydroxynaphthalen-1-yl)methyleneamino)-2-thioxo-4-p-tolyl-1,2-dihydropyrimidin-5-yl)(p-tolyl)methanone] dichloro cobalt(II) (6) was prepared using [(1-amino-2-thioxo-4-p-tolyl-1,2-dihydropyrimidin-5-yl)(p-tolyl)methanone], 2-hydroxy-1-naphthaldehyde, and cobalt(II) chloride. It was obtained as a dark-brown solid, yield 51%; m.p. 199 °C (disintegration); UV (EtOH) λ max (log ε) 320.2 (5.42) nm; FTIR γ max 1602, 1280, 751 cm−1. 1H NMR (DMSO-d6, 400 MHz,): δ: 11.99 (1 H, s), 11.80 (1 H, s), 9.84 (1 H, s), 9.71 (1 H, s), 9.16 (1 H, s), 8.88(1 H, s), 8.76 (2 H, t, J: 4 Hz), 8.73 (2 H, t, J: 12 Hz), 8.10 (2 H, t, J: 8 Hz), 8.08 (2 H, t, J: 8 Hz), 8.06 (4 H, t, J: 8 Hz), 8.02 (4 H, t, J: 16 Hz), 8.00 (4 H, t, J: 8 Hz), 7.91 (4 H, t, J: 36 Hz), 7.89 (2H, t, J: 12 Hz), 7.86 (2 H, t, J: 12 Hz), 7.84 (2 H, t, J: 8 Hz), 7.82 (2 H, t, J: 8 Hz) 7.79 (1 H, d, J: 16 Hz), 7.77 (4 H, d, J: 8 Hz), 7.73 (4 H, d, J: 24 Hz), 7.65 (4 H, d, J: 8 Hz), 7.63 (1 H, d, J: 8 Hz), 7.61 (2 H, d, J: 8 Hz), 7.59 (1 H, d, J: 12 Hz), 7.57 (1 H, d, J: 8 Hz), 7.55 (1 H, d, J: 8 Hz), 7.53 (1 H, d, J: 12 Hz), 7.44 (1 H, d, J: 4 Hz), 7.42 (1 H, d, J: 8 Hz), 7.40 (2 H, t, J: 4 Hz), 7.38 (2 H, t, J: 81 Hz), 7.36 (2 H, t, J: 8 Hz), 7.34 (2 H, t, J: 8 Hz), 2.53 (3 H, m), 2.49 (3 H, m), 2.36 (3 H, m), 2.32 (3 H, m). 13C NMR (DMSO-d6, 400 MHz,) δ: 191.80, 165.37, 147.94, 144.59, 141.23, 134.75, 132.34, 130.47, 130.24, 129.76, 129.59, 129.38, 129.15, 128.94, 128.35, 124.50, 123.72, 119.00, 116.04, 108.66, 107.73, 107.53, 39.93, 21.66, 21.39. LC/MZ(70 eV, APC1): 1038.10 [M+].
Biological activity evaluation
The procedures of antimicrobial activity study were performed according to the literature [12].
Minimal inhibitory concentration
Minimal inhibitory concentrations (MIC) [13,14,15,16,17] were determined by the microdilution broth method following the procedures recommended by the National Committee for Clinical Laboratory Standards (NCCLS, 2000). Trimethoprim and tetracycline were used as standard drugs.
General procedure for the Suzuki–Miyaura cross-coupling reaction
Compounds (0.03 mmol), aryl chloride (1 mmol), phenylboronic acid (1.5 mmol), base (2.0 mmol), and H2O/DMF (2/2 mL) were added to a small Schlenk tube, and the reaction mixture was heated up to various temperature for different time. At the end of the reaction, the mixture was cooled, extracted with ethyl acetate/hexane (1:5), and purified by flash chromatography on silica gel. The purity of the compounds was checked by GC–MS. Conversions were based on aryl chlorides.
