Abstract
Purpose
Bromodomain-containing protein 7 (BRD7) is downregulated and functions as a tumor suppressor in many types of cancers including breast cancer, and the dysregulation of BRD7 expression is closely related to the development and progression of breast cancer. Whereas little attention has been focused on the regulation of BRD7 protein levels in breast cancer, which needs to be further elucidated.
Methods
The protein stability of BRD7 in breast cancer cells and BRD7 protein level in breast cancer tissues was examined by Western Blotting. The potential E3 ubiquitin ligase proteins that interact with the BRD7 was screened by coimmunoprecipitation combined with mass spectrometry analysis in MDA-MB-231 cells. We proved the interaction between BRD7 and tripartite motif containing 28 (TRIM28) through Co-Immunoprecipitation (Co-IP) and immunofluorescence assays. Co-IP and ubiquitination assay were used to explore the specific binding domain between BRD7 and TRIM28 and the ubiquitination site of BRD7. The effects of TRIM28 on the BRD7 protein stability and ubiquitination level was investigated by qPCR, Western Blot and Co-IP assay. CCK-8 and clone formation assays were carried out to assess the effect of TRIM28 on proliferation ability of breast cancer ells. Transwell assay and wound healing assay were used to investigate the effect of TRIM28 on breast cancer cell invasion and migration. Flow cytometry was used to detect the effect of TRIM28 on cell cycle and apoptosis of breast cancer cells. In addition, we confirmed effect of TRIM28 on tumor growth and metastasis by xenograft and metastatic mouse models. We designed some recovery assays to explore the role of recovery BRD7 in TRIM28-mediated promotion of malignant progression of breast cancer in vivo and in vitro. Finally, the clinical significance of TRIM28 and BRD7 was proved by immunohistochemistry.
Results
In this study, we demonstrated that BRD7 was an unstable protein and might be regulated by ubiquitination in breast cancer; furthermore, we found that the Coiled-Coil region of TRIM28 could directly bind to N-terminal of BRD7, and TRIM28 mediates BRD7 ubiquitination and degradation dependent on K21 by acting as a potential E3 ubiquitin ligase. Moreover, TRIM28 promoted cell proliferation, migration, invasion, xenograft tumor growth and metastasis, thus playing an oncogenic role in breast cancer. Furthermore, the restoration of BRD7 expression in breast cancer significantly reversed the promotional effects of TRIM28 on malignant progression both in vitro and in vivo. In addition, TRIM28 was highly expressed in the biopsy tissues of breast cancer, and its expression was negatively correlated with BRD7 expression and positively correlated with TNM stage and poor prognosis of BC patients.
Conclusions
Our findings provide a novel mechanism by which TRIM28 significantly facilitates BRD7 ubiquitination and degradation, thus promoting breast cancer malignant progression. Targeting the TRIM28/BRD7 axis might be a novel potential strategy for the clinical diagnosis and treatment of breast cancer.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Breast cancer is a type of malignant tumor occurring in the ductal epithelium and lobular epithelium of the breast; among all malignancies, breast cancer has the highest morbidity and mortality rates in women, posing a serious threat to women’s health [1]. The pathogenesis of breast cancer is affected by genetic and nongenetic risk factors. The genetic factors include susceptibility genes such as Breast cancer susceptibility gene 1 (BRCA1), Breast cancer susceptibility gene 2 (BRCA2), checkpoint kinase 2 (CHEK2), and single-nucleotide polymorphisms (SNPs) associated with breast cancer [2]. The nongenetic factors include age, a personal history of breast disease, radiation, exogenous estrogen use, and reproductive factors [3,4,5]. However, the detailed mechanism of breast cancer is still unknown. Therefore, it is vital to explore the potential molecular regulatory mechanism in breast cancer progression, which will provide new treatment strategies for BC.
Bromodomain-containing protein 7 (BRD7) is a protein containing a single bromodomain (Brd), which was screened and identified by cDNA differential analysis and library screening in nasopharyngeal carcinoma in our lab in 2000 [6]. BRD7 is downregulated and functions as a tumor suppressor in multiple tumors, such as nasopharyngeal carcinoma, breast cancer, lung cancer, liver cancer, ovarian cancer, and endometrial cancer [7,8,9,10,11,12,13]. BRD7 has been reported to function as a transcriptional regulator that participates in the transcriptional regulation of downstream genes in tumors [14,15,16]. In addition, it has been established that ubiquitination, sumoylation and phosphorylation are involved in the posttranslational modification of BRD7 expression [17,18,19]. However, the posttranslational modification of BRD7 in breast cancer is still not well understood.
The tripartite motif-containing (TRIM) family, containing more than 80 members, is known as a subfamily of the RING-type E3 ubiquitin ligase family [20]. Accumulating evidence has demonstrated that TRIM family proteins play important roles in the occurrence and development of tumors, intracellular signal transduction, transcription regulation, innate immunity, autophagy and tumor progression [21, 22]. Tripartite motif containing 28 (TRIM28) is a member of the TRIM family, which is located on chromosome 19 and contains 17 exons [23]. Several studies have proven that TRIM28 is involved in the malignant progression of several tumors as an oncogene, including glioblastoma [24], hepatocellular carcinoma [25], pancreatic cancer [26], non-small cell lung cancer [27], and breast cancer [28]. Nevertheless, the detailed functional role of TRIM28 in BC and its underlying mechanism are still unclear.
In the present study, we found that BRD7 was confirmed to be an unstable protein and that TRIM28 could directly bind to BRD7 and promote its ubiquitination and degradation by functioning as a potential E3 ubiquitin ligase. Moreover, TRIM28 promoted cell proliferation, migration, invasion, xenograft tumor growth and metastasis by decreasing BRD7 protein stability in breast cancer. Targeting the TRIM28/BRD7 axis might be a novel potential strategy for the clinical diagnosis and treatment of breast cancer.
2 Materials and methods
2.1 Clinical data information
A total of 178 breast cancer and 33 normal breast paraffin-embedded samples were collected from the Second Xiangya Hospital of Central South University, and this study was approved by the Ethics Review Committees/Institutional Review Boards of Central South University. The clinicopathologic features of the breast cancer patients mainly included gender, age, tumor size, node metastasis, distant metastasis, clinical tumor node metastasis (TNM) stage, pathology diagnosis, survival time and molecular subtype. The immunohistochemical scores of clinical samples were based on the detailed procedures described in previous articles [29].
2.2 Antibodies
Anti-TRIM28 (1:1000), anti-BRD7 (1:2000), anti-GAPDH (1:5000) and anti-t-PARP (1:1000) were purchased from Proteintech Group, Inc. (Wuhan, China). Anti-Flag (1:1000) and anti-HA (1:1000) were purchased from Sigma‒Aldrich. Anti-Vimentin (1:1000) was purchased from Arigo (Taiwan, China). Anti-Snail (1:1000), anti-E-cadherin (1:1000) and anti-c-PARP (1:1000) were purchased from Cell Signaling Technology, Inc. (Beverly, MA). HRP-conjugated secondary antibodies were purchased from Beyotime Biotech (Wuhan, China) (Supplementary Table 1).
2.3 Cell lines and culture
MDA-MB-231 (RRID: CVCL_0062) and MCF7 (RRID: CVCL_0031) cells were obtained from ATCC (The Global Bioresource Center). All cells were cultured in high glucose Dulbecco’s modified Eagle’s medium (DMEM) containing 10% fetal bovine serum (FBS) (BI, Israel) and 1% penicillin/streptomycin and maintained in a humidified atmosphere of 5% CO2 at 37 °C. All experiments were performed with mycoplasma-free breast cancer cells.
2.4 TRIM28 expression level and relapse-free survival analysis
The expression of TRIM28 in cancer was analyzed by GEPIA (http://gepia.cancer-pku.cn/index.html), and the expression of TRIM28 in breast cancer and normal tissues was analyzed by the TCGA database. The prognostic value of TRIM28 expression was examined using the Kaplan‒Meier-Plotter database online.
2.5 Cell transfection
Specific small interfering RNAs (siRNAs) against TRIM28 or BRD7 were purchased from RiboBio (Guangzhou, China). Cells grew at a suitable density to attain 50–60% confluence by the time that transfection was performed after 12 h. All plasmids and siRNAs were transfected into the indicated cells using Polyplus transfection (INTERFERin) according to the manufacturer’s instructions.
