Abstract
Today, clinicians rely more and more on medical images for screening, diagnosis, treatment planning, and follow-up examinations. While medical images provide a wealth of information for clinicians, content information cannot be automatically integrated into advanced medical applications such as those for the clinical decision support. The implementation of advanced medical applications requires means for the automated post-processing of medical image annotations. In this article we describe how we made use of reasoning technologies to post-process medical image annotations in the context of the automated staging process of lymphoma patients. First, we describe how automatic anatomy detectors and OWL reasoning processes can be used to preprocess medical images automatically and in a way that makes accurate input to further, more complex reasoning processes possible. Second, we enhance and integrate patients’ image metadata by formalized practical clinical knowledge sources. The resulting combined data serve as input to an automatic reasoning process in order to stage lymphoma patients automatically.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
The range of current different imaging technologies and modalities spans 4D 64-slice computer tomography (CT), whole-body magnet resonance imaging (MRI), 4D ultrasound, and the fusion of positron emission tomography and CT (PET or CT). These modalities provide a detailed insight into the human anatomy, its function, and respective disease associations. Moreover, advanced techniques for analyzing imaging data generate additional quantitative parameters, thus paving the way for improved clinical practice and diagnosis.
Advanced medical applications rely on semantic descriptions of clinical data such as medical images or patient records. There are several existing approaches addressing the challenge of semantic medical image annotation. For example, Seifert et al (2009) introduced a new method for automatic image parsing (anatomy and specific tissue detectors) and Möller et al. (2009) an approach for information extraction from DICOM headers and DICOM structured reports. Channin et al. (2009) and Rubin et al. (2008) aimed at integrating manual image annotation into the reporting workflow of radiologists, and Hu et al. (2003) introduced an image annotation approach suitable for improving breast cancer diagnosis. All these approaches make an important contribution to improve the access to medical image information by specifying the “semantics” of specific image regions.
In our application, we rely on image metadata generated by Seifert et al. (2009) and combine it with additional clinical knowledge. Automatic anatomy detectors and OWL reasoning processes can be used to preprocess medical images automatically and in a way that makes accurate input to further, more complex reasoning processes possible. Our goal is to use medical image content information for the automatic staging of cancer patients. We concern ourselves with the explicit descriptive knowledge of how an image finding (e.g., the number of enlarged lymph nodes) relates to the patient staging degree. The staging information is paramount when clinicians assess an individual patient’s progress and decide on subsequent treatment steps. The medical application scenario is defined by the specific patient context: patients suffering from lymphoma. Lymphoma, a type of cancer originating in lymphocytes, is a systematic disease with manifestations in multiple organs. The stage of lymphoma patients is determined by the number, location, and distribution of lymphatic occurrences. Therefore, automated staging of patients requires (pre/post) processing steps that explicitly describe the precise number as well the spatial positions of lymphatic occurrences captured by medical images.
This article’s main contribution is to introduce a new medical application for the automated classification of lymphoma patients in well-defined categories that relies on image metadata information. Image metadata information are extracted from the DICOM headers and/or extracted from the image regions automatically (preprocessing). The post-processing steps of the image metadata information rely on specific OWL Footnote 1 reasoning steps: In particular, we
-
utilize automatic plausibility checks to learn about the spatial position of lymphoma occurrences; and
-
develop a formal and explicit representation of the Ann-Arbor staging system that allows us to discover new classification results by means of existing reasoning procedures.
The remainder of this article is organized as follows: In Sect. 2 we give an account on the three different knowledge resources required for the implementation of the staging scenario. Section 3 introduces our approach of anatomical reasoning to learn about the spatial position of lymphoma occurrences. In Sect. 4 we will describe the automated staging of lymphoma patients in more detail and introduce our approach for aligning and integrating various heterogeneous knowledge resources. Section 5 sketches a clinical evaluation of the application and Sect. 6 concludes this article with an outlook on future work.
2 Used knowledge resources
The implementation of the staging application relies on (a) the formalized clinical knowledge about disease stages (here the Ann-Arbor staging system), (b) the consistent processing of concepts used for labeling image information (the involved medical ontologies), and (c) the availability of semantic image annotations, i.e., the medical image metadata (e.g., about specific anatomical structures).
2.1 Ann-Arbor staging system
The Ann-Arbor staging system (Wittekind et al. 2005) establishes an explicit classification of lymphoma patients in terms of disease progression.