Results and discussion
This report presented the synthesis, characterization, biologic, catalytic, and spectral studies of the ligand (1) and its five new nickel, palladium, platinum, copper, and cobalt complexes. According to FTIR, 1H NMR, and X-ray powder diffraction’s data, an octahedral geometry is observed around the nickel, copper, cobalt complexes and a square planar geometry around the palladium and platinum complexes, in which the ligand acts as a neutral bidentate one, coordinating through its nitrogen and sulfur atoms. In reality, ligand has carbonyl and hydroxyl groups. These groups are more effective than C=S group. Conversely, these attachment points are expected for complexation reactions. But according to 1H NMR results, the phenolic –OH signals of all compounds are determined between 10.26 and 10.79 ppm, and C=O signals are determined between 163–171 ppm and 1H NMR spectra of the free ligand task to (C=O) is not shifted. In IR spectra, ט(C=N) and ט(C=S) groups signals are shifted to –35, −63 cm−1 and −21, −39 cm−1, respectively. However, there is no shifting by –OH and C=O signals. This situation was investigated using Gaussian software program. The total energies were calculated with the HF (3.21G) method for each case. We have calculated less energy values when connected via sulfur atoms.
The results showed 1:2 (metal/ligand) stoichiometric for all the complexes and two additional values for the chloride ligands which is coordinated to the metal atoms (only Ni, Cu, Co). Ni(II), Cu(II), and Co(II) complexes have an octahedral geometry and Pd(II), Pt(II) complexes have a square planar geometry. However, in our previous studies [18, 19], an octahedral geometry was observed around the Cu(II) and Co(II) complexes and a square planar geometry for Ni(II), Pd(II), and Pt(II) complexes. All complexes found to be stable at room temperature in their solid states and generally soluble in CH2Cl2, DMSO, and DMF. According to these observed results, the nickel, cobalt, and copper complexes have a non-electrolytic nature. The platinum and palladium complexes have an electrolytic nature.
FTIR spectra
The –NH stretching vibrations were occurred at 3300–3500 cm−1. In here, the –NH stretching vibration was observed at 3450 cm−1 for (1). The aromatic structure showed the presence of –CH stretching vibrations in the range 3000–3100 cm−1, which is the characteristic region for –CH stretching. The –CH asymmetric stretching vibrations were observed at 3072 cm−1 for compound 1. The IR spectrum of compound (1) showed characteristic stretching bands corresponding to the carbonyl groups. These bands were observed at 1647 cm−1. The IR spectra of the Schiff base ligand showed a broad band at 3616 cm−1 [20]. Due to intramolecular hydrogen bonding between the H of OH and the azomethine nitrogen, this band was observed in low frequency (3080 cm−1). The IR spectra of the free ligand observed strong bands at 1639 and 764 cm−1 assigned to ט(C=N) and ט(C=S). These bands shifted to −39, −21 cm−1, and −14, −5 cm−1, respectively. These are indicating coordination via the azomethine nitrogen and thiocarbonyl of the Schiff base.
NMR spectra
DMSO used as a solvent to measure the 1H NMR spectra of 1 and its complexes. The sharp singlet peak was observed at 10.76 ppm, which is assigned to the phenolic proton of the ligand. The phenyl was observed as multiplet between 7.83 and 6.95 ppm. The azomethine proton and the pyrimidine ring (C–H) proton assigned as singlet at 9.08 and 8.90 ppm, respectively. The chemical shift values of the different types of protons in the examined compounds (1 and 2) are reported in Figs. 3 and 4.
In 13C NMR spectrum, the aromatic C peaks were observed between 117.26 and 145.84 ppm. The azomethine C and the pyrimidine ring C in the spectrum of 1 assigned at 190.65–192.25 and 155.07–171.84 ppm, respectively. The methyl carbons were observed in between 19.04 and 21.91 ppm.
The theoretical studies of 1H NMR were conducted with the HF (3.21G) method by Gaussian software program, and it was compared with the experimental data (Fig. 5). There are good correlations between theoretical and experimental results. However, hydrogen belonging to pyrimidine rings, hydrogen belonging to salycyl aldehyde’s, and hydrogen belonging to hydroxyl group of these complexes chemical shifts differ from theoretical chemical shifts. These results can be explained by the presence of an intramolecular hydrogen bond. The electronic charge density around of these atoms changes rapidly due to the influence of the hydrogen bridge bond or solvent effect. The chemical shift values of these hydrogens have been subtracted from the graph because of the above-mentioned reasons.
