Abstract
Herpes simplex virus (HSV) is a double-stranded DNA virus that can cause lytic infections in epithelial cells of the skin and latent infections in neuronal cells of the peripheral nervous system. After virion attachment to the cell membrane, the capsid enters the cytoplasm and is transported to the nucleus. Following docking at the nuclear pore, the HSV DNA, and contents of the virion, are injected into the nucleus. The viral DNA that enters the nucleus is devoid of histones, but begins to be covered with them soon after entry. The covering of histones, in the form of nucleosomes, reaches a maximum during the early stages of infection and drops off during late infection (after DNA replication). However, during latency, the genome is saturated with nucleosomes. In this study, we examine the role of proliferating cell nuclear antigen (PCNA), a cellular DNA polymerase accessory protein (processivity factor), and cell DNA polymerases in histone deposition during the early stages of HSV infection. Using SiRNA knockdown, and a cytosine arabinoside (araC) chemical inhibitor, we conclude that PCNA is important for viral replication and histone deposition. However, cell DNA polymerases that bind PCNA do not appear to be required for these processes and PCNA does not appear to bind to the viral DNA polymerase (which has its own viral processivity factor).
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
HSV-1 is a large (152 bp) dsDNA virus capable of establishing latent infections in the sensory ganglia of humans. This latent infection lasts for the lifetime of individuals and can be reactivated periodically, in some people, to cause recurrent disease (for review of HSV, see Roizman et al. 2013).
At the start of the lytic cycle, the linear HSV-1 genome circularizes upon entering a host cell nucleus through the nuclear pore and starts transcription of its Immediate Early (IE) class of genes. Another event that occurs at this time is the deposition of nucleosomes on the viral genome (Kent et al. 2004), presumably replacing the polyamine found associated with the viral DNA in the virion capsid (Gibson and Roizman 1971). It has been shown that there is a relationship between HSV-1 replication and the infected cell replication cycle (Schang et al. 1996), supporting the hypothesis that cellular proteins are required in the gene expression and/or genome replication processes.
The viral genome encodes seven essential proteins for its own replication: the origin binding protein (UL9), a DNA polymerase with a processivity factor (UL30 and UL42), a heterotrimeric helicase-primase complex (UL5, UL8, and UL52), and a DNA binding protein ICP8 (UL29) (Olivo et al. 1989). Among the cellular proteins that might be involved in the HSV-1 replication cycle is proliferating cell nuclear antigen (PCNA). PCNA is a eukaryotic DNA replication accessory protein, also known as a DNA processivity factor (Oku et al. 1998). The complex of PCNA, pol d, and replication factor C (RF-C) are needed for leading strand DNA synthesis following initiation by DNA polymerase a (pol a) and for completion of lagging strand DNA synthesis during elongation. In these processes, PCNA functions as a DNA sliding clamp to facilitate the link between primer-ends and pol d. In addition to its function in cellular replication, PCNA interacts with multiple factors related to DNA repair and cell cycle control (Paunesku et al. 2001; Ulrich and Takahashi 2013). PCNA’s involvement in DNA repair includes recruitment of the chromatin assembly factors CAF-1 and Asf1, which are responsible for depositing new nucleosomes in the DNA regions devoid of them, at the DNA repair site (Huertas et al. 2009). PCNA is a toroidal molecule with a central channel large enough to fit double-stranded DNA without directly interacting with it, and it requires RF-C to be loaded onto the primer/template junction in an ATP-dependent process (Zuccola et al. 2000).
HSV-1 has its own processivity factor, similar in DNA replication function to PCNA, encoded by the UL42 gene (Zuccola et al. 2000). UL42 has similar crystal structure and “processivity fold” but shares no sequence homology or biochemical properties with PCNA. Because UL42 and PCNA share functional and structural homology, they may have a common ancestor (Zuccola et al. 2000). UL42 forms a complex with the HSV DNA polymerase UL30 and directly binds to the HSV DNA without requiring ATP (Zuccola et al. 2000). Some authors believe that UL42 can slide on DNA despite its high affinity, and this involves conformational changes resulting in a temporarily decreased DNA-binding affinity, allowing the polymerase to move along the template; others postulate that UL42 is held in close proximity to DNA via strong electrostatic interaction and that the addition of polymerase gives both motive force and a directionality to the movement (Zuccola et al. 2000).