2.6 Cell proliferation and colony formation assay
Cell proliferation was detected by Cell Counting Kit-8 (CCK-8) (Selleck, Houston, TX, USA). MDA-MB-231 and MCF7 cells (1000 cells/well) were plated into 96-well plates, and then 10 µL of CCK-8 reagent was added to each well at different time points (0, 1, 2, 3, 4, and 5 d). A microplate reader (Beckman, Brea, CA, USA) was used to obtain the cell absorbance at 450 nm after 2 h of incubation at 37 °C. For the colony formation assays, MDA-MB-231 and MCF7 cells were plated in a 6-well plate at a density of 1000 per well. The cells were further cultured at 37 °C for 2 weeks, and then the plates were washed three times with 1 × PBS. The colonies were fixed with 4% polyoxymethylene and stained with crystal violet (Beyotime, Beijing, China). The colony counts were normalized to the control and expressed as a percentage.
2.7 Wound-healing and transwell assays
For the wound-healing assay, MDA-MB-231 and MCF7 cells were seeded in 6-well plates after transfection and cultured to the sub confluence. The “scratch” wounds were created by a 10-µL pipette tip. Next, the cells were cultured with 2% FBS, and photos were taken under a microscope at different time points and the images were analyzed by ImageJ and statistically analyzed by GraphPad Prism 8.01. To perform the Transwell assay, Transwell chambers (8-µm pore size; Corning, NY, USA) were coated with Matrigel (BD Biosciences, NY, USA) diluted in a 1:8 proportion with serum-free medium. Then, 5 × 104 cells suspended in serum-free medium were plated on the top of the chamber, while medium supplemented with 20% FBS was placed in the lower chamber. After incubation for 48 h, cells were fixed with 4% polyoxymethylene and stained with crystal violet (Beyotime, Beijing, China), and then three random fields per group were photographed under a microscope.
2.8 Total RNA extraction and qRT‒PCR analysis
Total RNA was extracted from MDA-MB-231 and MCF7 cells by using TRIzol reagent (Invitrogen, Carlsbad, USA). First strand cDNA synthesis of total RNAs was performed using a Revert Aid first strand cDNA synthesis Kit according to the instructions (K1622, Thermo Scientific, Waltham, USA). Quantitative RT‒PCR analysis was conducted to measure the levels of mRNA using Platinum SYBR Green qPCR SuperMix-UDG reagents (Accurate Biotechnology, Hunan, China) with the Bio-Rad CFX96 Touch sequence detection system (Bio-Rad Laboratories Inc.). The relative mRNA levels were normalized to that of GAPDH. The relative fold changes in expression were analyzed using the 2−ΔΔCT method, and each sample was analyzed in triplicate. The sequences of primers used in this article are listed in Table 1.
2.9 Western blotting assay
Cells were lysed with Western and IP lysis buffer (25mM Tris HCl (pH7.4), 150mM NaCl, 1% NP-40 and 5% glycerol) (NCM Biotech) in the presence of a protease inhibitor cocktail on ice for 30 min. Protein concentration was determined using a Bicinchoninic Acid (BCA) protein assay kit (Thermo Fisher Scientific, Inc.). Protein samples were mixed with 1 × SDS‒PAGE protein loading buffer (NCM Biotech) at 95 °C for 5 min, separated by 10% SDS‒PAGE (NCM Biotech) and then transferred to PVDF membranes (Millipore, Billerica, USA). The membranes were blocked in 5% nonfat milk at room temperature for 1 h and incubated with the indicated primary antibodies at 4 °C overnight. After incubation with HRP-conjugated secondary antibodies for 1 h at 37 °C, the signals were detected using an ECL kit (Pierce, Rockford, IL, USA) and a chemiluminescence detection system (MiniChemi™ I, SAGECREATION, China).
2.10 Coimmunoprecipitation
For immunoprecipitation (IP), cell lysates were prepared with Western and IP lysis buffer (25mM Tris HCl (pH7.4), 150mM NaCl, 1% NP-40 and 5% glycerol) (NCM Biotech), and the protein A/G-agarose beads (Santa Cruz Biotechnology, USA) were incubated with the indicated antibodies for 2 h at room temperature. Protein fractions and Protein A/G beads were incubated at 4 °C overnight (12–14 h). Then, the beads were washed 3 times for 5 min each and boiled at 95 °C for 5 min in 1 × SDS loading buffer for western blotting analysis.
2.11 Coimmunoprecipitation and mass spectrometry analysis (Co-IP-MS)
MDA-MB-231 and HEK293 cells were transfected with the plasmid pIRES2-EGFP-BRD7/3Flag for 48 h. The protein A/G-agarose beads (Santa Cruz Biotechnology, USA) were incubated with the indicated antibodies for 2 h at room temperature. Then, the protein extracts were incubated with Protein A/G beads conjugated to anti-Flag or anti-IgG antibodies overnight (12–14 h). Then, the beads were washed 3 times for 5 min each by Western and IP lysis buffer (NCM Biotech) and the samples were denatured in 30 µL of 2 × SDS loading buffer at 95 °C for 5 min and resolved by 10% SDS‒PAGE. Following protein separation, the gel was stained using a Coomassie blue staining kit (Beyotime Biotechnology, China) and shaken gently overnight in double distilled water for decolorization. The bands were cut into tiny micelles, decolorized to transparency with decoloring solution (50% acetonitrile (ACN) and 25 mM NH4HCO3), and infiltrated with 250 µL of protein protection solution (55 mM IAA and 25 mM NH4HCO3) at RT for 30 min. The samples were further infiltrated with 250 µL of a protective solution (25 mM dithiothreitol (DTT) and 25 mM NH4HCO3) at RT for 30 min, dehydrated with 100% ACN and dried using a vacuum drier; then, an appropriate amount of trypsin was added for digestion at 37 °C overnight. The samples were dehydrated with solution buffer (0.1% trifluoroacetic acid and 70% ACN). Then, the peptides were further diluted with 0.1% formic acid and analyzed by nano-LC‒MS/MS using an LTQ Velos Orbitrap MS (Thermo Fisher Scientific, Waltham, MA, USA) coupled with an UltiMate RSLCnano LC system (Dionex, Sunnyvale, CA, USA).
2.12 Immunofluorescence
MDA-MB-231 and MCF7 cells were plated in a 12-well plate, and then the cells were washed with PBS three times and fixed in 4% paraformaldehyde for 30 min at RT. The fixed cells were permeabilized with a buffer containing 0.3% Triton X-100 for 30 min and then blocked in normal goat serum (AR0009, BOSTER Biological Technology) for 30 min. Next, the indicated primary antibodies, anti-TRIM28 (1:200) and anti-BRD7 (1:400), were incubated overnight at 4 °C. Then, the cells were incubated with secondary antibodies diluted in antibody dilution buffer at 1:1000 at 37 °C for 1 h, and nuclei were stained with DAPI (Beyotime Biotechnology, China) for 10 min at RT. Cells were observed with confocal laser microscopy and ImageJ was used to quantitatively analyze the co-localization of TRIM28 and BRD7 in breast cancer cells [30, 31].
2.13 Protein stability and ubiquitination assays
For the BRD7 protein stability experiment, MDA-MB-231 and MCF7 cells were transfected with siRNA or plasmids for 24 h and exposed to 50 nM cycloheximide (CHX, MCE) for the indicated timepoints (0, 1, 2 and 4 h). Then, the cell lysate was prepared and subjected to western blotting analysis.
For the ubiquitination assay, MDA-MB-231 and MCF7 cells were cotransfected with HA-Ub plasmid, TRIM28 plasmid, TRIM28△RING plasmid, siTRIM28 (siRNA#1 and siRNA#2). For the detection of ubiquitination site of BRD7, MCF7 cells were cotransfected with HA-Ub plasmid, TRIM28 plasmid, BRD7 lysine mutant plasmid. After 48 h, the cells were treated with 20 µM MG132 for 4 h and lysed in Western and IP lysis buffer (NCM Biotech) supplemented with protease inhibitors. Then, the cell lysates were incubated with anti-BRD7 antibody overnight at 4 °C, and the cells lysates were further incubated with 20 µl of protein A/G beads (MCE, New Jersey, USA) at 4 °C for 2 h. Next, western blotting analysis was performed to measure the levels of Ub, TRIM28 and BRD7.