The staging system depends on two criteria. The first criterion is the location, the number and distribution of the malignant tissue, which can be identified by located biopsy as well as medical imaging methods, such as CT scanning and positron emission tomography (here, we assume the manual examination of the image material by a radiology expert). Four different stages can be recognized: Stage I indicates that the cancer is located in a single region and Stage II that it is located in two separated regions confined to one side of the diaphragm. Footnote 2 Stage III denotes that the cancer has spread to both sides of the diaphragm and Stage IV shows diffuse or disseminated involvement of one or more extra lymphatic organs (see Fig. 1)
Ann-Arbor staging system (Source: http://training.seer.cancer.gov)
We established a staging ontology in OWL DL following the rational of Ann-Arbor staging system (for the detailed knowledge engineering steps and knowledge model we refer to Zillner (2010) that allows to determine the patient data classification within the reasoning process (see Sect. 4). Each staging class is described as a defined OWL class that specifies all necessary and sufficient conditions and enabled the patient to be classified in accordance with the above mentioned criteria, that is, according to the number, type, and relative position of lymphatic occurrences. All concepts relating to anatomical information were labeled with the appropriate medical ontology, i.e. Radlex or FMA, in order to provide the basis for seamless integration of various data sources. The ontological model that captures the rational of the Ann-Arbor staging system was modeled manually (for more details see Zillner 2009).
2.2 Medical ontologies
To achieve re-usability and interoperability, we required third-party taxonomies or ontologies to inform our application of ontological concepts describing possible regions of lymphatic occurrences: lymph node regions as well as extra lymphatic organs and sites. Two ontologies—the Foundational Model of Anatomy (FMA) and the Radiology Lexicon (Radlex)—describe anatomical entities and provide the required coverage of anatomical concepts for the staging scenario.
The FMA (Rosse and Mejino 2003) is a comprehensive specification of anatomy taxonomy, namely an inheritance hierarchy of anatomical entities with different kinds of relationships. It covers approximately 70,000 distinct anatomical concepts and more than 1.5 million relations instances of 170 relation types. It provides concepts that describe single lymph nodes, such as ‘axilliary_lymph_node’, as well as concepts that describe multiple lymph nodes, such as ‘set_of_ axilliary_lymph_node’. (It also contains 425 concepts representing singular lymph nodes and 404 concepts describing sets of lymph nodes which distinction can be relevant as input to the reasoning processes.)
RadlexFootnote 3 is a terminology developed and maintained by the Radiological Society of North America (RSNA) for the purpose of achieving a uniform mode of indexing and retrieving radiology information, including medical images. Radlex contains over 8,000 anatomical and pathological concepts, including imaging techniques, difficulties, and diagnostic image qualities. Its purpose is to provide a standardized terminology for radiological practice.
2.3 Medical image metadata
Our application takes patient data of the MEDICO Footnote 4 project, in particular image metadata, as input for deducing the patient’s progress of the disease. In MEDICO, multiple ways to generate semantic image annotations have been implemented.
For example, methods for automated image parsing, such as (Seifert et al. 2009), enable the hierarchical parsing of whole body CT images (by starting with the head and subsequently moving down the body) and the efficient segmentation of multiple organs while taking contextual information into account.
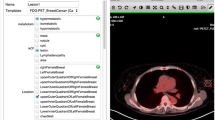
While automated image parsing remains incomplete (in many cases, crucial anatomy or disease information cannot be detected accurately), manual image annotation by the radiology expert is an important complement to automatic procedures. For example, MEDICO users can manually add semantic image annotation by selecting or defining arbitrary regions or respective volumes of interest by using the MEDICO image annotation tool (see Fig. 2) which can also be embedded into a more complex knowledge acquisition process for radiology images (Sonntag 2010). One of our aim is that clinical experts can indicate lymphatic occurrences by marking them on the image and subsequently labeling the body region with the corresponding Radlex or FMA concept. The semantic annotations are stored within a dedicated annotation ontology (Sascha et al. 2010) that links each semantic annotation to a corresponding concept of one of the two mentioned medical ontologies.
3 Spatial–anatomical reasoning
In lymphoma staging, one differentiates between patients that show lymphatic occurrences either only above, only below, or on both sides of the diaphragm. Thus, the relative position of lymphatic occurrences constitutes an important decision criteria of the staging system and needs to be considered in the reasoning procedure. In other words, the relative position of lymphatic occurrences in relation to the diaphragm need to be represented explictely. Our aim is to compute the spatial position of lymphatic occurrences by means of spatial–anatomical reasoning.