Conflicting information has been reported about the involvement of PCNA in the HSV-1 replication process. Some authors claimed that the HSV-1 replisome is functional in the absence of PCNA but dependent upon UL42 (Muylaert and Elias 2010). Others state that PCNA co-localizes with HSV replication proteins (Wilcock and Lane 1991) and ICP34.5 (Harland et al. 2003), suggesting its functional importance in viral replication. The latter research group showed that ICP34.5 forms a DNA-binding complex with PCNA and HSV replication proteins. Harland et al. (2003) also postulated that PCNA is recruited to aid in DNA repair at the early stage of HSV infection and that ICP34.5 helps to de-regulate the PCNA into replication mode at the later stage of HSV infection. They propose that ICP34.5 is a regulatory factor for PCNA and is required to subvert the cellular replication machinery to replicate HSV DNA. ICP34.5 is important for HSV growth in non-dividing cells, such as neurons of the nervous system, and ICP34.5 null virus has been shown to be avirulent (Chou et al. 1990; Thompson et al. 1983; Ward et al. 2003).
At the earliest stages in HSV replication, when the virion capsid docks at the nuclear pore, and the viral genome enters the cell nucleus, it is in the form of a linear double-stranded molecule that is devoid of nucleosomes (Gibson and Roizman 1971; Oh and Fraser 2008; Pignatti and Cassai 1980). It has been shown that this genome has single-stranded breaks (Wilkie 1973; Wilkinson and Weller 2004). During early times in the nucleus, the genome has the single-stranded breaks repaired, is circularized, and coated with nucleosomes (Garber et al. 1993; Poffenberg and Roizman 1985). Although a regular pattern of nucleosomes can be detected on latent DNA, HSV DNA from lytic infections appears to have an irregular pattern of distribution (Leinbach and Summers 1980; Muggeridge and Fraser 1986). The density of nucleosomes on lytic HSV DNA has been shown to be less than that on cellular DNA (Kutluay and Triezenberg 2009a). Furthermore, later in the infectious cycle, when newly replicated viral DNA is synthesized, nucleosomes do not appear to accumulate on the newly replicated and packaged viral DNA (Oh and Fraser 2008). The nucleosomes on HSV DNA contain modifications found on cellular nucleosomes. These modifications are believed to play a role in gene regulation (for review, see Knipe and Cliffe 2008; Kutluay and Triezenberg 2009b; Lu and Triezenberg 2010). Currently, the mechanism of nucleosome deposition on HSV DNA, and whether this process involves cellular, viral, or both types of protein, is not known.
In this study, we investigate the role of PCNA in the early stages of HSV infection and conclude that it has a role in early nucleosome deposition and efficient viral replication.
Materials and methods
Cell, viruses, and PCNA inhibitors
All experiments were performed in Vero cells grown in Dulbecco’s modified Eagle’s medium (DMEM) containing penicillin, streptomycin, and 5 % calf serum. For infections, we used HSV strain 1716 and wild-type 17+ viruses. For PCNA inhibition, we used siGENOME SMART pool, Human PCNA (Thermo Scientific Dharmacon), and cytosine β-d-arabinofuranoside (Sigma).
PCNA inhibition
Subconfluent monolayers (5 × 105cells) in six-well plastic tissue culture plates were transfected with siGENOME SMART pool Human PCNA using GenCarrier (Epoch Biolabs, Inc.) and OptiMem (Gibco). As a negative control, we used Silencer FAM Labeled Negative control siRNA (Ambion). After 5 h, OptiMem was removed and fresh DMEM media, containing penicillin, streptomycin, and 5 % calf serum, was added. The plates were incubated for 72 h at 37 °C.
For chemical inhibition, cytosine β-d-arabinofuranoside was added to subconfluent monolayers (5 × 105cells) in six-well plastic tissue culture plates containing (DMEM) media with penicillin, streptomycin, and 5 % calf serum. The plates were incubated for 72 h at 37 °C.
After 72 h of inhibition, the monolayers were infected with 1716 and 17+ viruses at MOI 5. Cells were scrapped off at 0 (mock infection), 1, 4, and 8 h post-infection and frozen at −70 °C. Each sample was divided into three parts to be used for viral titration, DNA extraction, and MNase digestion.