UV spectra
Experimental UV parameters and suggested geometries for complexes are presented in the Table 1 [21].
LC–MS spectra
LC/MS showed that Cu (5) and Co (6) complex give the molecular ion [M+] at 1038.7 and 1042.3 m/z, respectively. The Ni complex (2) gave the molecular ion [M+] + 2H+ at 1037.9 m/z. The Pd and Pt complexes (3, 4) gave the molecular ions, [M+] + H+ at the desired positions at 1089.59 and 1174.25 m/z. Separately, all complexes and ligand gave the fragmentation of the C6H5 group, which was observed at 79.2 m/z.
X-ray powder diffraction
The powder patterns of the ligand (Hnafmmp) were indexed in orthorhombic crystal systems with unit cell parameters of a: 12.91, b: 10.55, c: 9.878 Å. Compound 2 was indexed in an orthorhombic crystal system with unit cell dimensions of a: 13.03, b: 12.59, c: 8.75 Å. Compound 3 was indexed in an orthorhombic crystal system with unit cell dimensions of a: 6.39, b: 3.17, c: 2.00 Å. Compound 4 was indexed in a cubic crystal system with unit cell dimensions of a: 3.14 Å. Compound 5 was indexed in an orthorhombic crystal system with unit cell dimensions of a: 16.93, b: 10.27, c: 8.88 Å. Compound 6 was indexed in orthorhombic crystal system with unit cell dimensions of a: 15.78, b: 14.38, c: 6.66 Å.
Thermal studies
TG/DTA analyses for the compounds were taken in nitrogen at the temperature from 50 to 1200 °C. Compounds 1 and 3 were discussed as detail. The thermal decomposition of 1 (C30H23SO2N3) takes place in three stages. Firstly, –CHC10H6OH group degrades at 200 °C, shown as one maximum in the DTG curve at 195.26 °C with 30.00% mass loss (calc. = 31.84%). Secondly, one mole of the –C6H5CH3 and one mole of the –COC6H5CH3 groups have been degraded. It shows one peak at 207.57 °C in DTG curve for this stage. Third stage is related to the decomposition, and this stage finishes at 1200 °C. The [C60H46N6O4S2Cl2Pt] complex is thermally decomposed in four stages. Firstly, from 0 to 200 °C, two moles of chloride molecules degrade, shown as one maximum in the DTG curve at 200.00 °C with a 5.77% mass loss (calc. = 5.62%). Second stage’s mass loss of 25.34% is due to the degradation of two moles of –CHC10H6OH molecules, which is in agreement with the calculated value (calc. = 25.06%). The DTG curve shows one peak at 290.00 °C. The third stage’s mass loss of 70.94% is due to the degradation of two moles of –COC6H5CH3, two moles of –C6H5CH3, two moles of –N, and two moles of –S molecules, which is in agreement with the calculated value (calc. = 71.81%). The DTG curve shows one peak at 310.00 °C. The last stage is related to the decomposition of the remaining part of the complex molecule. The total residue (calc. = 15.67%) is Pt (ECPDS number: 04-0802). The residue indexed in a cubic crystal system with unit cell dimensions of a: 3.9231 Å. The thermal behaviors of the compounds are presented in Table 2.
The DTA/TG curves of the ligand (L) and its complexes are shown in Fig. 6.
Conductometric measurements
Measurements were taken at 25 °C in 10−3 M DMSO solutions using a conductivity meter. The complexes 2, 5, and 6 dissolve in DMSO to give a total one ion. These complexes did not have an electrolytic nature. The molar conductivity of complexes 2, 5, and 6 are 48, 32, and 70 S/M, respectively. The platinum and palladium complexes molar conductivity were found as 256 and 254 S/M, respectively.