We define spatio-anatomical reasoning as follows: spatial–anatomical reasoning means to use ontology-based knowledge to verify or falsify the spatial configurations that are found by independent automatic detectors.
For example, in a two-stage process, we augmented the FMA as the most comprehensive reference ontology for human anatomy with spatial relations (these relations were acquired inductively from a corpus of semantically annotated CT volume data sets). The first stage of this process abstracted relational information using a fuzzy set representation formalism. In the second stage we further abstracted from the fuzzy anatomical atlas to a symbolic level using an extension of the spatial relation model of the FMA (details can be found in Möller et al. 2011). This approach (Möller et al. 2011) augments medical domain ontologies and allows for an automatic detection of anatomically implausible constellations in the results of a state-of-the-art system for automatic object recognition in 3D CT scans. The output of this preprocessing step is a feedback on which anatomical entities are most likely to have been located incorrectly (thereby, the necessary spatio-anatomical knowledge is learned from a large corpus of annotated medical image volume data sets). Quantitative spatial models are the foundation of digital anatomical atlases.
The approach in Hudelot et al. (2008) is complementary to this work in so far as the authors also propose to add spatial relations to an existing anatomical ontology. Their use case is the automatic recognition of brain structures in 3D MRI scans. Fuzzy logic has been proven as an appropriate formalism which allows for quantitative representations of spatial models (Bloch 2005). Krishnapuram et al. (1993) expressed spatial features and relations of object regions using fuzzy logic. Bloch and Ralescu (2003) and Bloch (1999) describe generalizations of this approach and compare different options to express relative positions and distances between 3D objects with fuzzy logic. From this atlas we then abstract the information further onto a purely symbolic level to generate a generic qualitative model of the spatial relations in human anatomy. In our first evaluation we describe how this model can be used to check the results of a state-of-the-art medical object recognition system for 3D CT volume data sets for spatial plausibility.
The usage of the resulting medical object recognition system for 3D CT volume data sets in the staging use case is straightforward. Specific lymph node affection at a specific body part has a direct influence on the correct staging process. The anatomical configuration we can reconstruct according to the learned quantitative anatomical atlas and the image organ detectors of a specific patient case lets us determine the positions of the affected lymph nodes with great accuracy.
During our inspection we found that the quality of the detector results exhibits a high variability. Accordingly, we distinguished three quality classes: clearly incorrect, sufficiently correct, and perfectly correct. The visualizations in Fig. 3 show one example for each class. For the staging cases, we rely on the configurations which are “sufficiently correct.” This means we run the detectors and check the resulting anatomical atlas for plausibility according to our own empirical model. It is important to note that the staging reasoning process does not need a perfect anatomical model. The decision whether or not a specific lymph node is on a specific side of the diaphragm can be done accurately with only a few indicators in the anatomical proximity such as the bronchial bifurcation and the urinary bladder.
3.1 Example reasoning case
From the knowledge model (FMA), we know that the organ kidney is located below the diaphragm. In addition, we know that the relationship is_below is transitive.
By means of spatial reasoning, we can infer that the lymphnode occurence X is below the organ kidney (see Fig. 4). Addional evidence is provided by the configurations of other landmarks such as the urinary bladder or the bronchial bifurcation.
Therefore, we can automatically infer that the lymph node occurrence is below the diaphragm and provide further input for the consequential staging reasoning step explained further down (the reasoning step of how to combine the individual landmarks to a combined estimate of the location of the suspicious lymph node is left out for simplicity).
We performed a systematic evaluation of the positions on our manually labeled corpus (anatomy detections) using four-fold cross evaluation. In total, 1,119 detector results have been inspected and classified manually: there are 388 sufficiently correct detector results and 147 perfect detector results (Möller et al. 2011). Our results show that the average difference in percent between the spatial relation instances in the learned model and the instances generated for an element from the evaluation set is an appropriate measure for the spatial positions (showing that in our case, lymphatic occurrences are either only above, only below, or on both sides of the diaphragm).
Therefore, the detection of “above diaphragm” can be performed with a precision of 85% and a recall of 65% (according to the systematic evaluation of the positions on our manually labeled corpus of anatomy detections. The values were obtained by comparing the results to the supervised test set, whereby we considered an individual anatomy classification as either correct or incorrect. This binary classification allowed us to create the full truth table with all false positives and false negatives. Given a direct anatomy detector result (a sufficiently correct detector result or perfect detector result), the reasoning and detection of “above diaphragm” is straightforward and does not introduce a classification bias or noise.