Viral titration
Frozen samples were thawed and frozen three times and centrifuged (1000×g, 10 min, 4 °C) to get rid of cell debris. A serial dilution for each sample was made in DMEM media containing penicillin and streptomycin. Confluent Vero cells in 48-well plastic tissue culture plates were infected with 100 μl of the diluted samples in triplicates and incubated at 37 °C for 1 h. After 1 h, 300 μl of DMEM media containing penicillin, streptomycin, and 5 % calf serum was added to each well and plates were incubated overnight at 37 °C. Cells were fixed using a methanol/acetone (2:1) mixture and dried. The next day, 48-well plates were washed with PBS/1 % BSA, incubated with 100 μl of anti-HSV antibodies for 1 h, washed with PBS/1 % BSA, incubated with 100 μl of goat anti-rabbit biotinylated antibodies for 1 h, washed with PBS/1 % BSA, incubated with ABC substrate (Vector Laboratories) for 1 h, and washed with PBS. Plaques were visualized using Vector VIP kit (Vector Laboratories) and counted.
DNA extraction
Cells were thawed and centrifuged for 5 min at 1700 rpm at 4 °C. The pellets were incubated in lysis buffer (in 5 ml = 50 μl Tris, pH 8, 100 μl 0.5 M EDTA, 150 μl 20 % SDS, 82 μl Proteinase K) overnight at 37 °C. DNA was extracted using phenol/chloroform method followed by ethanol precipitation. DNA was re-suspended in water.
Micrococcal nuclease (MNase) digestion
Cells were thawed and re-suspended in RSB buffer (10 mM Tris, pH 7.5, 10 mM NaCl, 3 mM MgCl2). The membranes were disrupted using 1.0 % NP40 made in RSB buffer. Samples were underlayed with an equal volume of 0.33 M sucrose made in RSB buffer and centrifuged for 10 min at 1700 rpm at 4 °C. Precipitated nuclei were re-suspended in MNase buffer with 0.1 mM PMSF (Spiker et al. 1983). Mnase was added (5 U/μl for complete digestion) for 30 min at room temperature for 200–500 μg of DNA. The reaction was stopped by adding 0.5 M EDTA at a final concentration of 12 mM on ice. Proteinase K at a final concentration of 100 μg/ml was added to all samples, which were then incubated at 37 °C overnight. DNA was purified using Wizard DNA clean-up kit (Promega) and re-suspended in water. After measuring all concentrations, samples were run on 2 % agarose gel. Bands corresponding to 150 bp size were cut from the gel and DNA was purified using Gel Extraction Kit (Qiagen). DNA was re-suspended in water.
RT-PCR detection
RT-PCR was performed in 10-μl volumes containing 10 μg of DNA, 5 μl of SYBR green (Applied Biosystems), and 1 μl of forward and reverse TK, ICP0, VP16, and GAPDH primers. GAPDH F: ATCTCTGCCCCCTCTGCTG; GADPH R: ATGGTTCACACCCATGACGA; VP16 F: GCCGCCCCGTACCTCGTGAC; VP16 R: CAGCCCGCTCCGCTTGTCG; TK F: GTATGATGACACAAACCCCG; TK R: GAGTTTCACGCCACCAAGAT.
Reactions were run on an Applied Biosystems StepOne system.
Immunoprecipitation (ChIP)
Confluent monolayers (1.75 × 107) in T75 plastic tissue culture plates were infected with 17+ HSV-1 virus at MOI 5. Before harvesting cells at 0 (mock), 1, 4, and 8 hpi, cells were cross-linked using 1 % HCHO for 15 min at RT; this reaction was stopped by adding glycin at a final concentration of 0.125 M for 5 min. Later, cells were washed twice with cold 1× PBS and collected by centrifugation at 2500 rpm for 15 min. First, cells were lysed by ChIP buffer I (0.25 % Triton X-100, 10 mM EDTA, 0.5 mM EGTA, 10 mM Tris pH 8.0, 1 mM PMSF) by incubating on a rotator for 15 min at RT. Cells were collected by centrifugation at 2500 rpm for 15 min. Second, cells were lysed by ChIP buffer II (10 mM Tris pH 8.0, 200 mM NaCl, 10 mM EDTA, 0.5 mM EGTA, 1 mM PMSF) and collected by centrifugation at 2500 rpm for 15 min. Finally, the pellets were re-suspended in IP buffer (20 mM Tris pH 8.0, 200 mM NaCl, 0.5 % Triton X-100, 0.05 % DOC-Na, 0.5 % NP40, 1 mM PMSF). The mixture was sonicated to obtain the size fragment 250–350 bp. The insoluble matter was discarded by centrifugation. Ten percent of the mixtures were saved as the inputs and the rest was divided into two parts: one was incubated with IgG antibodies (Santa Cruz Biotechnology) and the other was incubated with anti-PCNA or anti-DNA polymerase antibodies (Santa Cruz Biotechnology), overnight. Next day, the samples were incubated with protein A agarose beads and washed with a series of buffers: IP buffer (three times), IP buffer + 0.5 M NaCl (one time), LiCl (one time), and TE + PMSF (two times). The DNA-protein complexes were eluted from beads at 65 °C. The supernatants (and inputs) were cross-linked by heat at the presence of 200 mM NaCl overnight. Next day, all samples were treated with Proteinase K and DNA was purified using phenol-chloroform extraction and precipitated using ethanol. DNA was re-suspended in water and was used to run RT-PCRs.