2.14 Analysis of apoptosis and cell cycle
Cell apoptosis and cell cycle analyses were all performed with a flow cytometry system (BD Biosciences, Franklin Lakes, NJ, USA) according to the manufacturer’s instructions. For apoptosis analysis, BC cells under different treatments were collected and suspended in 1× binding buffer and then incubated with annexin V-FITC and propidium iodide (PI) at RM for 15 min. Then, the apoptotic ratio was detected by the above flow cytometry system. For cell cycle analysis, cells were harvested and fixed in 70% ethanol for 24 h at −20 °C. Next, the cells were treated with RNase A, stained with 25 µg/ml propidium iodide (PI), and then analyzed by flow cytometry.
2.15 Mouse model
Four-week-old female BALB/c nude mice were purchased from Hunan Slake Jingda Experimental Animal Co., Ltd. Animal experiments were divided into three groups: the negative control, TRIM28 knockdown (shTRIM28#1), and TRIM28 knockdown (shTRIM28#1) plus BRD7 knockdown (shBRD7) groups. For the breast cancer xenograft model, 4 × 106 stably-transfected MCF7 cells in 150 µL of saline were subcutaneously inoculated into the shoulders of the nude mice, respectively, with five mice in each group (n = 5). Tumor size was observed and measured every 4 days. Tumor volume was evaluated using the following formula: volume = (length × width2) × 1/2. All mice were sacrificed 30 days after subcutaneous inoculation under anesthesia, and the tumor samples were then collected, fixed with 4% formalin, and embedded in paraffin for IHC analysis. For tumor metastasis experiments, 2 × 106 stably-transfected MDA-MB-231 cells in 200 µL of saline were injected into nude mice via the tail, and the experiment was also divided into three groups as described above, with 5 mice in each group. The mice were sacrificed 35 days after transplantation, the lung tissues were isolated and embedded in 4% paraffin for H&E staining assays, and the micronodules were counted.
2.16 Immunohistochemistry (IHC) staining
For the IHC assays, the procedures were described in a previously published article [29]. Briefly, after deparaffinization and rehydration, the sections were blocked with 5% goat serum blocking solution for 30 min and incubated overnight with the indicated primary antibodies at 4 °C and then incubated with the secondary antibody at RT for 30 min. The primary antibodies used in this article included anti-TRIM28 (1:1600, Proteintech), anti-BRD7 antibody (1:400, Proteintech), anti-c-PARP (1:100, CST), anti-Vimentin (1:600, Arigo), anti-Ki67 (1:400, Bioworld), anti-E-cadherin (1:1000, CST), anti-c-PARP (1:300, CST), and anti-CDK4 (1:400, ABclonal). The IHC score was calculated based on the staining intensity and the percentage of stained cells [29]. Briefly, the staining intensity was graded as 0 (negative) to 3 (strong). The percentage of positive cells was scored 0–1.0 (0–100%). The final staining score (0–3) was calculated as the multiplication of the intensity score and extent score.
2.17 Statistical analysis
The relationships between the BRD7 and TRIM28 expression levels and clinicopathological characteristics of patients with breast cancer were assessed using the chi-squared test. Spearman’s rank correlation analysis was used to assess the significance of the association between BRD7 and TRIM28 expression in breast cancer. Kaplan‒Meier analysis was performed to generate RFS curves, and statistical significance was assessed using the log-rank test. Comparisons between two groups of data were analyzed using Student’s t test, and multiple sets of data were analyzed with one-way ANOVA; data are presented as the means ± SDs using GraphPad Prism 8.01. P values less than 0.05 indicate statistical significance (ns, P > 0.05; *, P < 0.05; **, P < 0.01; and ***, P < 0.001).
3 Results
3.1 BRD7 is an unstable protein in breast cancer
As BRD7 is dysregulated in many tumors and its low expression is a critical mechanism leading to the malignant progression of many tumors, it is necessary to investigate the molecular mechanism of dysregulated expression of BRD7 in breast cancer. We randomly collected 6 pairs of breast cancer patient tumor tissues and adjacent tissues, then detected the protein expression of BRD7 in these tissues by western blotting. The results showed that the expression of BRD7 in breast cancer tissues was significantly lower than that in adjacent tissues (Fig. 1A). Then, we set up a CHX treatment group (CHX treatment for 4 h) and an MG132 (an inhibitor of the proteasome pathway) treatment group (MG132 treatment for 4 h) in MDA-MB-231 and MCF7 cells. As shown in Fig. 1B, with the prolongation of CHX treatment time, the protein level of BRD7 was gradually reduced and almost completely degraded at the 4 h timepoint, indicating that BRD7 was a very unstable protein with a half-life of 0.5–1 h. Furthermore, the protein level of BRD7 was restored after MG132 treatment. Taken together, these results indicate that BRD7 is an unstable protein in BC and that the ubiquitin‒proteasome degradation pathway might be involved in the posttranslational regulation of BRD7.
BRD7 is proved to be an unstable protein in breast cancer. A Western blotting analysis of the protein expression level of BRD7 in tumor tissue and paired adjacent tissues of breast cancer patients. B MDA-MB-231 and MCF7 cells were treated with cycloheximide (CHX) (50 nM), or treated with cycloheximide (CHX) (50 nM) and MG132 (20 µM) for 4 h, the expression of BRD7 were detected by western blotting. The line chart was further quantitatively analyzed BRD7 expression
3.2 TRIM28 promotes the degradation of the BRD7 protein by the ubiquitin‒proteasome degradation pathway in breast cancer
Since BRD7 was identified as an unstable protein in BC, its degradation might be regulated by a ubiquitin-proteasome pathway. To identify the regulatory mechanism, we screened potential E3 ubiquitin ligase proteins that interact with the BRD7 protein by mass spectrometry in MDA-MB-231 cells transfected with Flag-tagged BRD7, followed by immunoprecipitation using an anti-Flag antibody. The results showed that the E3 ubiquitin ligases with score values greater than 9 were screened, including tripartite motif containing 21 (TRIM21), tripartite motif containing 4 (TRIM4), TRIM28 and other E3 ubiquitin ligases (Fig. S1A, B) (Supplementary Table 2). Similarly, we also screened the potential interaction proteins with BRD7 in HEK293 cells (70 molecules) (Supplementary Table 3). To our interest, TRIM28 was found to potentially interact with BRD7 in both of the cell lines as potential E3 ubiquitin ligase. Additionally, as BRD7 was determined to be a tumor suppressor gene in breast cancer, to investigate the degradation of BRD7, we focused on potential oncogenes from the TRIM family. As shown in Fig. S1C–E, it was found that TRIM28 was highly expressed in many cancers through GEPIA online. Moreover, the expression level of TRIM28 was significantly higher in BC tissues (n = 1099) than in normal tissues (n = 292) using RNA-seq data from the TCGA database. In addition, the Kaplan‒Meier plotter database showed that TRIM28 is an independent predictor of poor prognosis in patients with BC. These results indicated a potential oncogenic role of TRIM28 in the development of BC and suggested that TRIM28 may be a valuable predictive factor for poor prognosis in BC. Considering that TRIM28 might be involved in the malignant progression of BC, we chose TRIM28 for further investigation.