4 Automated patient staging
Beside the automatic detection of the spatial location of suspicious lymphoma occurrences (as described further up), the automated lymphoma patient staging requires further post-processing of the knowledge resources: (a) the efficient alignment of the used medical knowledge resources (FMA and RadLex) and (b) the semantic integration of the various heterogenous knowledge resources by means of reasoning. Both tasks will be detailed in the following:
4.1 Aligning medical ontology fragments
Within the MEDICO project, the patient’s text and image annotation relies on Radlex and FMA. Thus, for the consistent processing of the data, we needed to establish alignments between concepts of the two ontologies. As both knowledge models are large in size, we restricted the scope of the mapping to the part of the information which is of relevance for the specific lymphoma staging application, i.e., the concepts that relate to lymph node occurrences. We use text mining methods for extracting all concepts containing the concept “lymph node” or “node” as part of their preferred name. The resulting list encompasses 104 Radlex and 807 FMA concepts representing relevant lymph node occurrences (in singular form).
A variety of methods for ontology alignments in the medical domain have been proposed (e.g., Sonntag et al. 2009) and have been reported in general surveys (Euzenat and Shvaiko 2007; Doan et al. 2003; Rahm and Bernstein 2001; Noy 2004). However, complex methods for ontology alignment in the medical domain turned out to be infeasible due to the large size and complexity of medical ontologies. There exist pragmatic approaches for handling the complexity of the medical domain. For instance, Baumgartner et al. (2006) take an information retrieval approach to discover relationships between different medical ontologies by indexed ontology concept using Lucene (http://lucene.apache.org/) and, subsequently, matching them against the search queries (the concepts of the target ontologies). Although this approach is efficient and easy to implement (it can successfully applied to large medical ontologies), it does not account for the complex linguistic structure typically observed in the concept labels of the medical ontologies and may result in inaccurate matches.
For discovering the relationships between Radlex and FMA concepts, we extended the alignment approach of Baumgartner et al. (2006) by incorporating mapping rules that reflect linguistic features of the natural language phrases which describe a particular concept (Zillner and Sonntag 2011). As most concept labels in medical ontologies are multi-word terms that are usually rich with implicit semantic relations (such as the FMA concept “Superior deep lateral cervical lymph node”), we can (only) rely on more complex linguistic rules to exploit the implicit semantics for identifying automatically ontology alignment correspondences. In addition, the analysis of user feedback results provided us guidance in fine-tuning the initial mapping results. For improving the precision of our alignment, we formalized relevant context information (e.g., information about antonym terms) which has been used to filter out incorrect mappings.
4.2 Knowledge integration by means of reasoning
The seamless integration of the three different knowledge resources requires specific pre-processing steps. Figure 5 provides an overview of the five steps to achieve automatic patient staging. The five steps are indicated by a number:
As first step, we established a staging ontology in OWL DL following the rational of Ann-Arbor staging system (for the detailed knowledge engineering steps we refer to Zillner (2010) that allows us to determine the patient data classification within the reasoning process. The Ann-Arbor ontology consists of a set of defined classes (OWL classes described by necessary and sufficient constraints) capturing the information that leads to a patient’s lymphoma stage (i.e., the number, types and distribution of patients’ indicated lymphatic occurrences). As already mentioned, in order to provide data items with precise semantics, any concept in the ontology that is related to anatomical knowledge, was labeled by the corresponding concept of the RadLex or FMA ontology. According to the semantics of each patient class, it will be classified within the reasoning process (by using the OWL reasoner Pellet Footnote 5). In addition, the concepts describing anatomical information are labeled by the
In a second step, we transformed the patient data of the annotation procedure in Seifert et al. (2009) into an OWL representation. Emanating from a flat view of the patient data, i.e., the patient identifier and the patient’s list of lymphatic occurrences, we create an OWL view with each patient being a class and each elected lymphatic occurrences represented as a restriction class axiom.
Within the third step we identified the relevant ontology fragments and established the required ontology alignments as described in the preceding subsection.
The fourth step focuses on the integration of all created ontologies, i.e., the aligned medical ontology fragments, the patient ontology, and the Ann-Arbor ontology as well as the execution of the reasoning process on top of the integrated ontological model for staging. The seamless integration of the information stored in all created ontologies basically relies on two factors: First, the concepts of all created ontologies are linked by one of two medical ontologies, and secondly we established a mapping between the FMA and RadLex concepts (see Sect. 4) that are relevant for our application. By providing precise semantics of each data item integrated, the integration of data can be realized seamlessly.