Antimicrobial activity
Ligand and its complexes were evaluated by the microdilution method procedure for their in vitro antibacterial activity. All the compounds displayed activity against M. luteus, P. mirabilis, P. aeruginosa, and C. renale. On the other hand, none of the compounds displayed activity against E. aerogenes and S. epidermidis bacteria. E. aerogenes and S. epidermitis are pathogenic bacterium. They cause opportunistic infections; these include most types of infections. E. aerogenes quickly become resistant to standard antibiotics during treatment. S. epidermitis lives on the human skin and mucosa. It has ability to form biofilms on plastic devices and the most common infections on catheters and implant. The Cu, Ni, Pt complexes and Hnafmmp generally seemed to be more active against gram-positive bacteria. But, the Co and Pd complexes displayed poor activity against all gram-positive and gram-negative bacteria. This situation is contrary to with previous studies, where the ligand and its Pd and Pt complexes were found to be more active against gram-negative and gram-positive bacteria [18, 19]. The biological activity results displayed that complex 5(Cu(II)) provided good protection against convulsions while complex 2(Ni(II)) is significantly less active. The differences between the high and the low active complexes consist of the amount of distribution of the negative electrostatic potential around the sulfur and nitrogen atoms. Complex 5 had a high value for the negative electrostatic potential around the sulfur and nitrogen atoms, which is the most potent compound. On the other hand, complex 2 had lower negative electrostatic potential around its sulfur and nitrogen atoms, which is the very least active compound. This is evident in the electrostatic maps of the two complexes in Fig. 7.
The molecular electrostatic potential maps for the two complexes are shown in Fig. 7. The optimized geometries of complexes are shown in Fig. 8. The antimicrobial activity results are given in Table 3.
SAR analysis
The parameters of the synthesized compounds were selected. The chemical structures of the compounds are set up with the GAUSSIAN 3.0 program and optimized with the HF (3.21G) method. HOMO and LUMO were calculated and founded values are given in Table 4.
The treatment of the HOMO and LUMO is based on the general principles governing the nature of reactions. The LUMO energy is related to the electron affinity susceptibility of the molecule toward attack by nucleophiles. On the other hand, hard and soft nucleophiles have been directly related to the energy of HOMO–LUMO: hard electrophiles have a high energy. LUMO and soft electrophiles have a low energy. There was a linear relationship between LUMO-L. monocytogenes and HOMO-L. monocytogenes where R 2 was obtained as 0.992 and 0.993, respectively (Figs. 9, 10).
Catalytic activities in Suzuki–Miyaura reaction
First experiment was performed with phenylboronic acid and 4-chloroacetophenone as model compounds to find optimum conditions. The coupling product 1 was obtained in 70% conversion in 4 catalyst (Table 5, entry 1). After that, to assure this tendency, two catalysts were used in the same reaction, and good conversions were obtained such as 61%. Then, 4-chloroanisole was reacted with phenylboronic acid by using these novel complexes (Table 5, entries 4–8). For this transformation, complexes 5, 6, and 4 were found to be effective catalysts at 80 °C for 2 h. The coupling of phenylboronic acid with chlorobenzene was observed to proceed under a variety of conditions. Two different bases (K2CO3 and NaOH) and solvents (DMF and H2O) as well as synthesized novel compound (6) were employed. When two bases were used, good results were obtained. At low time and temperature, NaOH was used as a base and achieved very good result such as 80% conversion. Therefore, both K2CO3 and NaOH were a good choice in DMF/H2O systems as a base. Water was used as solvent in this coupling reaction. When Table 5 is examined, it is seen that the results of the Suzuki–Miyaura cross-coupling reaction are between 34 and 95. As a result of this study, we concluded that 4 and 5 complexes in the Suzuki–Miyaura coupling reactions showed high activity in comparison with other complexes. As a result, synthesized complexes are found to be effective Suzuki–Miyaura cross-coupling reactions. In our previous study, the catalytic activities in the end Suzuki–Miyaura coupling (DMF-H2O) reactions of synthesized compounds were examined [22, 23] and obtained similar results in this study.

