Abstract
Purpose
Uveal melanoma (UM) with BAP1 inactivating mutations has a high risk of metastasis, but the mechanism behind BAP1 deficiency driving UM metastasis is unknown.
Methods
We analyzed the single-cell RNA sequencing (scRNA-Seq) data comprised primary and metastatic UM with or without BAP1 mutations (MUTs) to reveal inter- and intra-tumor heterogeneity among different groups. Then, an immune-competent mouse liver metastatic model was used to explore the role of ITGB2-ICAM1 in BAP1-associated UM metastasis.
Results
Cluster 1 tumor cells expressed high levels of genes linked to tumor metastasis, such as GDF15, ATF3, and CDKN1A, all of which are associated with poor prognosis. The strength of communication between terminally exhausted CD8+ T cells and GDF15hiATF3hiCDKN1Ahi tumor cells was enhanced in BAP1-mutated UM, with CellChat analysis predicting strong ITGB2-ICAM1 signaling between them. High expression of either ITGB2 or ICAM1 was a worse prognostic indicator. Using an immune-competent mouse liver metastatic model, we indicated that inhibiting either ICAM1 or ITGB2 prevented liver metastasis in the BAP1-mutated group in vivo. The inhibitors primarily inhibited hypoxia- and ECM-related pathways indicated by changes in the expression of genes such as ADAM8, CAV2, ENO1, PGK1, LOXL2, ITGA5, and VCAN. etc.
Conclusion
This study suggested that the ITGB2-ICAM1 axis may play a crucial role for BAP1-associated UM metastasis by preserving hypoxia- and ECM- related signatures, which provide a potential strategy for preventing UM metastasis in patients with BAP1 mutation.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Up to 50% of uveal melanoma (UM) cases develop metastases in a median of 2.4 years with liver as the first metastatic site [1, 2], and 90% of metastatic patients result in death within six months [3, 4]. Determining the precise mechanism underlying UM metastasis becomes essential. Class 2 UM, which normally contains BRCA1-associated protein 1 (BAP1) inactivating mutation, is linked to a short-term survival and unknown contributions to UM metastasis [5, 6].
BAP1 is a two-hit tumor suppressor gene that frequently mutated in various sporadic human cancers, including uveal melanomas [7], cutaneous melanomas [8], meningiomas [9], malignant mesothelioma [10, 11], cholangiocarcinoma [12] and renal cell carcinomas [13, 14]. BAP1 regulates DNA repair through homologous recombination, and its inactivation, impairs ferroptosis, cell apoptosis and other cell death mechanisms leading to the accumulation of cells with DNA damage, and eventually becoming malignant cells [15, 16]. Interestingly, the deletion of BAP1 in UM was thought to not significantly affect the DNA damage repair mechanism [17]. Therefore, proteins related to BAP1 loss in UM could be potential targets for therapy.
The tumor microenvironment (TME) is composed of tumor cells and their surroundings such as immune cells, fibroblasts, vascular endothelial cells [18]. Previous studies have shown that the interactions between cells in the TME can impact tumor initiation [19], progression and metastasis [20], as well as the responsiveness to treatment and the development of drug resistance [21, 22]. The interactions may be mediated by various mechanisms, such as receptor signaling, release of small molecules, mechanical forces, gap junctions, and tunneling nanotubes [23]. However, the precise mechanisms through which BAP1 deficiency promotes tumor promotion and regulates communication between tumor cells and TME remain unclear.
This study analyzed 115,225 single cells from 19 samples obtained from 18 UM patients. Both primary and metastatic samples were tested for the presence of a BAP1 mutation. The objective of this study was to identify the possibility of BAP1-mutation in driving UM metastasis and to discover potential molecules for the prevention or treatment of metastatic UM.
2 Materials and methods
2.1 Processing of data for scRNA-seq
Raw data were obtained from the Gene Expression Omnibus database (GSE138433 [24], GSE139829 [25] and GSE176029 [26]) (Table S1). The object was generated using the Seurat R package (version 4.0.3) and low-quality cells were filtered out. Genes with fewer than three detected cells were excluded, and low-quality cells were eliminated if the following conditions were met: (a) expressed less than 300 genes, (b) the ratio of mitochondrial genes to all expressed genes exceeds 20%, (c) the percentage of expressed genes with > 2% of hemoglobin genes. Potential doublets were distinguished and removed using the DoubletFinder package (version 2.0.3).
2.2 Data integration and dimension reduction
The “NormalizedData” function was used to normalize the filtered expression matrix, and then the “FindVariableFeatures” function was used to identify 2000 genes with high intercellular variation. The “RunPCA” function was used to perform Principal Component Analysis, and the Harmony R package (version 0.1.0) was used to remove the batch effects. Using the “FindNeighbors” and “FindClusters” functions, the first 15 principal components were then used to cluster the cells. Using the “RunUMAP” function, Uniform Manifold Approximation and Projection (UMAP) was executed on the top 15 PCs (resolution = 0.6).
2.3 Identification of the cell type
Markers for each of the identified clusters were identified using the “FindAllMarkers” function, and then the clusters were annotated based on SingleR (version 1.4.1) and well-established cellular markers.
2.4 Single-cell copy-number variation (CNV) analysis
Initial CNVs were computed for each UM tumor cell using the inferCNV R package (version 1.6.0). CNVs of UM tumor cells were calculated using T/NK cells as the reference.
2.5 Analysis of the path of CD8+ T cells
The R package Monocle2 (version 2.18.0) was utilized for pseudotime trajectory analysis to characterize the potential process of CD8+ T functional changes and establish the potential lineage differentiation.
2.6 Gene set enrichment analysis
Using the Gene Set Enrichment Analysis (GSEA), multiple pathways within distinct subclusters were analyzed based on the GO term enrichment analysis.
2.7 Comparative analysis of gene expression
The function “FindAllMarkers” was used to perform differential gene expression analysis. A difference was statistically significant when the adjusted P-value was less than 0.05.
2.8 Ligand-receptor expression and cell interactions
Cell-to-cell communication (CellChat) (version 1.1.3) R package was utilized to analyze the potential ligand-receptor pairs in cell populations.
2.9 Survival analysis
The correlation between the gene expression of UM patients and their overall survival (OS) was estimated using the GEPIA database (http://gepia.cancer-pku.cn/).