The automatic reasoning process enables us to automatically classify the stage of a patient by integrating knowledge captured by the medical ontologies. The resulting ontological model then explicitly captures the inferred staging of individual patient records.
Fifth, the knowledge captured in the inferred model, in particular the deduced staging information of a patient database, can now be queried by means of SPARQL. Footnote 6
5 Clinical evaluation
We conducted a proof-of-concept study on the basis of more than 10 real patient records to analyze the practical potential of the automatic anatomy detection and staging approach. With the help of our clinical experts, the corresponding medical images were manually annotated by Radlex as well as FMA concepts and the semantic annotations integrated into our knowledge base. In addition, information in discharge letters covering diagnose and findings was fed into the knowledge model.
We presented and discussed the automated patient staging scenarios with our clinical project partners at the Friedrich-Alexander-University of Erlangen. The response towards the staging applications was very positive. It is important to note that the automatic reasoning procedure does not give the clinician new information that he or she would not know after (re)examining the CT images—yet the doctors acknowledged the relevance of inferred staging knowledge for the purposes of quality control in clinical diagnosis, e.g. the analysis of real patient records often reveals the fact that there are patient cases with a clinician’s diagnosis in the discharge letter that contradicts the automated staging result based on the image annotation.
5.1 Automatic reasoning case
In the context of one particular patient, we can identify a contradiction between the clinicians manual diagnosis (Stage I–IV) and the automated staging results based on the image annotation: consider advanced stage lymphoma patient who was already treated with three chemotherapies using the CHOP-protocol. As the accomplished treatments did not help to improve the patient’s health condition, he or she has been referred to a specialist hospital.
-
The patient’s discharge letter covers details about the diagnose, past medical diagnose, as well as findings, assessment, progression, and recent therapy. Our example patient’s discharge letter indicates a diagnose of Ann-Arbor IV-Stage.
-
In the specialist hospital, the patient was screened using CT. By analyzing and annotating the medical images, 16 enlarged and pathological lymph nodes on both sides of the diaphragm could be identified. However, no indication of the involvement of extra lymphatic organs was noted. Relying on the formal Ann-Arbor Classification criteria, the ontology-based staging approach classifies Patient Speck as Ann-Arbor III-Stage.
In our clinical, albeit realistic example, the clinician’s diagnosis contradicts the results of the automated reasoning application (Fig. 6).
As the staging grade strongly influences clinicians in their sequential treatment decisions, this issue is of considerable importance for achieving effective diagnostics and treatments. By highlighting such contradictory results, special clinical cases can be spotted out and potential medical treatment errors can be reduced. In our interviews, the clinicians emphasized that, in medical settings in particular, contradictions do not necessarily have to be considered as mistakes that need to be corrected. Instead, contradictions provide either a second opinion to be considered, or an indication that a more detailed analysis is required. The doctors explained that usually two different explanations can be given as “good” reasons for a contradiction:
-
1.
The patient’s health condition has improved significantly, but the changes were not explicitly documented in the findings. Such incidents are not exceptional, as clinicians typically avoid to make definite statements about the patient’s health condition, but concentrate on the documentation of related indications.
-
2.
The discharging physician has come to a different conclusion after intensive investigation.
In both cases, the patients’ symptoms and findings need to be analyzed again. By highlighting clinical contradictions, the quality of medical care can be improved. The patient examples show the impact of the automatic processing of image metadata to improve clinical decision support systems.
6 Conclusions
There is a growing interest in the automatic processing of medical image content and the semantic integration of explicitly expressed content into clinical applications. In this paper, we introduced an application for the automated staging of lymphoma patients using image metadata information. In our future work, we aim to formalize and integrate related staging systems, such as the TNM classification, into advanced medical applications.
Notes
As a formal representation language, we use the Web Ontology Language (OWL). More precisely, we rely on the sublanguage OWL DL that is based on description logics (Baader et al. 2003). description logics, a family of formal representation languages for ontologies, are designed for classification-based reasoning.
A sheet-form-like internal skeletal muscle that extends across the bottom of the rib cage. The diaphragm separates the thoracic cavity (heart, lungs, and ribs) from the abdominal cavity.
http://www.w3.org/TR/rdf-sparql-query/on the semantic RDF counterpart.