To further confirm the interaction between TRIM28 and BRD7, co-transfection with HA-BRD7-expressing and Flag-TRIM28-expressing plasmids was conducted in BC cells, and then a coimmunoprecipitation (Co-IP) assay was performed by using an antibody against Flag or HA. The results showed that TRIM28 interacted with BRD7 in BC cells (Fig. 2A). Moreover, TRIM28 and BRD7 colocalized in the nucleus of MDA-MB-231 and MCF7 cells (Fig. 2B). We next determined the regulatory relationship between TRIM28 and BRD7 expression in BC cells. As shown in Fig. 2C, TRIM28 knockdown by small interfering RNAs (siRNA) significantly increased the protein expression of BRD7 in MDA-MB-231 and MCF7 cells, while TRIM28 overexpression reduced the protein expression of BRD7 (Fig. S2A). However, overexpression or knockdown of TRIM28 did not alter the mRNA level of BRD7 (Fig. S2B). These data suggested that TRIM28 may regulate BRD7 in a translational or posttranslational manner. We further investigated whether TRIM28 impacted the stability of the BRD7 protein. We treated BC cells with CHX and monitored BRD7 expression by western blotting at different time points. The results indicated that TRIM28 knockdown increased the half-life of the BRD7 protein in both MDA-MB-231 and MCF7 cells (Fig. 2D). Additionally, the inhibitory effect of TRIM28 overexpression on BRD7 protein was dramatically reversed in BC cells treated with MG132 (Fig. 2E). These results indicated that TRIM28 promoted BRD7 degradation by the proteasomal pathway. To further illustrate the importance of the E3 ligase activity of TRIM28, we construct the expression plasmids encoding TRIM28 with deletion of the RING domain. Notably, overexpression of full-length TRIM28 reduced the protein stability of BRD7, whereas deletion of the RING domain of TRIM28 abrogated the capability (Fig. 2F). As TRIM28 is a potential E3 ubiquitin ligase, we next explored the effect of TRIM28 on BRD7 ubiquitination. We found that TRIM28 knockdown decreased the polyubiquitination of BRD7 in MDA-MB-231 and MCF7 cells (Fig. 3A). However, the overexpression of TRIM28 presented the opposite result (Fig. S2C). We further characterized which type of ubiquitination of BRD7 was regulated by TRIM28. MCF7 cells were cotransfected with TRIM28 and K48 or K63 ubiquitin plasmids. As shown in Fig. 3B, TRIM28 promoted the K48-linked ubiquitination and K63-linked ubiquitination of BRD7. The overexpression of full-length TRIM28 promoted ubiquitination of BRD7, while deletion of the RING domain of TRIM28 eliminated this ability the capability (Fig. 3C).These data revealed that TRIM28 serves as a potential E3 ubiquitin ligase promotes the degradation of BRD7 protein by ubiquitination in BC cells.
E3 ligase TRIM28 promotes the degradation of BRD7 protein by ubiquitination in BC. A Co-immunoprecipitation (Co-IP) assay was performed by using the Flag or HA antibody and western blotting analysis of Flag-TRIM28 and HA-BRD7 in BC cells. B Co-localization of TRIM28 and BRD7 was performed by immunofluorescence analysis. Endogenous TRIM28 and BRD7 were stained by anti-TRIM28 (red) and anti-BRD7 (green) antibodies. ImageJ was used to quantitatively analyze the co-localization of TRIM28 and BRD7 in breast cancer cells. Scale bar, 10 μm. C Western blotting analysis of TRIM28 and BRD7 in MDA-MB-231 and MCF7 cells transfected with siTRIM28#1, siTRIM28#2, or control siRNA. Equal loading is confirmed by GAPDH levels. D MDA-MB-231 and MCF7 cells transfected with siTRIM28 or control siRNA were treated with cycloheximide (CHX) (50nM) for 0, 1, 2, 4 h, the expression of TRIM28 and BRD7 were detected by western blotting. The line chart was further quantitatively analyzed BRD7 expression. E Western blotting analysis of TRIM28, BRD7 and GAPDH in TRIM28 overexpressed MDA-MB-231 and MCF7 cells treated with or without MG132 (20 µM) for 4 h. F MCF7 cells transfected with Flag-TRIM28 or Flag-TRIM28△RING were treated with cycloheximide (CHX) (50nM) for 0, 1, 2, 4 h, the expression of TRIM28 and BRD7 were detected by western blotting. The line chart was further quantitatively analyzed BRD7 expression
TRIM28 mediates BRD7 ubiquitination at lysine 21 in BC cells. A MDA-MB-231 and MCF7 cells transfected with HA-Ub and siTRIM28#1, siTRIM28#2 or control siRNA treated with MG132 (20 µM) for 4 h, the level of ubiquitin conjugates of BRD7 was detected by western blotting assay. B The MCF7 cells were co-transfected with TRIM28, Ub-K48, and Ub-K63 plasmids, then the cells lysates were pulled down with BRD7 antibodies and analyzed by western blotting. C Evaluation of BRD7 ubiquitination in MCF7 cells co-transfected with Flag-TRIM28 or Flag-TRIM28△RING, HA-BRD7 and HA-Ub. D Schemes of BRD7 deletion mutants. E Schemes of TRIM28 deletion mutants. F Co-IP using anti-Flag antibodies in MCF7 cells co-transfected with HA-BRD7 deletion mutants and Flag-TRIM28 and western blotting analysis of HA and Flag. G Co-IP using anti-HA antibodies in MCF7 cells co-transfected with Flag-TRIM28 deletion mutants and HA-BRD7 and western blotting analysis of HA and Flag. H The ubiquitination sites of N-terminal of BRD7 were predicted by UBPRED (http://ubpred.org/). I MCF7 cells were co-transfected with the lysine mutant HA-BRD7, Flag-TRIM28 and HA-Ub, then Co-IP using anti-BRD7 antibodies and analyzed by western blotting
To further investigated the interaction region of TRIM28 and BRD7, a series of BRD7 deletion mutants and TRIM28 deletion mutants were generated (Fig. 3D, E), then Co-IP was performed to explored the interaction domain between TRIM28 and BRD7. As shown in Fig. 3F, G, there was an interaction between the N-terminal domain of BRD7 and the Coiled-Coil domain of TRIM28. In order to explore the lysine site of BRD7 ubiquitination mediated by TRIM28, we further predicted that there were 6 possible ubiquitination sites of the N-terminal domain of BRD7 by using UBPRED (http://www.ubpred.org/) website (K21, K28, K52, K103, K119, and K127) (Fig. 3H). We then mutated these lysine residues to arginine (K21R, K28R, K52R, K103R, K119R, and K127R) and performed the ubiquitination assay to explore which lysine is required for TRIM28-mediated BRD7 ubiquitination. As shown in Fig. 3I, overexpression of Flag-TRIM28 in MCF7 cells obviously elevated the ubiquitination level of wild-type HA-BRD7. The increase in ubiquitination level of BRD7 could also be observed in K28R, K52R, K103R, K119R, and K127R mutants of HA-BRD7, but not in K21R mutant. Taken together, these data reveal that K21 is critical for TRIM28-mediated ubiquitination of BRD7.
3.3 TRIM28 promotes BC progression and functions as an oncogene in vitro
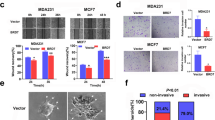
To further clarify the effects of TRIM28 in BC malignant progression, we knocked down TRIM28 by RNA interference technology with two different siRNAs against TRIM28, and the knockdown efficiency of TRIM28 was confirmed by western blotting (Fig. 4A). Then, Cell Counting Kit-8 (CCK-8) and colony formation assays were performed to assess the effect of TRIM28 knockdown on cell proliferation, and the results showed that the knockdown of TRIM28 decreased cell proliferation and colony formation in BC cells (Fig. 4B, C). We next analyzed the effect of TRIM28 knockdown on cell apoptosis by conducting annexin V-FITC/PI double-staining and flow cytometry analysis after the cells were serum-starved for 24 h. As shown in Fig. S3A, the knockdown of TRIM28 increased the number of apoptotic MDA-MB-231 cells. Additionally, the effect of TRIM28 knockdown on cell cycle progression was determined by flow cytometry assay. As a result, compared with the negative control, TRIM28 knockdown reduced the cell population in G2/M phase in both MDA-MB-231 and MCF7 cells (Fig. S3B). In addition, Transwell assays and wound healing assays demonstrated that the downregulation of TRIM28 remarkably suppressed the invasive and migratory capacities of MDA-MB-231 and MCF7 cells (Fig. 4D, E).
TRIM28 knockdown inhibit BC progression in vitro. A Western blotting analysis of TRIM28 protein level in MDA-MB-231 and MCF7 cells transfected with siNC, siTRIM28#1 and siTRIM28#2, and GAPDH served as an internal control. B CCK-8 assays in MDA-MB-231 and MCF7 cells transfected with TRIM28 siRNAs or siNC. C Colony forming assay was performed in MDA-MB-231 and MCF7 cells transfected with TRIM28 siRNAs or siNC. D Scratch wound healing analysis of cell migration in MDA-MB-231 and MCF7 cells with transfection of TRIM28 siRNAs or siNC. E Transwell invasion assay of cell invasive capabilities was performed in MDA-MB-231 and MCF7 cells transfected with control siRNA and TRIM28 siRNAs. Data are shown as the mean ± SD of at least three independent experiments, and the significant level was identified by *P < 0.05, **P < 0.01, and ***P < 0.001
Correspondingly, we further explored the effect of TRIM28 overexpression on the tumor progression of BC. TRIM28-overexpressing BC cell lines were constructed by using MDA-MB-231 and MCF7 cell lines, and western blotting assays indicated that TRIM28 was successfully upregulated (Fig. 5A). Accordingly, upregulation of TRIM28 promoted BC cell proliferation and colony formation as determined by CCK-8 and colony formation assays (Fig. 5B, C). Moreover, TRIM28 overexpression promoted the invasion and migration of BC cells (Fig. 5D, E). Therefore, these results demonstrated that TRIM28 acts as a potential oncogene to promote the proliferation, migration and invasion of breast cancer cells.