2.10 Development of BAP1-defective cells
To generate BAP1-loss cells, the CRISPR-Cas9 system was utilized. sgRNAs were designed using CRISPR-ERA (http://crispr-era.stanford.edu/) and cloned into lentiCRISPRv2 (Addgene plasmid, #52961). The sequences of BAP1 sgRNAs were 5’-CACCGTCAAATGGATCGAAGAGCGC-3’, 5’-CACCGGGATCGAAGAGCGCCGGTCC-3’, 5’-CACCGAGAGCGCCGGTCCCGGCGAA-3’ and 5’-AGAGCGCCGGTCCCGGCGAA-3’. A scrambled sgRNA (5’-GCACTCACATCGCTACATCA-3’) was used as a control. Plasmids and packaging plasmids (psPAX2 and PMD2G) are cotransfected into 293T cells. At 48 and 72 h, lentiviral particles were isolated and used to infect MUM2B cells in a medium containing 10 µg/mL polybrene. Puromycin (2 µg/mL) was used to select cells for two weeks, and the knockout (Ko) was confirmed by immunoblotting.
2.11 Transwell
Chambers (8-mm pore, Corning, USA) were precoated with a mixture of Matrigel (BD Biosciences, USA) and serum-free RPMI 1640 (GIBCO, USA) at a ratio of 1:7 followed by solidifying at 37 °C for 2 h. A total of 5 × 104 (5 × 104/100 µL) transfected cells were resuspended in serum-free culture medium and seeded in the upper chambers. The bottom chambers were filled with 550 µL of RPMI 1640 containing 30% fetal bovine serum (FBS). After 24 h, cells were stained with crystal violet and quantified. Five fields per well were counted with an inverted microscope system.
2.12 Cell lines
Cell lines used: MUM2B (established human UM cell line) and 293T (established human embryonic kidney cell line). The MUM2B was grown at 37 °C and 5% CO2 in RPMI 1640 medium (GIBCO, USA) supplemented with 10% FBS (GIBCO, USA) and 20mM HEPES buffer (Corning, USA). The 293T cell line was cultivated in Dulbecco’s modified Eagle’s medium (GIBCO, USA) supplemented with 10% FBS (GIBCO, USA) at 37 °C in a 5% CO2 atmosphere.
2.13 In vivo tumor studies
Eighteen-week-old female huHSC-NCG mice (purchased from GemPharmatech) were used (n = 6 per cell line) in this study. MUM2B cells (3 × 105) were gathered and suspended in 50 µL of cold PBS buffer. 3 × 105 cells were injected into the spleens of huHSC-NCG mice (18 weeks old). Anti-ITGB2 mAb (BioLegend, USA; 302,116; RRID:AB_2561483), anti-ICAM-1 mAb (Bio X Cell, USA; BE0020-2; RRID:AB_1107659), or an IgG isotype control (IgG2a) (Bio X Cell, USA; BE0085; RRID:AB_1107771) were injected intraperitoneally into tumor-bearing mice for 3 times at an interval of three days. The quantity of liver metastases was then analyzed by ultrasound examination (GE LOGIC™ E20, USA).
2.14 Immunoblotting
Transfected cells were lysed in RIPA lysis buffer containing protease and phosphatase inhibitors (Selleck, USA), and protein concentrations were measured with BCA according to the instruction manual. A total of 50 µg protein was used for Western blot analysis. Proteins were separated using 10% SDS-PAGE and transferred to PVDF membranes. The membranes were then incubated with antibodies against BAP1 (1:500; Santa Cruz; sc-28,383; RRID:AB_626723) and GAPDH (1:5000; Proteintech; 60004-1‐lg; RRID:AB_2107436). Finally, the blots were imaged with an image analysis system (Bio-Rad, USA).
2.15 Flow cytometry
Mice liver metastases were collected and cut into pieces (1–3 mm in diameter) and digested with collagenase II (2 mg/mL) mixed with DNase-I (1 mg/mL) for 30 mins at 37 °C. Cell suspensions were blocked and stained with antibodies as indicated at 4 °C for 30 mins protecting from light. Antibodies used in this experiment were as follows and subjected for FACS analysis: CD45 (BD Horizon, #563204), CD14 (BD Horizon, #564054), CD8 (BioLegend, #344710), CD11b (BioLegend, #301322), CD3 (BioLegend, #317318).
2.16 Bulk RNA sequencing (RNA-seq)
Liver metastases were sent to Shanghai Mingma Technologies Sequencing Company for RNA-seq.
2.17 Quantitative real-time PCR analysis
RNA from the section “Bulk RNA sequencing (RNA-seq)” were stored at -80℃ for qRT-PCR. All PCR primers used in this study were provided in Table S2. The qRT-PCR assays were performed on a real-time PCR system (Applied Biosystems, USA). The data were analyzed by using the 2−△△CT values.
2.18 Immunohistochemistry
Formalin-fixed, paraffin-embedded liver sections were baked at 65 °C, dewaxed with turpentine, rehydrated through an alcohol gradient, and immersed in peroxidase followed by blocking with goat serum to block endogenous peroxidase activity. The sections were then incubated with primary antibody VCAN (1:200, HUABIO, #ET7107-09, RRID:AB_2938780), LOX (1:100, HUABIO, #ET1706-31, RRID:AB_2938781), ENO1 (1:8000, abcam, #ab227978, RRID:AB_2827927) and HIF-2 alpha (1:2000, HUABIO, #ET7107-32; RRID:AB_2938782) at 4 °C overnight. Sections were then incubated with the secondary antibody for 30 mins and stained with 3,3′-diaminobenzidine (DAB). Images were acquired with an inverted microscope system and positive indexes were analyzed by Image J (version 2.9.0).
2.19 Quantification and statistical analysis
GraphPad Prism (version 8.0.1) was utilized for all statistical analysis, with a statistically significant difference of p < 0.05, α = 0.05 is the inspection level. To compare the signature scores of different cell groups, the Student’s t-test and one- or two‐way ANOVA was applied. In the Figure Legends and Methods, the statistical tests performed for each individual analysis are detailed.