References
Baader F, Calvanese D, McGuinness DL, Nardi D, Patel-Schneider PF (eds) (2003) The description logic handbook: theory, implementation, and applications. Description logic handbook. Cambridge University Press, Cambridge
Baumgartner WA, Johnson HL, Johnson HL, Cohen KB, Cohen KB, Lu Z, Bada M, Kester T, Kim H, Hunter L (2006) Evaluation of lexical methods for detecting relationships between concepts from multiple ontologies. In: Pacific Symposium on Biocomputing, pp 28–39
Bloch I (1999) On fuzzy distances and their use in image processing under imprecision. Pattern Recognitionit 32:1873–1895
Bloch I (2005) Fuzzy spatial relationships for image processing and interpretation: a review. Image and Vision Computing 23:89–110
Bloch I, Ralescu A (2003) Directional relative position between objects in image processing: a comparison between fuzzy approaches. Pattern Recognit 36:1563–1582
Channin D, Mongkolwat P, Kleper V, Sepukar K, Rubin D (2009) The cabib annotation and image markup project. J Digital Imaging (online)
Doan A, Madhavan J, Domingos P, Halevy A (2003) Ontology matching: a machine learning approach. In: Handbook on ontologies in information systems. Springer, pp 397–416
Euzenat J, Shvaiko P (2007) Ontology matching. Springer, Heidelberg
Hu B, Dasmahapatra S, Lewis PH, Shadbolt N (2003) Ontology-based medical image annotation with description logics. In: ICTAI
Hudelot C, Atif J, Bloch I (2008) Fuzzy spatial relation ontology for image interpretation. Fuzzy Sets Syst. 159:1929–1951
Krishnapuram R, Keller JM, Ma Y (1993) Quantitative analysis of properties and spatial relations of fuzzy image regions. IEEE Trans Fuzzy Syst 1:222–233
Möller M., Regel S., Sintek M (2009) Radsem: semantic annotation and retrieval for medical images. In: The 6th annual European semantic web conference (ESWC)
Möller M, Sonntag D, Ernst P (2011) A spatio-anatomical medical ontology and automatic plausibility checks. In: IC3K 2010 selection, communications in computer and information science (CCIS)
Noy NF (2004) Tools for mapping and merging ontologies. In: Handbook on ontologies, pp 365–384
Rahm E, Bernstein PA (2001) A survey of approaches to automatic schema matching. VLDB J 10:334–350
Rosse C, Mejino JJ (2003) A reference ontology for biomedical informatics: the foundational model of anatomy. J Biomed Inform 36
Rubin D, Mongkolwat P, Kleper V, Supekar K, Channin D (2008) Medical imaging on the semantic web: annotation and image markup. In: AAAI spring symposium series, semantic scientific knowledge integration, Stanford
Seifert S, Kelm M, Möller M, Mukherjee S, Cavallaro A, Huber M, Comaniciu D (2010) Semantic annotation of medical images. In: Proceedings of SPIE medical imaging
Seifert S, Barbu A, Zhou S, Liu D, Feulner J, Huber M, Suehling M, Cavallaro A, Comaniciu D (2009) Hierarchical parsing and semantic navigation of full body ct data. In: SPIE medical imaging
Sonntag D (2010) Intelligent interaction and incremental knowledge acquisition for radiology images. In: Proceedings of the international conference on semantic and digital media technologies (SAMT). Springer
Sonntag D, Wennerberg P, Buitelaar P, Zillner S (2009) Cases on semantic interoperability for information systems integration: practices and applications. IGI Global
Wittekind C, Meyer H, Bootz F (2005) TNM Klassifikation maligner Tumoren. Springer, Heidelberg
Zillner S (2010) Reasoning-based patient classification for enhanced medical image annotations. In: Proceedings of the 7th extended semantic web conference, Heraklion
Zillner S (2009) Towards the ontology-based classification of lymphoma patients using semantic image annotations. In: Ceur Proceedings of SWAT4LS semantic web applications and tools for life sciences (SWAT4LS), Amsterdam
Zillner S, Sonntag D (2011) Aligning medical ontologies by axiomatic models, corpus linguistic syntactic rules and context information. In: Proceedings of the 24th international symposium on computer-based medical systems (CMBS), Bristol
Acknowledgments
This research has been supported in part by the THESEUS Program in the MEDICO Project, which is funded by the German Federal Ministry of Economics and Technology under grant number 01MQ07016. The responsibility for this publication lies with the authors.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Zillner, S., Sonntag, D. Image metadata reasoning for improved clinical decision support. Netw Model Anal Health Inform Bioinforma 1, 37–46 (2012). https://doi.org/10.1007/s13721-012-0003-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13721-012-0003-9