Southern blot and radioactive labeling
Ten micrograms of DNA, with and without siRNA treatment, obtained from the MNase digestion experiment was subjected to agarose gel electrophoresis. The gel was incubated for 30 min in 1.5 M NaCl, 0.5 M NaOH to denature the DNA, followed by 2 × 20 min in 1.5 M NaCl, 0.5 M Tris–Cl to neutralize the gel. The transfer was done overnight. Next day, the membrane was UV cross-linked in UV Stratalinker 2400 and pre-incubated in Rapid-hyb buffer (GE Healthcare) for 2 h. We used five cosmids covering the entire HSV-1 genome as probes (Cunningham and Davison 1993), labeled using the nick translation technique. In brief, 250 ng of DNA, 5 μl dNTPs, and 5 μl 32P-dCTP were mixed to obtain a final volume of 45 μl, and 5 μl of Pol I/DNase was added to the mix. The mix was incubated at 15 °C for 1 h; the reaction was stopped by adding 5 μl of Stop buffer. The unincorporated nucleotides were purified from the mix using Quick Spin Columns for radiolabeled DNA purification (Roche); the incorporation was about 25 %. The probe was boiled for 5 min and added to the hybridization solution and membrane. The membrane was incubated with the radioactive probe for 2 h at 62 °C. After hybridization, the membrane was washed, using the following series of buffers: 2× SSC + 0.5 % SDS for 10 min at room temperature, 2× SSC + 0.1 % SDS for 20 min at room temperature, and 0.1× SSC + 0.1 % SDS for 1 h (×2) at 62 °C (SSC = 150 mM NaCl, 15 mM Na citrate, pH = 7.0). The membrane was placed in a phosphorus screen overnight and scanned using a Typhoon instrument (Amersham).
Results
PCNA inhibition affects HSV DNA synthesis
In order to inhibit PCNA, we used two types of treatment: siRNA (Gehen et al. 2007) and a chemical agent cytosine arabinoside (araC) (Marchion et al. 2003). At the doses used, the effects of both inhibitors on cell viability were not significant—as determined by counting live cell numbers (Fig. 1). The number of live cells was measured prior to, and at 24, 48, and 72 h post-siRNA transfection, drug treatment, to confirm that all experiments were performed using the same number of cells and to test for toxicity. The data show that araC treatment is slightly more toxic than siRNA treatment, which does not appear to be toxic to the Vero cells (Fig. 1).
SiRNA and araC treatment does not affect cell growth. Subconfluent Vero cell monolayers were transfected with siGENOME SMART pool Human PCNA using GenCarrier, and GenCarrier alone and treated with araC. Cells were harvested prior to treatment and at 1, 2, 3, and 4 days post-treatment. After dilution with Trypan Blue dye, the number of cells was counted using TC10 Automatic Cell Counter. This experiment was repeated three times with similar results. a With and without siRNA treatment. b With and without araC treatment
To determine the effect of PCNA inhibition on HSV-1 replication, we examined the replication of HSV-17+ before and after PCNA inhibition using siRNAPCNA or araC treatment, by analyzing plaque formation using an immunoperoxidase-based assay (Fig. 2a). The results show that the titer of HSV-1 was decreased by 3 logs at 20 h PI.