3 Results
3.1 scRNA-seq profiling
To investigate the TME of UM at the single-cell level, we analyzed scRNA-seq data from public databases for 19 human UM samples, which were classified into primary BAP1 wide type (P-WT) (n = 4), primary BAP1-mutated (P-MUT) (n = 10), metastatic BAP1 wide type (M-WT) (n = 1), and metastatic BAP1-mutated (M-MUT) (n = 4) (Fig. 1a-d). Using the UMAP approach, we identified eight major cell types, including tumor cells (MLANA and MITF), cycling cells (STMN1, MKI67, CDK1 and TOP2A), myeloid cells (C1QB, AIF1, CD68 and CD14), endothelial cells (TM4SF1, RAMP2, PECAM1 and ENG), T/NK cells (CD3E, CD3D, CD2 and CD7), plasma cells (MZB1, IGHG1 and IGLL5), fibroblasts (COL1A2, COL3A1, COL1A1 and DCN) and B cells (CD79A, MS4A1 and BANK1) (Fig. 1B and Fig. S1a,b). The infer-copy number variants (infer CNV) analysis confirmed the presence of tumor cells (Fig. S2). The proportion of tumor cells was higher in the BAP1-WT group than in the MUT group in both primary and metastatic UM (Fig. 1e). An inflammatory phenotype characterized by massive infiltration of T cells and macrophages is associated with increased UM metastasis [27,28,29]. The proportion of T and myeloid cells was greater in BAP1-MUT than in the other groups, indicating more pronounced inflammatory characteristics in BAP1-MUTs (Fig. 1f,g). BAP1-MUTs also showed an increase of the endothelial cell ratio (Fig. 1h), which might be a key player in tumor cell migration and metastatic formation [30, 31].
scRNA-seq profiling of UM samples. a Schematic illustration for experimental design of this study. b UMAP plot showing the annotation and color codes for cell types in the UM ecosystem. c UMAP plot showing the sample origin of cells by color. d UMAP plot showing cell types by color in P-WT, P-MUT, M-WT or M-MUT. e-h Sector chart indicating the proportions of tumor cells e, T/NK cells f, myeloid cells g and endothelial cells h in each group
3.2 Augmentation of the macrophage matrix in BAP1-MUT
We further analyzed myeloid cells and their variation among groups in the TME. Myeloid cells were classified to ten distinct subgroups, including four subtypes of macrophages (Macro1-Macro4), two monocytes (Mono1-Mono2), two dendritic groups (DC1-DC2), one group of cycling, and a Myeloid-EEF1A1 subtype (Fig. 2a-c). Except for EEF1A1 cells, the immune checkpoints and immune evasion escape-related genes (LGALS9, VSIG4, SIGLEC10, CD274, PDCD1LG2) were highly expressed in the other myeloid subpopulations (Fig. 2d). The two most prevalent macrophages are pro-inflammatory M1 (CD86 and TLR4) and anti-inflammatory M2 (MSR1, HLA-DRA and CD163), which are difficult to distinguish using standard macrophage markers, and Macro3 expressed both sets of genes (Fig. 2e). CD68 and HLA-DRA were highly expressed in all subtypes (Fig. 2e). Notably, the extracellular matrix (ECM) pathway was significantly enriched in BAP1-MUT macrophages relative to their WT control, this might be a link to UM metastasis (Fig. 2f). This was supported by the highly expressed genes related to ECM in BAP1-MUTs (Fig. 2g).
Detailed characterization of myeloid cells in UM. a UMAP plot showing the subtypes of myeloid cells derived from P-WT, P-MUT, M-WT and M-MUT samples. Each cluster is color-coded according to subtype. b Bar plot illustrating the fraction of subgroups in P-WT, P-MUT, M-WT and M-MUT groups. c Heatmap showing the expression of marker genes in each subtype of myeloid cells. d Dot plot showing the percentage of cells and expression level in each cell type expressing immune checkpoint and evasion-related genes. e Violin plots representing the expression levels of M1 and M2 marker genes in macrophages. f Heatmap of GSEA scores indicating the pathways significantly enriched in BAP1-mutated groups. g Violin and box plots showing the expression variation of ECM-related genes among P-WT, P-MUT, M-WT and M-MUT groups
3.3 Clustering and state analysis of T/NK cells
Since T cells are critical for controlling tumor progression, we subsequent unsupervised clustering of T/NK cells and identified 9 clusters, including 3 subclusters of CD4+ T cells (CD4T.C1- CD4T.C3), 4 subclusters of CD8+ T cells (CD8T.C1- CD8T.C4), and 1 cluster each of NK and NKT cells (Fig. 3a). CD4+ T cells were further categorized into naive (CD4T.C3; CCR7+), helper (CD4T.C1; IL2+), and suppressive regulatory Treg (CD4T.C2; FOXP3+) CD4+ T cells. CD8+ T cells are classified as tissue-resident (CD8T.C4; CD69+), terminally exhausted (CD8T.C1; LAG3+), and cytotoxic (CD8T.C2 and CD8T.C3; GNLY+ and GZMK+) CD8+ T cells (Fig. 3a-d). These subtypes were observed in both BAP1-MUTs and WT tumors (Fig. 3c). Based on distinct transcriptional characteristics, CD8+ T cells can be separated into progenitor- and terminally-exhausted subpopulations [32]. Terminally-exhausted tumor infiltrating cells (TILs) are incapable of responding to immune checkpoint blockade therapy, whereas progenitor-exhausted TILs can respond to it. It has been reported that a BAP1-mutation can lead to chemotherapeutic resistance in metastatic UM [32, 33]. We aimed to ascertain whether a BAP1 mutation influences the progenitor- and terminally-exhausted characteristics of CD8+ TILs (Table S3). The data indicated that CD8T.C1 has the lowest progenitor exhaustion and the highest terminal exhaustion scores (Fig. 3e). CD8T.C2 and CD8T.C3 have lower terminally exhausted scores compared to CD8T.C1 and CD8T.C4 (Fig. 3e). The signature of progenitor exhausted genes was enriched in BAP1-WTs than that of in BAP1-MUTs (Fig. 3f). The trajectory analysis showed that CD8T.C3 was in an early state, CD8T.C4 and CD8T.C1 stood in the terminal, while CD8T.C2 was in transitional states (Fig. 3g). Furthermore, BAP1 mutation appears to promote a favorable niche for exhausted CD8+ T cell transformation and infiltration (Fig. 3h).