TRIM28 overexpression promotes BC progression in vitro. A Western blotting assay of TRIM28 overexpression in MDA-MB-231 and MCF7 cells. B CCK-8 assays in MDA-MB-231 and MCF7 cells transfected with vector or TRIM28 plasmid. C Colony forming assay was performed in BC cells after TRIM28 overexpression. D Scratch wound healing analysis of cell migration in BC cells with transfection of vector or TRIM28 plasmid. E Representative images of transwell invasion assay using MDA-MB-231 and MCF7 cells transfected with vector or TRIM28. Quantification of transwell invasion assay is shown. Data are shown as the mean ± SD of at least three independent experiments, and the significant level was identified by *P < 0.05, **P < 0.01, and ***P < 0.001
3.4 Restoring the expression of BRD7 abrogates the promoting effect of TRIM28 on the malignant phenotype of BC cells
As BRD7 has been previously reported as a tumor suppressor that inhibits cell proliferation, invasion, tumor growth and metastasis in breast cancer [9, 32], while we demonstrated that TRIM28 exerts an oncogenic role in BC and promotes the degradation of BRD7 protein, we sought to determine whether BRD7 was required for TRIM28-mediated tumor oncogenic roles in BC. First, we restored BRD7 expression in TRIM28-silenced BC cells and then examined the effects of BRD7 restoration on TRIM28 knockdown-mediated tumor inhibitory function. Western blotting assays showed that TRIM28 was effectively knocked down and that BRD7 was successfully restored (Fig. 6A). Next, CCK-8, colony formation, Transwell, and wound healing assays were performed to investigate the effects of BRD7 on TRIM28-mediated BC cell malignant progression. The results showed that knockdown of TRIM28 inhibited the proliferation, invasion and migration of BC cells, which was consistent with the above results, while the restoration of BRD7 rescued the oncogenic phenotypes of TRIM28 (Fig. 6B–E). Furthermore, a flow cytometry assay was performed to explore the roles of BRD7 in TRIM28-mediated apoptosis and cell cycle progression in BC, and the results showed that knockdown of TRIM28 arrested the cell cycle in G2/M and promoted apoptosis, whereas these phenotypes were reversed when BRD7 was restored (Fig. S4A, B). To further investigate the molecular mechanisms of BRD7 in TRIM28-mediated malignant progression, we detected the expression of cell cycle-, apoptosis- and epithelial-mesenchymal transition (EMT)-related molecules by western blotting assays. As a result, the downregulation of TRIM28 reduced the expression of the cell cycle marker Cyclin Dependent Kinase 4 (CDK4) and the mesenchymal molecular markers Vimentin and Snail and increased the expression of the apoptosis marker cleaved-PARP and the epithelial molecular markers Zona Occludens 1(ZO-1) and E-cadherin, whereas the expression levels of these cell proliferation-, apoptosis- and EMT-related markers were significantly reversed after BRD7 restoration (Fig. 6F).
Restoring the expression of BRD7 eliminated the inhibitory effect of silent expression of TRIM28 on cell proliferation, migration, invasion and epithelial-mesenchymal transition process. A Western blotting analysis indicated the protein level of TRIM28 and BRD7 in MDA-MB-231 and MCF7 cells co-transfected with siTRIM28 and siBRD7. Equal loading is confirmed by GAPDH levels. B CCK-8 analysis of cell proliferation in MDA-MB-231 and MCF7 cells with TRIM28 knockdown, TRIM28 and BRD7 simultaneous knockdown or negative control. C Colony forming assay of cell proliferation in MDA-MB-231 and MCF7 cells with TRIM28 knockdown, TRIM28 and BRD7 simultaneous knockdown or negative control. D Representative images of wound healing assay using MDA-MB-231 and MCF7 cells with TRIM28 knockdown, TRIM28 and BRD7 simultaneous knockdown or negative control. E Transwell invasion assay of cell invasive capabilities was performed in MDA-MB-231 and MCF7 cells with TRIM28 knockdown, TRIM28 and BRD7 simultaneous knockdown or negative control. F Western blotting analysis of the expression of TRIM28, BRD7, ZO-1, E-cadherin, Vimentin, Snail, t-PARP, c-PARP and CDK4 in MDA-MB-231 and MCF7 cells with TRIM28 knockdown and BRD7 restoration. Equal loading is confirmed by GAPDH levels. Data are shown as the mean ± SD of at least three independent experiments, and the significant level was identified by *P < 0.05, **P < 0.01, and ***P < 0.001
Consistently, we also restored BRD7 expression in TRIM28-overexpressing MDA-MB-231 cells. As shown in Fig. 7A, TRIM28 was successfully overexpressed, and BRD7 was restored. Overexpression of TIRM28 significantly enhanced cell proliferation in MDA-MB-231 cells, as determined by CCK-8 and colony formation assays, while BRD7 overexpression rescued the promoting effect of TRIM28 on BC cell proliferation (Fig. 7B, C). We also conducted Transwell invasion assays and wound healing assays to determine the function of BRD7 in TRIM28-dependent invasion and migration. The results showed that overexpression of BRD7 rescued the promoting effect of TRIM28 on cell invasion and migration in MDA-MB-231 cells (Fig. 7D, E). In addition, western blotting assays were performed to detect the expression of apoptosis-, cell cycle- and EMT-related molecules. As expected, upregulation of Vimentin, Snail and CDK4 and downregulation of ZO-1, E-cadherin and cleaved-PARP were found in TRIM28-overexpressing MDA-MB-231 cells, while the expression levels of these molecules were reversed by BRD7 overexpression (Fig. 7F). Therefore, these results demonstrated that TRIM28 exerts an oncogenic role in BC by negatively regulating BRD7 protein stability.
Restoring the expression of BRD7 abrogated the promoting effect of TRIM28 overexpression on cell proliferation, migration, invasion, and epithelial-mesenchymal transition process. A The MDA-MB-231 cells were transfected with TRIM28 and BRD7 plasmid, and the expression of TRIM28 and BRD7 were detected by western blotting. Equal loading is confirmed by GAPDH levels. B Cell proliferation was performed in MDA-MB-231 through CCK-8 analysis after TRIM28 overexpression, TRIM28 and BRD7 simultaneous overexpression or control group. C Colony forming assay was performed in TRIM28 overexpression and BRD7 restoration MDA-MB-231 cells. D Wound healing analysis of cell migration with MDA-MB-231 cells transfected with TRIM28 or TRIM28 and BRD7 plasmids. E Transwell invasion assay of cell invasive capabilities was performed in MDA-MB-231 cells with TRIM28 overexpression, TRIM28 and BRD7 overexpression or control. F Western blotting analysis of the expression of TRIM28, BRD7, ZO-1, E-cadherin, Vimentin, Snail, t-PARP, c-PARP and CDK4 in MDA-MB-231 cells with TRIM28 overexpression and BRD7 restoration. Equal loading is confirmed by GAPDH levels. Data are shown as the mean ± SD of at least three independent experiments, and the significant level was identified by *P < 0.05, **P < 0.01, and ***P < 0.001
3.5 TRIM28 promotes tumor growth and metastasis through negative regulation of BRD7 expression in vivo
Since TRIM28 promotes the malignant progression of breast cancer by inhibiting the expression of BRD7 in vitro. Xenograft and metastasis models were further generated in BALB/c nude mice to investigate whether BRD7 is critical for the TRIM28-mediated malignant tumor phenotype in vivo. We first constructed stably-transfected MDA-MB-231 and MCF7 cell lines by using shRNA (Fig. S5). Then, three different MCF7 cell groups, including the negative control, TRIM28 knockdown and BRD7 recovery groups, were used to construct nude mouse xenograft tumor models. As a result, the volume and weight of tumors in the TRIM28 knockdown group were significantly inhibited compared with those in the negative control group, while the volume and weight of tumors were recovered after the expression of BRD7 was restored, as shown in Fig. 8A–D. Moreover, immunohistochemistry (IHC) of xenograft tumor tissues was performed to detect the expression of TRIM28, BRD7, cleaved-PARP, E-cadherin, Vimentin, Ki67 and CDK4, and the results showed that TRIM28 was successfully knocked down and BRD7 was decreased in the TRIM28-knockdown group, and the expression of BRD7 was successfully restored in the BRD7 recovery group. Additionally, in contrast to the negative control group, upregulation of cleaved- PARP and E-cadherin expression and reduction of Vimentin, Ki67 and CDK4 expression were observed in the TRIM28 knockdown group. As expected, these changes were markedly reversed after BRD7 restoration (Fig. 8E). These data clarified that the knockdown of TRIM28 inhibits the xenograft tumor growth of BC, and this effect is rescued by BRD7 restoration.