Conclusion
As conclusion, one ligand and its complexes were synthesized and characterized. From the spectroscopic characterization, it was concluded that compound 1 acts as a bidentate ligand, coordinating through –C=N and –S. The Ni(II), Co(II), and Cu(II) complexes had an octahedral geometry, and Pd(II), Pt(II) complexes had a square planar geometry. Compound 1 (Hnafmmp) was indexed in orthorhombic crystal systems with unit cell parameters of a: 12.91, b: 10.55, c: 9.878 Å. Complexes 2, 3, 4, 5 and 6 were indexed in an orthorhombic, an orthorhombic, a cubic, an orthorhombic, and an orthorhombic crystal systems, respectively. The thermal decomposition of compound 1 (C30H23SO2N3) takes place in three stages. The complexes with the molecular formula [C60H46N6O4S2Cl2M] (M, Ni(II), Pd(II), Pt(II), Cu(II), Co(II)) were thermally decomposed in four stages. All compounds demonstrated biological activity against all the tested strains. Tested compounds in Suzuki–Miyaura cross-coupling reactions for catalytic activities gave very good conversions. 1H NMR studies of these complexes were calculated as theoretically.
References
D.N. Dhar, C.L. Taploo, J. Sci. Ind. Res. 41, 501 (1982)
P. Przybyslki, A. Huczynski, K. Pyta et al., Curr. Org. Chem. 13, 124 (2009)
N.K. Kaushik, A.K. Mishra, Ind. J. Chem. 42A, 2762 (2003)
B. Dede, F. Karipcin, M. Cengiz, J. Hazard. Mater. 163, 1148 (2009)
A.K. Mishra, N. Manav, N.K. Kaushik, Spectrochim. Acta A 61, 3097 (2005)
M.G. Abd El Wahed, E.M. Nour, S. Teleb, S. Fahim, J. Therm. Anal. Calorim. 76, 343 (2004)
M. Ak, D. Taban, H. Deligoz, J. Hazard. Mater. 154, 51 (2008)
S. Karabocek, S. Güner, N. Karabocek, J. Inorg. Biochem. 66, 57 (1997)
Y.M. Al-Hıari, B.A. Sweileh, Asian J. Chem. 18, 2285 (2006)
N. Zanatta, S.H. Alues, H.S. Coelho et al., Bioorg. Med. Chem. 15, 1947 (2007)
Z. Önal, İ. Yıldırım, Heterocycl. Commun. 13, 113 (2000)
H.G. Aslan, Z. Onal, Med. Chem. Res. 23, 2596–2607 (2014)
K.K. Anderson, in Comprehensive Organic Chemistry, vol. 3, ed. by D.N. Jones (Pergamon Press, Oxford, 1979), p. 345
S.L. Graham, T.H. Scholz, Synthesis 12, 1031 (1986)
A. Boruah, M. Boruah, D. Prajapati, J.S. Sandhu, Synlett 1997, 1253 (1997)
S. Iyer, A.K. Sattar, Synth. Commun. 28, 1721 (1998)
W. Chan, Y.C. Berthelette, Tetrahedron Lett. 43, 4537 (2002)
H.G. Aslan, S. Özcan, N. Karacan, Inorg. Chem. Commun. 14, 1550 (2008)
H.G. Aslan, S. Özcan, N. Karacan, Spectrochim. Acta A Mol. Biomol. Spectrosc. 98, 329 (2012)
A.G. Newcombe, Can. J. Chem. 33, 1250–1255 (1955)
A.B.P. Lever, Inorganic Electronic Spectroscopy, 2nd edn. (Elsevier, Amsterdam, 1984)
S. Akkoç, Y. Gök, J. Iran. Chem. Soc. 11, 1767–1774 (2014)
S. Akkoç, Y. Gök, İ.Ö. İlhan, V. Kayser, Beilstein J. Org. Chem. 12, 81–88 (2016)
Acknowledgements
The authors are grateful to Erciyes University’s Research Foundation for financial support under project number EUBAP-FBA-09-975.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Aslan, H.G., Akkoç, S., Kökbudak, Z. et al. Synthesis, characterization, and antimicrobial and catalytic activity of a new Schiff base and its metal(II) complexes. J IRAN CHEM SOC 14, 2263–2273 (2017). https://doi.org/10.1007/s13738-017-1163-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13738-017-1163-4