scRNA-seq revealed heterogeneity of T/NK cells in UM. a UMAP plot showing the subtypes of T/NK cells, labeled in different colors. b UMAP projection showing T/NK cells, clustered and color-coded, according to the samples. c Bar plot illustrating the proportion of T/NK subtypes among P-WT, P-MUT, M-WT and M-MUT groups. d Heatmap indicating the expression of selected gene sets in T/NK subtypes, including naive, resident, inhibitory, cytokines, co-stimulatory, transcriptional factors (TF) functions. e Violin and box plots showing the signature score distribution for progenitor exhausted (left) and terminally exhausted (right) CD8+ T cells within CD8T.C1 to CD8T.C4 subsets. f Heatmap showing the log2 fold change in the expression of the progenitor exhausted gene set of CD8T.C1 to CD8T.C4 cells in BAP1-MUT compared with BAP1-WT groups. g Pseudotime analysis of CD8+ T cells. h Pseudotime analysis of CD8+ T cells from BAP1-MUT and BAP1-WT samples
3.4 BAP1-MUT malignant cells exhibited increased cell migration
To gain a better understanding of the activities of BAP1-mutated malignant cells that may communicate with immune cells, we investigated their expression patterns and identified five clusters (Fig. 4a-c). Cluster 1 expresses high levels of factors which may linked to tumor metastasis, including GDF15, ATF3, and CDKN1A [34, 35], and its proportion in P-MUTs appears greater than in P-WTs (Fig. 4d,e). In addition, MUTs show higher GDF15 and ATF3 expression than WTs (Fig. 4f). In a TCGA UM cohort, high levels of GDF15, ATF3, and CDKN1A expression were associated with poor patient survival (Fig. 4g). Tumor metastatic-related pathways typically include angiogenesis, ECM, and positive regulation of cell migration. UM with a BAP1 mutation exhibited significantly higher levels of these signatures than BAP1-WTs in both primary and metastatic malignancies (Fig. 4h,i), as well as upregulation of less cell death in BAP1-mutated UM (Fig. S3a,b). In both two conditions, BAP1-MUT UM showed upregulation of hypoxia-related pathways, including response to decreased oxygen levels, to hypoxia and to oxygen (Fig. 4h,i and Fig. S3c,d). Given that Cluster 1 contains markers related to poor prognosis, we compared the differential pathways between Cluster 1 (GDF15hiATF3hiCDKN1Ahi) and the rest of clusters and found that Cluster 1 exhibited significantly higher migration-related signatures in both primary and metastatic conditions (Fig. S4a,b). These data together reveal that UM with a BAP1 mutation has a higher rate of tumor metastasis that may be involved with hypoxia- and ECM- related pathways.
Identification and characterization of malignant cells in UM. a UMAP projection showing the subtypes of tumor cells. b UMAP plot showing tumor cells derived from P-WT, P-MUT, M-WT and M-MUT samples. c Bar plot illustrating the average proportion of each tumor cell subtype. d Heatmap indicating the expression of selected gene sets in tumor cell subtypes. e Box plots illustrating the proportion of Cluster1 in P-WTs and P-MUTs. The significance of differences was evaluated using Student’s t-test (*p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001). f Violin plots representing the expression levels of GDF15 and ATF3 among P-WT, P-MUT, M-WT and M-MUT groups. g Kaplan-Meier plot showing the worse OS of UM patients with high expression of GDF15, ATF3, and CDKN1A. Patient sample size is 78, and the cut-off criteria for high/low gene expression was 50%. h GSEA of the angiogenesis, positive regulation of cell migration, response to hypoxia and ECM enrichment scores in tumor cells from P-MUTs compared with in P-WTs. i GSEA of the angiogenesis, positive regulation of cell migration, response to hypoxia and ECM enrichment scores in tumor cells from M-MUTs compared with in M-WTs
3.5 The interaction between tumor cells and CD8T.C1 is associated with a poor prognosis
To further elucidate the mechanisms underlying pro-metastasis of BAP1 mutation in UM, we used CellChat to analyze the cell-cell interactions. As depicted in Fig. 5a, the strength of interactions between tumor and T cells were greater in P-MUTs than in P-WTs (Fig. 5a, left panel), with CD8T.C1 showing the strongest interaction (Fig. 5a, right panel). The heatmap in Fig. 5b revealed complex networks of cell-cell interactions between cancerous and T/NK cells. We investigated several signaling pathways and found that the ITGB2 signaling was the primary source of ligands in CD8+ T cells, acting on Cluster1 malignant cells (Fig. 5b). This signaling pathway was obviously upregulated and stronger between CD8T.C1 and Cluster1 malignant cell communication in P-MUT compared with P-WT (Fig. 5c). More importantly, in this ITGB2 signaling we observed that ITGB2 and ICAM1 were the unique ligand and the only receptor, respectively (Fig. 5d). Despite the expression level of ITGB2 being stable compared to P-WT counterparts, the expression level of ICAM1 in P-MUTs T cells was higher (Fig. 5e). Similar CellChat outcomes were observed in metastasizing UMs (Fig. 5f,g). The expression level of ITGB2 in M-MUTs T cells was higher than that of M-WTs (Fig. 5h). Furthermore, higher expression of either ITGB2 or ICAM1 was associated with a poor prognosis in UM patients (Fig. 5i).