TRIM28 inhibits tumor growth and metastasis through regulation of BRD7 expression in vivo. A The tumor growth curve (n = 5). B Macroscopic mouse of xenograft model, n = 5 per group. C Representative images of tumors of the MCF7 xenograft model in nude mice. D The tumor weight (n = 5). E IHC staining of TRIM28, BRD7, Cleaved-PARP, E-cadherin, Vimentin, Ki67 and CDK4 in MCF7 xenograft model. Scale bar, 100 μm. F H&E staining is shown in negative control, TRIM28 knockdown, and BRD7 restoration group. Red arrows indicate metastatic tumors. Scale bar, 200 μm. G The number of metastatic lung nodules in lung tissue section of every mouse per group were counted in microscopy (n = 5). Error bars represent the mean ± SD. *P < 0.05, **P < 0.01, and ***P < 0.001
To further evaluate the effects of TRIM28 on tumor metastasis, BALB/c nude mice were injected with the stably-transfected MDA-MB-231 cells by tail vein injection. After five weeks, all mice were sacrificed, and the lung nodules were calculated. We found that TRIM28 knockdown inhibited the number and size of lung metastasis foci in breast cancer, and the number and size of lung metastasis foci were significantly recovered in the group with restored BRD7 expression (Fig. 8F, G). Overall, these results demonstrate that TRIM28 promotes xenograft tumor growth and metastasis of breast cancer by negatively regulating BRD7 expression in vivo.
3.6 Expression and clinical correlation of TRIM28 and BRD7 in biopsy tissues of breast cancer at different stages
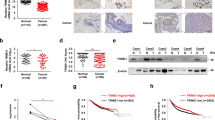
To further explore the clinical correlation between TRIM28 and BRD7 in clinical tissue samples, we detected the expression of TRIM28 and BRD7 in 33 noncancerous breast samples and 178 BC samples by IHC. The expression of TRIM28 in BC tissues was significantly higher than that in breast noncancerous tissues, and its expression in clinical stages III and IV was significantly higher than that in stages I and II, suggesting the potential oncogenic activity of TRIM28 in BC (Fig. 9A–C), while the expression of BRD7 presented the exact opposite result (Fig. S6A). In addition, there was a negative correlation between TRIM28 and BRD7 in BC tissues (Pearson correlation coefficient r = −0.3089, P < 0.0001) (Fig. 9D). Next, we explored whether the higher expression of TRIM28 in BC was related to poor prognosis. Survival analysis revealed that TRIM28 high expression predicted reduced overall survival times compared with TRIM28 low, while the patients with low expression of BRD7 had a longer total survival time (Fig. S6B). Additionally, the patients with a combination of high TRIM28 expression and low BRD7 expression had a worse prognosis (Fig. 9E). Furthermore, we analyzed the correlation between TRIM28 expression and the clinicopathological features of BC patients. The results showed that high TRIM28 expression and high TRIM28 expression combined with low BRD7 expression both correlated with clinical stage, tumor size, lymph node metastasis, and distant metastasis, whereas no significant correlation was found with age (Table 2). Collectively, these findings indicate that TRIM28 is negatively correlated with BRD7 and that high TRIM28 combined with low BRD7 may be a predictor for diagnosis and prognosis in BC patients.
TRIM28 and BRD7 are negatively correlated in breast cancer prognosis. A Representative images of TRIM28 expression in breast cancer tissues and normal tissues using IHC. B The expression of TRIM28 was detected in normal (n = 33) and tumor samples (n = 178) by IHC. C TRIM28 expression in different T stages of breast cancer. D The correlation between TRIM28 expression and BRD7 expression in breast cancer tumor tissues. E Kaplan-Meier curves showed the overall survival of breast cancer patients with high and low expression levels of TRIM28, and high TRIM28 plus low BRD7 level and low TRIM28 plus high BRD7 level. F Schematic model of molecular mechanism of TRIM28/BRD7 axis involved in oncogenesis and tumor progression of breast cancer
4 Discussion
BRD7 was confirmed to be a critical tumor suppressor and is expressed at low levels in a variety of tumors, and epigenetic modifications, transcriptional regulation, and posttranslational modifications were confirmed to be involved in the regulation of BRD7 expression. For example, hypermethylation of the BRD7 promoter results in low expression of BRD7 in nasopharyngeal carcinoma and highly differentiated oral squamous cell carcinoma [33, 34]. C-Myc could bind to the BRD7 promoter and negatively regulate BRD7 promoter activity and mRNA expression [35]. In addition, the SUMO modification of BRD7 in osteosarcoma cells plays an important role in DNA damage repair [17]. In our previous study, BRD7 was identified to inhibit cell proliferation, migration, invasion, EMT and in vivo xenograft tumor growth and metastasis, thus exerting a tumor suppressor role in breast cancer by regulating the expression of its target genes P53 and Y-box binding protein 1 (YB1) [9, 29, 32], whereas the upstream regulatory mechanism of low BRD7 expression in breast cancer remains unclear and needs to be further elucidated.
In the current study, we found that the protein level of BRD7 was lower in breast cancer tissues than in precancerous tissues. As few studies have reported on the regulation of BRD7 protein levels in breast cancer, we investigated the potential mechanism resulting in the low expression of BRD7 at the posttranslational level. In this study, BRD7 was found to be an unstable protein in breast cancer, and the ubiquitin‒proteasome pathway might be involved in the regulation of BRD7 protein instability. Therefore, we screened and identified a potential E3 ubiquitin ligase of BRD7, TRIM28, which could lead to the ubiquitination and subsequent proteasomal degradation of BRD7. TRIM28 has been reported to regulate proteasome-mediated protein stability, thus playing a critical role in tumorigenesis and tumor progression by functioning as an E3 ubiquitin ligase. For example, TRIM28 promotes ubiquitination and degradation of Transcription factor enhancer 3 (TFE3) as an E3 ubiquitin ligase, thus playing a critical role in kidney cancer progression [36]. TRIM28 could combine with Ubiquitin Conjugating Enzyme E2 S (UBE2S) in the nucleus, and both together enhance the ubiquitination and degradation of p27, thus facilitating cell cycle progression and hepatocellular carcinoma progression [25]. Therefore, our results demonstrated that TRIM28 ubiquitinates and degrades BRD7 proteien depend on its K21 site and highlight a new posttranslational regulatory mechanism of BRD7. In addtion to directly participating in protein stability regulation through ubiquitin proteasome pathway as E3 ubiquitin enzyme, TRIM28 was also reported to participate in protein function and stability regulation through SUMOlylation pathway [37, 38]. Therefore, the detailed regulatory mechanism by which TRIM28 decreases BRD7 protein stability through the direct ubiquitin-mediated proteasome pathway or indirect SUMOylation still needs to be further clarified.
TRIM28 has been reported to be highly expressed in many cancers, such as gastric cancer, ovarian cancer, lung cancer, and cervical cancer, and is related to the malignant pathological features of tumors [36,37,38,42]. In breast cancer, TRIM28 has been shown to be closely associated with the proliferation, metastasis and stemness of breast cancer cells [40,41,45], whereas the function and mechanism still need to be further clarified. Consistent with these results, we demonstrated that TRIM28 promotes cell proliferation, migration, invasion, tumor growth and metastasis as well as the expression of phenotype-related molecules, suggesting that TRIM28 exerts an oncogenic role in breast cancer. As BRD7 was identified as a tumor suppressor in breast cancer in our previous study and TRIM28 negatively regulates protein stability by the ubiquitin-mediated proteasome pathway, we tried to clarify the mechanism by which TRIM28 plays an oncogenic role through the TRIM28/BRD7 axis. As a result, the restoration of BRD7 expression could reverse the promoting effect of TRIM28 on the malignant phenotype of breast cancer cells, suggesting that TRIM28 promotes tumor progression and metastasis of breast cancer at least partly by negatively regulating BRD7 protein stability.