Poor prognostic indicator of ITGB2 and ICAM1 in UM patients. a Heatmap showing the differential interaction strength between indicated cell types in P-MUTs compared with in P-WTs. b Heatmap showing outgoing and incoming signaling patterns in indicated cell types in P-WTs (left) and P-MUTs (right). c Circle plots showing the varied cell-cell communication in indicated cell types between P-WTs and P-MUTs mediated by ITGB2 signaling axis. d Contribution of each L-R pair in ITGB2 signaling. e Discrepancy analysis showing upregulated (blue) and downregulated (red) genes in tumor cells in comparison with P-MUTs and P-WTs groups. f Heatmap showing outgoing and incoming signaling patterns in indicated cell types in M-WTs (left) and M-MUTs (right). g Circle plots showing the varied cell-cell communication in indicated cell types between M-WTs (left) and M-MUTs (right) mediated by ITGB2 signaling axes. h Discrepancy analysis showing upregulated (blue) and downregulated (red) genes in T cells in comparison with M-MUTs and M-WTs groups. i Kaplan-Meier plot representing OS in UM with variant ITGB2 (left) and ICAM1 (right) expression. Patient sample size is 78, and the cut-off criteria for high/low gene expression was 50%
3.6 Enhanced metastasis in BAP1-MUT UM is attenuated by blocking ICAM1 and ITGB2
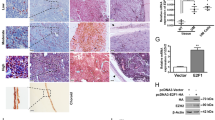
To further investigate the role of BAP1 in UM metastasis, CRISPR/Cas9 gene-editing was used to knockout (Ko) BAP1 in a BAP1-WT UM cell line, MUM2B. The results showed that the loss of BAP1 led to a significant increase in cell migration and invasion (Fig. 6a-c). Since a stronger ITGB2 signaling activation (ITGB2-ICAM1 pair) between CD8+ T and malignant cells was observed in both P-MUTs and M-MUTs groups. We next examined the effect of blocking ICAM1 and ITGB2 on UM metastasis induced by BAP1-loss, a liver metastasis mouse model was created in immune competent humanized-mice (huHSC-NCG) by injecting BAP1-deleted or -WT MUM2B cells into spleens. The liver metastatic status was monitored by using the ultrasonic scanning starting at around day 16 post cell injection. Once the liver metastasis was detected, these mice were treated with either anti-ITGB2 mAb, anti-ICAM-1 mAb, or an IgG isotype control (IgG2a) (Fig. 6d). The results showed that treatment with either anti-ITGB2 mAb or anti-ICAM-1 mAb obviously inhibited UM liver metastasis with BAP1 mutations, while no significant difference was observed between treated and untreated groups in BAP1-WT samples (Fig. 6e-g). These findings suggest that blocking ICAM1 and ITGB2 may be a promising strategy for inhibiting liver UM metastasis induced by BAP1-loss. Moreover, exhausted CD8+ T cells (PD1+CD8+) and tumor cells (HMB45+) were detected by immunofluorescence staining (Fig. 6h). To investigate the potential molecular mechanism linked to TME, we examined immune cell diversity in liver metastases via flow cytometry (Fig. 6i). Treatment with anti-ICAM1 (group MIC) increased the infiltration of T (CD45+CD3+) and CD8+ T cells (CD45+CD3+ CD8+) in BAP1-loss groups (Fig. 6j). There were decreased infiltration of myeloid cells (CD45+CD11b+) and monocytes (CD45+CD11b+CD14+) in both MIT (the BAP1- loss group treated with anti-ITGB2 mAb) and MIC group compared to controls (Fig. 6j). These data suggest that inhibition of ITGB2-ICAM1 axis may promotes more T infiltration and decreased myeloid cells.
Inhibition of UM liver metastasis by blocking ICAM1 and ITGB2. a Western blotting confirming the BAP1-Ko in MUM2B cells. b-c Transwell assay showing the invasion capability b and its quantification c of MUM2B cells with and without BAP1-Ko. Data represent the means (n = 5) ± SD. The significance of differences was evaluated using one-way ANOVA (*p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001). d Schematic of the in vivo study in immune competent humanized-mice (huHSC-NCG) model. e Representative images of liver with UM metastasis isolated from huHSC-NCG mice receiving the indicated treatments. f Representative ultrasound scanning images of each group. Boxes indicate liver metastases. g Number of UM metastases in huHSC-NCG mice receiving the indicated treatments. Data represent the means (n = 6) ± SD of each group. The significance of differences was evaluated using two‐way ANOVA (*p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001). h Immunofluorescence staining for indicated markers of mouse liver metastasis of MUT group. i Representative images of cell gating. j The percentage of various cell types in CD45+ immune cells. The p-value was calculated by one way ANOVA (*p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001). Data are presented as means (n = 5) ± SD
3.7 BAP1-MUT-induced liver metastases exhibit upregulated hypoxia and ECM signatures that can be inhibited by either anti-ITGB2 or anti-ICAM1 treatment
To gain a deeper understanding of the role of ITGB2-ICAM1 in promoting liver metastasis in BAP1-MUTs UM, we conducted bulk RNA-seq on liver metastases following antibody treatment. The data showed that 1090 and 1696 genes were upregulated and downregulated in MUT compared with WT, respectively. After treatment with anti-ITGB2 and anti-ICAM1, 1752 and 1796 genes were upregulated while 1021 and 932 genes were downregulated, respectively (Fig. 7a). Gene Ontology (GO) analysis revealed that the upregulated genes in MUT (vs. WT) were mainly involved in hypoxia- and ECM-related pathways (Fig. 7b). Following ITGB2 or ICAM1 inhibition, the above-mentioned pathways were downregulated (Fig. 7c,d). However, inhibition of ITGB2-ICAM1 axis in BAP1-WT groups had no obvious changes in these pathways (vs. WT). The varied expression of differentially expressed genes (DEGs) related to hypoxia- and ECM-related pathways were showed in Fig. 8e. We then validated the key DEGs by qRT-PCR. Consistent with the RNA-seq data, the alteration of several hypoxia-related genes (ADAM8, ANGPTL4, SLC2A1, STC2, CAV2, ENO1, EPAS1, PDK1, ETS1, NDRG1, FAM162A and HK2) and ECM-related genes (PGK1, LOXL2, LOX, SEPRINE1, COL5A1, CTSL, FAP, ITGA5, TGFBI, TGFBR1 and VCAN) were confirmed (Fig. 7f,g). In addition, the expression of hypoxia-associated proteins (ENO1 and HIF2A (EPAS1)) and ECM-related proteins (LOX and VCAN) was significantly increased in MUT-UM compared to WT-UM, while they were reduced in MIT and MIC groups (Fig. 7h,i). These results indicate that ITGB2-ICAM1 plays a key role in promoting liver metastasis in BAP1-MUTs UM partially by upregulation of hypoxia and ECM pathways (Fig. 7j).
Positive correlation of liver metastasis in UM-MUTs with hypoxia and ECM signatures. a Limma package of differential expression genes (DEGs). b-d GO analysis showing the representative positively enrichment and negatively enrichment pathways (MUT vs. WT b, MIT vs. MUT c, MIC vs. MUT d). The x- and y-axis indicate gene ratio and enriched pathways, respectively. The red number indicates the P-value. e Heatmap showing the expression levels of differentially expressed hypoxia and ECM-related genes. f-g qRT‐PCR analysis of hypoxia- f and ECM-related g genes. Data are presented as the mean (n = 3) ± SD of each group, asterisks indicate significant differences using one‐way ANOVA (*p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001). h Representative IHC staining of ENO1 and HIF2A in liver metastasis (left) and quantification of ENO1 and HIF2A index (% positive/total) (right). Data are presented as the means (n = 5) ± SD, asterisks indicate significant differences using one‐way ANOVA (*p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001) (right). i Representative IHC staining of LOX and VCAN in liver metastasis (left) and quantification of LOX and VCAN index (% positive/total) (right). Data are presented as the means (n = 5) ± SD, asterisks indicate significant differences using one‐way ANOVA (*p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001). j Schematic diagram of the mechanism of ITGB2-ICAM1driving UM metastasis
4 Discussion
Recent advances in understanding tumor biology and TME have improved cancer prognostication. However, the clinical outcomes of UM therapy remain unsatisfactory due to its high metastasis rate [36]. As of April 2023, no systematic treatment has been approved for patients with metastatic UM, resulting a dismal prognosis [36].
Loss of BAP1 has been linked to UM metastasis, it is still unclear how BAP1-loss in UM contributes to metastasis [17]. To gain insight into this, we analyzed scRNA-seq data from 19 UM tissues, including primary and metastatic UM tissues with and without abnormal BAP1. Malignant cells in BAP1-MUT UM significantly showed upregulation of metastasis-related pathways, including angiogenesis, cell adhesion, ECM, and hypoxia-related pathways (i.e., response to hypoxia and oxygen level variation). We also observed a receptor-ligand pair (ITGB2-ICAM1) between terminally exhausted CD8+ T cells and tumor cells. To further investigate the role of ITGB2-ICAM1 in promoting UM metastasis, we generated an immune competent humanized-mouse liver metastasis model. Our findings revealed that blocking either ICAM1 or ITGB2 effectively inhibited UM metastasis, which may be attributed to reduced hypoxia- and ECM- related signals.
Tumor metastasis can be categorized into four stages, including infiltration, circulation, extravasation, and colonization [37]. Tumor colonization is facilitated by the adhesion of cancer cells to the ECM and to each other, which allow them to acquire increased viability, proliferative capacity, stemness and build supportive niche [38]. The ECM serves as a scaffold for tumor growth and amplifies carcinogenic signals [39]. Previous studies have indicated that ECM remodeling promoted UM liver colonization [37, 40]. Hypoxic UM tumors have been associated with aggressive phenotypes, an increased risk of tumor metastasis and unfavorable clinical outcomes [41]. Furthermore, BAP1 somatic point mutations were enriched in hypoxic UM tumors, whereas in normoxic UM tumors more EIF1AX and SF3B1 mutations were observed [41]. Glycolysis has also been reported as a risk factor for worse OS in UM patients [42].
5 Conclusions
This study suggests that activation of ITGB2-ICAM1 axis plays a crucial role in promoting metastasis in BAP1-mutated UM. These findings could aid in the development of new therapeutics aimed at reducing UM metastasis. To support the clinical application of ITGB2 and ICAM1 antagonists, the safety must be tested in vivo, including evaluating their toxicology, metabolism and impact on immune evasion or activation.
Data Availability
Sequence data that support the findings of this study have been deposited in NCBI database with the primary accession code PRJNA975671.
References
N. Amirouchene-Angelozzi, M. Schoumacher, M.H. Stern, N. Cassoux, L. Desjardins, S. Piperno-Neumann, O. Lantz, S. Roman-Roman, Br. J. Cancer. 113, 1249–1253 (2015). https://doi.org/10.1038/bjc.2015.269
W. Shu, X. Zhu, K. Wang, S. Cherepanoff, R.M. Conway, M.C. Madigan, H. Zhu, L. Zhu, M. Murray, F. Zhou, Cell. Oncol. (Dordr). 45, 601–619 (2022). https://doi.org/10.1007/s13402-022-00686-5
C. Pandiani, G.E. Beranger, J. Leclerc, R. Ballotti, C. Bertolotto, Genes Dev. 31, 724–743 (2017). https://doi.org/10.1101/gad.296962.117
J. Yang, D.K. Manson, B.P. Marr, R.D. Carvajal, Ther. Adv. Med. Oncol. 10, 1758834018757175 (2018). https://doi.org/10.1177/1758834018757175
M.D. Onken, L.A. Worley, J.P. Ehlers, J.W. Harbour, Cancer Res. 64, 7205–7209 (2004). https://doi.org/10.1158/0008-5472.CAN-04-1750
K.N. Smit, M.J. Jager, A. de Klein, E. Kili, Prog Retin Eye Res. 75, 100800 (2020). https://doi.org/10.1016/j.preteyeres.2019.100800
J.W. Harbour, M.D. Onken, E.D. Roberson, S. Duan, L. Cao, L.A. Worley, M.L. Council, K.A. Matatall, C. Helms, A.M. Bowcock, Science. 330, 1410–1413 (2010). https://doi.org/10.1126/science.1194472
T. Wiesner, A.C. Obenauf, R. Murali, I. Fried, K.G. Griewank, P. Ulz, C. Windpassinger, W. Wackernagel, S. Loy, I. Wolf, A. Viale, A.E. Lash, M. Pirun, N.D. Socci, A. Rutten, G. Palmedo, D. Abramson, K. Offit, A. Ott, J.C. Becker, L. Cerroni, H. Kutzner, B.C. Bastian, M.R. Speicher, Nat. Genet. 43, 1018–1021 (2011). https://doi.org/10.1038/ng.910
G.M. Shankar, M. Abedalthagafi, R.A. Vaubel, P.H. Merrill, N. Nayyar, C.M. Gill, R. Brewster, W.L. Bi, P.K. Agarwalla, A.R. Thorner, D.A. Reardon, O. Al-Mefty, P.Y. Wen, B.M. Alexander, P. van Hummelen, T.T. Batchelor, K.L. Ligon, A.H. Ligon, M. Meyerson, I.F. Dunn, R. Beroukhim, D.N. Louis, A. Perry, S.L. Carter, C. Giannini, W.T. Jr. Curry, D.P. Cahill, F.G. 2 Barker nd, P.K. Brastianos, S. Santagata, Neuro Oncol. 19, 535–545 (2017). https://doi.org/10.1093/neuonc/now235
J.R. Testa, M. Cheung, J. Pei, J.E. Below, Y. Tan, E. Sementino, N.J. Cox, A.U. Dogan, H.I. Pass, S. Trusa, M. Hesdorffer, M. Nasu, A. Powers, Z. Rivera, S. Comertpay, M. Tanji, G. Gaudino, H. Yang, M. Carbone, Nat. Genet. 43, 1022–1025 (2011). https://doi.org/10.1038/ng.912