In addition to revealing the mechanism of dysregulation of BRD7 expression at the posttranslational level and the biological importance of the TRIM28/BRD7 axis in breast cancer, our work indicates the potential of TRIM28 and BRD7 in the clinical prognosis of breast cancer patients. Several articles have reported that TRIM28 is highly expressed in a variety of tumors and can be used as a molecular target for tumor diagnosis and prognosis prediction. For example, in non-small cell lung cancer, patients with high TRIM28 expression have lower recurrence and survival rates, which could be a predictor of non-small cell lung cancer (NSCLC) patient prognosis [46]. In addition, the TRIM28/ Twist Family BHLH Transcription Factor 1 (TWIST1) axis could be a potential therapeutic target for breast cancer [45]. Consistent with these findings, we showed evidence in this study that TRIM28 was highly expressed in breast cancer biopsy specimens and that TRIM28 was correlated with poor clinical outcome. Additionally, TRIM28 was negatively correlated with BRD7 expression, and the combination of low TRIM28 expression and high BRD7 expression was significantly associated with a better prognosis. For this reason, targeting the TRIM28/BRD7 axis might be a novel potential strategy for the clinical diagnosis and treatment of breast cancer. For examples, development of small molecule activators to BRD7, inhibitors to TRIM28 or blocking peptides that inhibit the binding of TRIM28 to BRD7 protein could theoretically exert antitumor effects in breast cancer and thus it could be used as the drug discovery of breast cancer. Therefore, the TRIM28/BRD7 axis can be used as a potential target to guide the early diagnosis and treatment of breast cancer patients, providing a promising strategy for improving their prognosis.
5 Conclusions
In summary, our work reveals the mechanism of posttranslational modification of BRD7 in breast cancer for the first time. We demonstrated that TRIM28 destabilized the BRD7 protein by ubiquitylating and degrading BRD7 depend on K21 site to promote breast cancer progression in vitro and in vivo. Moreover, the high expression of TRIM28 was positively correlated with poor survival, higher TMN stage, and metastasis in breast cancer patients. Therefore, the TRIM28/BRD7 axis plays a critical role in oncogenesis and tumor progression of breast cancer, and targeting the TRIM28/BRD7 axis might be a novel strategy for the diagnosis and treatment of breast cancer (Fig. 9F).
Data availability
No datasets were generated or analysed during the current study.
Abbreviations
- TRIM28:
-
tripartite motif protein 28
- BC:
-
breast cancer
- BRD7:
-
bromodomain-containing protein 7
- DMEM:
-
Dulbecco’s modified Eagle’s medium
- Brd:
-
bromodomain
- TNM:
-
tumor node metastasis
- EMT:
-
epithelial-mesenchymal transition
- siRNA:
-
small interfering RNA
- BCA:
-
Bicinchoninic Acid
- IP:
-
immunoprecipitation
- HE:
-
hematoxylin-eosin staining
- SNP:
-
single-nucleotide polymorphism
- FBS:
-
Fetal bovine serum
- IHC:
-
immunohistochemistry
References
F. Bray, J. Ferlay, I. Soerjomataram et al., Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA. Cancer J. Clin. 68, 394–424 (2018). https://doi.org/10.3322/caac.21492
A. Budny, E. Staroslawska, B. Budny et al., Epidemiology and diagnosis of breast cancer. Pol. Merkur Lekarski. 46, 195–204 (2019)
C. He, P. Kraft, D.I. Chasman et al., A large-scale candidate gene association study of age at menarche and age at natural menopause. Hum. Genet. 128, 515–527 (2010). https://doi.org/10.1007/s00439-010-0878-4
J. Stone, G.S. Dite, A. Gunasekara et al., The heritability of mammographically dense and nondense breast tissue. Cancer Epidemiol. Biomarkers Prev. 15, 612–617 (2006).https://doi.org/10.1158/1055-9965.EPI-05-0127
L. Stolk, J.R. Perry, D.I. Chasman et al., Meta-analyses identify 13 loci associated with age at menopause and highlight DNA repair and immune pathways. Nat. Genet. 44, 260–268 (2012).https://doi.org/10.1038/ng.1051
J. Zhou, J. Ma, B.C. Zhang et al., BRD7, a novel bromodomain gene, inhibits G1-S progression by transcriptionally regulating some important molecules involved in ras/MEK/ERK and Rb/E2F pathways. J. Cell. Physiol. 200, 89–98 (2004). https://doi.org/10.1002/jcp.20013
X. Yu, Z. Li, J. Shen, BRD7: a novel tumor suppressor gene in different cancers. Am. J. Transl. Res. 8, 742–748 (2016)
Y. Liu, R. Zhao, H. Wang et al., miR-141 is involved in BRD7-mediated cell proliferation and tumor formation through suppression of the PTEN/AKT pathway in nasopharyngeal carcinoma. Cell. Death Dis. 7, e2156 (2016). https://doi.org/10.1038/cddis.2016.64
W. Niu, Y. Luo, Y. Zhou et al., BRD7 suppresses invasion and metastasis in breast cancer by negatively regulating YB1-induced epithelial-mesenchymal transition. J. Exp. Clin. Cancer Res. 39, 30 (2020). https://doi.org/10.1186/s13046-019-1493-4
Y. Gao, B. Wang, S. Gao, BRD7 acts as a tumor suppressor gene in Lung Adenocarcinoma. Plos One. 11, e0156701 (2016).https://doi.org/10.1371/journal.pone.0156701
M.D. Kaeser, A. Aslanian, M.Q. Dong et al., BRD7, a novel PBAF-specific SWI/SNF subunit, is required for target gene activation and repression in embryonic stem cells. J. Biol. Chem. 283, 32254–32263 (2008). https://doi.org/10.1074/jbc.M806061200
Q. Zhang, L. Wei, H. Yang et al., Bromodomain containing protein represses the Ras/Raf/MEK/ERK pathway to attenuate human hepatoma cell proliferation during HCV infection. Cancer Lett. 371, 107–116 (2016).https://doi.org/10.1016/j.canlet.2015.11.027
Y.A. Park, J.W. Lee, H.S. Kim et al., Tumor suppressive effects of bromodomain-containing protein 7 (BRD7) in epithelial ovarian carcinoma. Clin. Cancer Res. 20, 565–575 (2014). https://doi.org/10.1158/1078-0432.CCR-13-1271
A.E. Burrows, A. Smogorzewska, S.J. Elledge, Polybromo-associated BRG1-associated factor components BRD7 and BAF180 are critical regulators of p53 required for induction of replicative senescence. Proc. Natl. Acad. Sci. U S A 107, 14280–14285 (2010). https://doi.org/10.1073/pnas.1009559107
J. Drost, F. Mantovani, F. Tocco et al., BRD7 is a candidate tumour suppressor gene required for p53 function. Nat. Cell. Biol. 12, 380–389 (2010). https://doi.org/10.1038/ncb2038
M. Li, Y. Wei, Y. Liu et al., BRD7 inhibits enhancer activity and expression of BIRC2 to suppress tumor growth and metastasis in nasopharyngeal carcinoma. Cell. Death Dis. 14, 121 (2023). https://doi.org/10.1038/s41419-023-05632-3
Z. Xiao, J.G. Chang, I.A. Hendriks et al., System-wide analysis of SUMOylation dynamics in response to replication stress reveals Novel Small Ubiquitin-like modified target proteins and acceptor lysines relevant for genome stability. Mol. Cell. Proteom. 14, 1419–1434 (2015). https://doi.org/10.1074/mcp.O114.044792
J.V. Olsen, B. Blagoev, F. Gnad et al., Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell. 127, 635–648 (2006).https://doi.org/10.1016/j.cell.2006.09.026
K. Hu, W. Wu, Y. Li, et al., Poly(ADP-ribosyl)ation of BRD7 by PARP1 confers resistance to DNA-damaging chemotherapeutic agents. EMBO Rep. 20 (2019). https://doi.org/10.15252/embr.201846166
A. Reymond, G. Meroni, A. Fantozzi et al., The tripartite motif family identifies cell compartments. EMBO J. 20, 2140–2151 (2001). https://doi.org/10.1093/emboj/20.9.2140