M. Bott, M. Brevet, B.S. Taylor, S. Shimizu, T. Ito, L. Wang, J. Creaney, R.A. Lake, M.F. Zakowski, B. Reva, C. Sander, R. Delsite, S. Powell, Q. Zhou, R. Shen, A. Olshen, V. Rusch, M. Ladanyi, Nat. Genet. 43, 668–672 (2011). https://doi.org/10.1038/ng.855
Y. Jiao, T.M. Pawlik, R.A. Anders, F.M. Selaru, M.M. Streppel, D.J. Lucas, N. Niknafs, V.B. Guthrie, A. Maitra, P. Argani, G.J.A. Offerhaus, J.C. Roa, L.R. Roberts, G.J. Gores, I. Popescu, S.T. Alexandrescu, S. Dima, M. Fassan, M. Simbolo, A. Mafficini, P. Capelli, R.T. Lawlor, A. Ruzzenente, A. Guglielmi, G. Tortora, F. de Braud, A. Scarpa, W. Jarnagin, D. Klimstra, R. Karchin, V.E. Velculescu, R.H. Hruban, B. Vogelstein, K.W. Kinzler, N. Papadopoulos, L.D. Wood, Nat. Genet. 45, 1470–1473 (2013). https://doi.org/10.1038/ng.2813
T. Popova, L. Hebert, V. Jacquemin, S. Gad, V. Caux-Moncoutier, C. Dubois-d’Enghien, B. Richaudeau, X. Renaudin, J. Sellers, A. Nicolas, X. Sastre-Garau, L. Desjardins, G. Gyapay, V. Raynal, O.M. Sinilnikova, N. Andrieu, E. Manie, A. de Pauw, P. Gesta, V. Bonadona, C.M. Maugard, C. Penet, M.F. Avril, E. Barillot, O. Cabaret, O. Delattre, S. Richard, O. Caron, M. Benfodda, H.H. Hu, N. Soufir, B. Bressac-de, D. Paillerets, Stoppa-Lyonnet, M.H. Stern, Am. J. Hum. Genet. 92, 974–980 (2013). https://doi.org/10.1016/j.ajhg.2013.04.012
S. Pena-Llopis, S. Vega-Rubin-de-Celis, A. Liao, N. Leng, A. Pavia-Jimenez, S. Wang, T. Yamasaki, L. Zhrebker, S. Sivanand, P. Spence, L. Kinch, T. Hambuch, S. Jain, Y. Lotan, V. Margulis, A.I. Sagalowsky, P.B. Summerour, W. Kabbani, S.W. Wong, N. Grishin, M. Laurent, X.J. Xie, C.D. Haudenschild, M.T. Ross, D.R. Bentley, P. Kapur, J. Brugarolas, Nat. Genet. 44, 751–759 (2012). https://doi.org/10.1038/ng.2323
A. Bononi, C. Giorgi, S. Patergnani, D. Larson, K. Verbruggen, M. Tanji, L. Pellegrini, V. Signorato, F. Olivetto, S. Pastorino, M. Nasu, A. Napolitano, G. Gaudino, P. Morris, G. Sakamoto, L.K. Ferris, A. Danese, A. Raimondi, C. Tacchetti, S. Kuchay, H.I. Pass, E.B. Affar, H. Yang, P. Pinton, M. Carbone, Nature. 546, 549–553 (2017). https://doi.org/10.1038/nature22798
Y. Zhang, J. Shi, X. Liu, L. Feng, Z. Gong, P. Koppula, K. Sirohi, X. Li, Y. Wei, H. Lee, L. Zhuang, G. Chen, Z.D. Xiao, M.C. Hung, J. Chen, P. Huang, W. Li, B. Gan, Nat. Cell. Biol. 20, 1181–1192 (2018). https://doi.org/10.1038/s41556-018-0178-0
K.N. Smit, M.J. Jager, A. de Klein, E. Kiliҫ, Prog Retin Eye Res. 75, 100800 (2020). https://doi.org/10.1016/j.preteyeres.2019.100800
C.G. Ioannides, T.L. Whiteside, Clin. Immunol. Immunopathol. 66, 91–106 (1993). https://doi.org/10.1006/clin.1993.1012
F.R. Greten, S.I. Grivennikov, Immunity. 51, 27–41 (2019). https://doi.org/10.1016/j.immuni.2019.06.025
D.F. Quail, J.A. Joyce, Nat. Med. 19, 1423–1437 (2013). https://doi.org/10.1038/nm.3394
J.M. Pitt, A. Marabelle, A. Eggermont, J.C. Soria, G. Kroemer, L. Zitvogel, Ann. Oncol. 27, 1482–1492 (2016). https://doi.org/10.1093/annonc/mdw168
O. Tredan, C.M. Galmarini, K. Patel, I.F. Tannock, J. Natl. Cancer Inst. 99, 1441–1454 (2007). https://doi.org/10.1093/jnci/djm135
M. Li, P. Jiang, S. Wei, J. Wang, C. Li, Front. Immunol. 14, 1113312 (2023). https://doi.org/10.3389/fimmu.2023.1113312
C. Pandiani, T. Strub, N. Nottet, Y. Cheli, G. Gambi, K. Bille, C. Husser, M. Dalmasso, G. Beranger, S. Lassalle, V. Magnone, F. Pedeutour, M. Irondelle, C. Maschi, S. Nahon-Esteve, A. Martel, J.P. Caujolle, P. Hofman, K. LeBrigand, I. Davidson, S. Baillif, P. Barbry, R. Ballotti, C. Bertolotto, Cell. Death Differ. 28, 1990–2000 (2021). https://doi.org/10.1038/s41418-020-00730-7
M.A. Durante, D.A. Rodriguez, S. Kurtenbach, J.N. Kuznetsov, M.I. Sanchez, C.L. Decatur, H. Snyder, L.G. Feun, A.S. Livingstone, J.W. Harbour, Nat. Commun. 11, 496 (2020). https://doi.org/10.1038/s41467-019-14256-1
W. Lin, A.B. Beasley, N.M. Ardakani, E. Denisenko, L. Calapre, M. Jones, B.A. Wood, L. Warburton, A.R.R. Forrest, E.S. Gray, Cold Spring Harb Mol Case Stud. 7 (2021). https://doi.org/10.1101/mcs.a006111
Z. Souri, A.P.A. Wierenga, C. van Weeghel, P.A. van der Velden, W.G.M. Kroes, G.P.M. Luyten, S.H. van der Burg, A.G. Jochemsen, M.J. Jager, Cancers (Basel). 11 (2019). https://doi.org/10.3390/cancers11081102
C. Ericsson, S. Seregard, A. Bartolazzi, E. Levitskaya, S. Ferrone, R. Kiessling, O. Larsson, Invest. Ophthalmol. Vis. Sci. 42, 2153–2156 (2001)