S. Hatakeyama, TRIM proteins and cancer. Nat. Rev. Cancer. 11, 792–804 (2011). https://doi.org/10.1038/nrc3139
L.M. Napolitano, G. Meroni, TRIM family: Pleiotropy and diversification through homomultimer and heteromultimer formation. IUBMB Life. 64, 64–71 (2012).https://doi.org/10.1002/iub.580
C. International Human Genome Sequencing, Finishing the euchromatic sequence of the human genome. Nature. 431, 931–945 (2004).https://doi.org/10.1038/nature03001
Z. Qi, S. Cai, J. Cai et al., miR-491 regulates glioma cells proliferation by targeting TRIM28 in vitro. BMC Neurol. 16, 248 (2016).https://doi.org/10.1186/s12883-016-0769-y
R.Y. Zhang, Z.K. Liu, D. Wei et al., UBE2S interacting with TRIM28 in the nucleus accelerates cell cycle by ubiquitination of p27 to promote hepatocellular carcinoma development. Signal. Transduct. Target. Ther. 6, 64 (2021). https://doi.org/10.1038/s41392-020-00432-z
C. Yu, L. Zhan, J. Jiang et al., KAP-1 is overexpressed and correlates with increased metastatic ability and tumorigenicity in pancreatic cancer. Med. Oncol. 31, 25 (2014).https://doi.org/10.1007/s12032-014-0025-5
L. Chen, T. Munoz-Antonia, W.D. Cress, Trim28 contributes to EMT via regulation of E-cadherin and N-cadherin in lung cancer cell lines. Plos One. 9, e101040 (2014). https://doi.org/10.1371/journal.pone.0101040
J.B. Addison, C. Koontz, J.H. Fugett et al., KAP1 promotes proliferation and metastatic progression of breast cancer cells. Cancer Res. 75, 344–355 (2015). https://doi.org/10.1158/0008-5472.CAN-14-1561
J. Ma, W. Niu, X. Wang et al., Bromodomain–containing protein 7 sensitizes breast cancer cells to paclitaxel by activating Bcl2–antagonist/killer protein. Oncol Rep. 41, 1487–1496 (2019).https://doi.org/10.3892/or.2018.6951
M. Stiekema, F.C.S. Ramaekers, D. Kapsokalyvas et al., Super-resolution imaging of the A- and B-Type Lamin networks: a comparative study of different fluorescence labeling procedures. Int. J. Mol. Sci. 22 (2021). https://doi.org/10.3390/ijms221910194
C.E. O’Brien, L. Bonanno, H. Zhang et al., Beclin 1 regulates neuronal transforming growth factor-beta signaling by mediating recycling of the type I receptor ALK5. Mol. Neurodegener. 10, 69 (2015).https://doi.org/10.1186/s13024-015-0065-0
Y. Luo, X. Wang, W. Niu et al., BRD7 stabilizes P53 via dephosphorylation of MDM2 to inhibit Tumor growth in breast Cancer harboring wild-type P53. J. Cancer. 13, 1436–1448 (2022).https://doi.org/10.7150/jca.67447
A. Balasubramanian, R. Subramaniam, V. Narayanan et al., BRD7 promoter hypermethylation as an indicator of well differentiated oral squamous cell carcinomas. Asian. Pac. J. Cancer Prev. 16, 1615–1619 (2015). https://doi.org/10.7314/apjcp.2015.16.4.1615
H. Liu, L. Zhang, Z. Niu et al., Promoter methylation inhibits BRD7 expression in human nasopharyngeal carcinoma cells. BMC Cancer. 8 (2008). https://doi.org/10.1186/1471-2407-8-253
H. Liu, M. Zhou, X. Luo et al., Transcriptional regulation of BRD7 expression by Sp1 and c-Myc. BMC Mol. Biol. 9, 111 (2008). https://doi.org/10.1186/1471-2199-9-111
T. Song, S. Lv, X. Ma et al., TRIM28 represses renal cell carcinoma cell proliferation by inhibiting TFE3/KDM6A-regulated autophagy. J. Biol. Chem. 299 (2023). https://doi.org/10.1016/j.jbc.2023.104621
X. Ma, S. Jia, G. Wang et al., TRIM28 promotes the escape of gastric cancer cells from immune surveillance by increasing PD-L1 abundance. Signal Transduct. Target. Ther. 8(1), 246 (2023). https://doi.org/10.1038/s41392-023-01450-3
K. Li, H. Wang, B. Jiang, X. Jin, TRIM28 in cancer and cancer therapy. Front. genet. 15, 1431564 (2024). https://doi.org/10.3389/fgene.2024.1431564
T. Yokoe, Y. Toiyama, Y. Okugawa et al., KAP1 is associated with peritoneal carcinomatosis in gastric cancer. Ann. Surg. Oncol. 17, 821–828 (2010). https://doi.org/10.1245/s10434-009-0795-8
Y. Cui, S. Yang, X. Fu et al., High levels of KAP1 expression are associated with aggressive clinical features in ovarian cancer. Int. J. Mol. Sci. 16, 363–377 (2014). https://doi.org/10.3390/ijms16010363
L. Chen, D.-T. Chen, C. Kurtyka et al., Tripartite motif containing 28 (Trim28) can regulate cell proliferation by bridging HDAC1/E2F interactions. J. Biol. Chem. 287, 40106–40118 (2012).https://doi.org/10.1074/jbc.M112.380865
F. Li, Z. Wang, G. Lu, TRIM28 promotes cervical cancer growth through the mTOR signaling pathway. Oncol. Rep. 39, 1860–1866 (2018).https://doi.org/10.3892/or.2018.6235
J. Li, Y. Xi, W. Li et al., TRIM28 interacts with EZH2 and SWI/SNF to activate genes that promote mammosphere formation. Oncogene. 36, 2991–3001 (2017). https://doi.org/10.1038/onc.2016.453
Y. Zhou, B. Wang, Y. Wang et al., Mir-140-3p inhibits breast cancer proliferation and migration by directly regulating the expression of tripartite motif 28. Oncol. Lett. 17, 3835–3841 (2019). https://doi.org/10.3892/ol.2019.10038
C. Wei, J. Cheng, B. Zhou et al., Tripartite motif containing 28 (TRIM28) promotes breast cancer metastasis by stabilizing TWIST1 protein. Sci. Rep. 6, 29822 (2016).https://doi.org/10.1038/srep29822
L. Liu, E. Zhao, C. Li et al., TRIM28, a new molecular marker predicting metastasis and survival in early-stage non-small cell lung cancer. Cancer Epidemiol. 37, 71–78 (2013). https://doi.org/10.1016/j.canep.2012.08.005
Acknowledgements
Not applicable.
Funding
This study was supported by the grants from the National Natural Science Foundation of China (grant nos. 82172592), the Hunan Provincial Key Research and Development Program (2023SK2008), the Free Exploration Program of Central South University (grant nos. 2021zzts0934), the program of Introducing Talents of Discipline to Universities (grant no. 111-2-12).
Author information
Authors and Affiliations
Contributions
MZ, CX and HM designed this study, wrote and revised the manuscript. CX, HM and WN mainly performed the experiment. ML and JW were involved in providing guidance on experimental techniques. SC and LZ helped with cell culture, solution preparation and reagents ordering. YD performed the nude mouse samples of IHC. SF and HD contributed to collection and grading of clinical samples. WX, MT and FT provided important comments and opinions for the manuscript. All authors have read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
This study was approved by Ethics Review Committees/Institutional Review Boards of Central South University (Changsha, China). And all the animal experiments in this study were approved by the Institutional Animal Care and Use Committee (IACUC) of central south university (Changsha, China). Clinical samples of breast cancer and normal breast samples were all collected from the Second Xiangya Hospital of Central South University. All patients involved in this article signed informed consent.
Consent for publication
All authors agree to submit the article for publication.
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Xue, C., Meng, H., Niu, W. et al. TRIM28 promotes tumor growth and metastasis in breast cancer by targeting the BRD7 protein for ubiquitination and degradation. Cell Oncol. (2024). https://doi.org/10.1007/s13402-024-00981-3
Accepted:
Published:
DOI: https://doi.org/10.1007/s13402-024-00981-3