M.J. Jager, H.M. Hurks, J. Levitskaya, R. Kiessling, Hum. Immunol. 63, 444–451 (2002). https://doi.org/10.1016/s0198-8859(02)00389-0
F. De Sanctis, S. Ugel, J. Facciponte, A. Facciabene, Semin Immunol. 35, 35–47 (2018). https://doi.org/10.1016/j.smim.2018.02.002
S. Soleimani, M. Shamsi, M.A. Ghazani, H.P. Modarres, K.P. Valente, M. Saghafian, M.M. Ashani, M. Akbari, A. Sanati-Nezhad, Biotechnol. Adv. 36, 880–893 (2018). https://doi.org/10.1016/j.biotechadv.2018.01.013
B.C. Miller, D.R. Sen, R. Al Abosy, K. Bi, Y.V. Virkud, M.W. LaFleur, K.B. Yates, A. Lako, K. Felt, G.S. Naik, M. Manos, E. Gjini, J.R. Kuchroo, J.J. Ishizuka, J.L. Collier, G.K. Griffin, S. Maleri, D.E. Comstock, S.A. Weiss, F.D. Brown, A. Panda, M.D. Zimmer, R.T. Manguso, F.S. Hodi, S.J. Rodig, A.H. Sharpe, W.N. Haining, Nat. Immunol. 20, 326–336 (2019). https://doi.org/10.1038/s41590-019-0312-6
L. Ny, H. Jespersen, J. Karlsson, S. Alsen, S. Filges, C. All-Eriksson, B. Andersson, A. Carneiro, H. Helgadottir, M. Levin, I. Ljuslinder, R. Olofsson Bagge, V.R. Sah, U. Stierner, A. Stahlberg, G. Ullenhag, L.M. Nilsson, J.A. Nilsson, Nat. Commun. 12, 5155 (2021). https://doi.org/10.1038/s41467-021-25332-w
R. Folberg, Z. Arbieva, J. Moses, A. Hayee, T. Sandal, S. Kadkol, A.Y. Lin, K. Valyi-Nagy, S. Setty, L. Leach, P. Chevez-Barrios, P. Larsen, D. Majumdar, J. Pe’er and, A.J. Maniotis, Am J Pathol 69, 1376–1389 (2006) https://doi.org/10.2353/ajpath.2006.060223
Z. Lin, D. Susskind, Cancers (Basel). 13 (2021). https://doi.org/10.3390/cancers13102464
R.D. Carvajal, J.J. Sacco, M.J. Jager, D.J. Eschelman, R. Olofsson Bagge, J.W. Harbour, N.D. Chieng, S.P. Patel, A.M. Joshua, S. Piperno-Neumann, Nat. Rev. Clin. Oncol. 20, 99–115 (2023). https://doi.org/10.1038/s41571-022-00714-1
W. Dai, S. Liu, S. Wang, L. Zhao, X. Yang, J. Zhou, Y. Wang, J. Zhang, P. Zhang, K. Ding, Y. Li, J. Pan, Signal. Transduct. Target. Ther. 6, 176 (2021). https://doi.org/10.1038/s41392-021-00563-x
J.A. Eble, S. Niland, Clin. Exp. Metastasis. 36, 171–198 (2019). https://doi.org/10.1007/s10585-019-09966-1
F. Gattazzo, A. Urciuolo, P. Bonaldo, Biochim. Biophys. Acta. 1840, 2506–2519 (2014). https://doi.org/10.1016/j.bbagen.2014.01.010
M. Berube, M. Talbot, C. Collin, C. Paquet-Bouchard, L. Germain, S.L. Guerin, E. Petitclerc, Int. J. Oncol. 26, 405–413 (2005)
J. Zhao, Q. Yi, K. Li, L. Chen, L. Dai, J. Feng, Y. Li, M. Zhou, J. Sun, Comput. Struct. Biotechnol. J. 20, 3182–3194 (2022). https://doi.org/10.1016/j.csbj.2022.06.034
J. Liu, J. Lu, W. Li, Front. Immunol. 12, 738068 (2021). https://doi.org/10.3389/fimmu.2021.738068
Acknowledgements
We are grateful to Prof. Hong Tu and Dr. Qian Li for their assistance in generating the liver metastatic mouse model. The work was supported by grants from National Natural Science Foundation of China (32170924), Science and Technology Commission of Shanghai Municipality (21140901600, 23ZR1438400), Shanghai Jiaotong University School of Medicine (02.101005.001.29.38 A), State Key Laboratory of Oncogenes and Related Genes (SB21-22, ZZ-21-13rcpy, ZZ-GCYJ-23-02, ZZ-94-2313), China Postdoctoral Science Foundation (2022M722115).
Author information
Authors and Affiliations
Contributions
J.L., S.S., Y.Z., A.Z. and D.X. designed and performed experiments; J.L., D.C., L.J., and Y.Z. analyzed the data; J.L. and D.X. wrote and edited the manuscript; D.X. supervised the study, obtained funding, and provided critical review. All authors approved the final version of the manuscript.
Corresponding authors
Ethics declarations
Ethics approval
Animal experiments were performed in accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals and relevant Chinese laws and regulations. The protocol was approved by the Institutional Animal Care and Use Committee (IACUC) of Shanghai Jiao Tong University, the Animal Protocol number is RJ2022-1010. Ethical approval was obtained from the Research Ethics Committee of Renji Hospital, School of Medicine, Shanghai Jiao Tong University.
Competing interests
The authors Dongxi Xiang, Jiaoduan Li, Lixin Jiang have submitted a patent (Application NO: 202310685260.6) related to this work. Dongxi Xiang is an editorial board member. Siyuan Shao is an employee of Shanghai OneTar Biomedicine.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Li, J., Cao, D., Jiang, L. et al. ITGB2-ICAM1 axis promotes liver metastasis in BAP1-mutated uveal melanoma with retained hypoxia and ECM signatures. Cell Oncol. 47, 951–965 (2024). https://doi.org/10.1007/s13402-023-00908-4
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13402-023-00908-4