AraC or SiRNAPCNA treatment affects HSV replication. Vero cells were incubated with and without araC or transfected with siGENOME SMART pool Human PCNA and incubated for 72 h followed by infection with 17+ virus at a MOI of 5. a Cells were harvested at 0 (mock infection), 1, 4, 8, and 20 h post-infection and each sample was titered in triplicate on Vero cells using an immunoperoxidase-based assay. Each data point represents the mean ± standard deviation of three independent triplicate determinations. b, c Cells were harvested at 0 (mock infection), 1, 4, and 8 h post-infection and the genomic DNA extracted. Quantitative real-time PCR was used to determine the level of HSV and cell DNA. Each data point represents the mean ± standard deviation of three independent triplicate determinations. b 17+ virus before and after araC. c 17+ virus before and after siRNAPCNA
To confirm the inhibition of HSV DNA synthesis after PCNA inhibition, as determined by plaque assay, we performed RT-PCR for nuclear HSV DNA purified from untreated, siRNAPCNA-treated, and araC-treated cells, at various times post-infection with HSV-1 strain 17, using TK promoter primers. The results indicate that both PCNA-specific siRNA and araC treatments lead to significant inhibition of 17+ replication (Fig. 2b, c). Additionally, after negative control siRNA treatment, we observed the same pattern of 17+ DNA replication as in untreated cells (Fig. 2a, b). Thus, we conclude that PCNA is required for HSV replication.
As it is known that HSV replicates its DNA using its own DNA polymerase and processivity factor (UL30 and UL42), it is likely that PCNA is involved in some other aspects of the viral growth cycle.
PCNA inhibition affects histone deposition on HSV DNA
In order to determine if PCNA, which plays a role in tethering the factors involved in cellular nucleosome assembly on cellular DNA, plays a role in viral nucleosome assembly and deposition on the viral genome, we determined the amount of nucleosome-protected DNA in total viral DNA in untreated and PCNA-inhibited cells.
Vero cells were treated with SiRNAPCNA, SiRNAcontrol, AraC, or subjected to no treatment. After 72 h, the cells were infected with HSV-1 (MOI = 5), and, at various times, the cells were harvested and nuclei prepared. The chromatin within the nuclei was digested with Micrococcal nuclease (MNase), which cuts chromatinized DNA between nucleosomes, but leaves the nucleosome-wrapped DNA (150 bp) intact. Protected fragments were isolated (as described in the “Materials and methods” section) and size separated by agarose gel electrophoresis before transferring to nylon membranes (Southern Blot). The membranes were UV cross-linked and probed using radioactively labeled HSV DNA, comprising all regions of the genome, as described in the “Materials and methods” section. Figure 3 shows a typical blot and Table 1 shows the quantitated data from blot analysis using a Typhoon phosphorimager. The maximum amount of nucleosomes can be detected at 8 hpi, but when PCNA is inhibited by siRNA, nucleosome-protected DNA is diminished. A significant difference was noted at 4 h post-infection but not at the later time (8 hpi) using a one-tail t test (p = 0.001). From the data in Table 1, the PCNA SiRNA knockdown nucleosome-protected HSV DNA had an approximately 23 % reduction in signal strength compared to the negative siRNA control at 4 hpi (p = 0.001)..
Measurement of total nucleosome-protected HSV-1 DNA by Southern blot. Vero cells were transfected with and without siRNA followed by infection with 17+ virus at a MOI of 5. Cells were harvested at 0 (mock infection), 1, 4, and 8 hpi and subjected to MNase digestion of their DNA. Digests were subjected to agarose gel electrophoresis and Southern blotting. After hybridizing with a probe to the total HSV-1 genome, the quantity of nucleosome protected DNA was determined using a Typhoon phosphorimager (P PCNA SiRNA, N negative control SiRNA). The data are presented in Table 1
The relative amounts of MNase fragments from select regions of the HSV-1 genome were determined by PCR, using TK gene and VP16 promoter specific primers (Fig. 4a–d). Vero cells were treated with SiRNAPCNA, SiRNAcontrol, AraC, or subjected to no treatment, as before. A fraction of DNA was retained for determination of the total HSV DNA in each sample and the remainder used to prepare nucleosomes fractions, as before. The results support the conclusion that, after PCNA inhibition with either siRNA or araC, histones are no longer deposited on virus DNA.
Measurement of nucleosome-protected HSV-1 DNA regions by PCR. Vero cells were transfected with and without siRNA followed by infection with 17+ virus at a MOI of 5. Cells were harvested at 0 (mock infection), 1, 4, and 8 hpi and subjected to MNase digestion of their DNA. This DNA was used for RT-PCR using TK or VP16 promoter primers. Each data point represents the mean ± standard deviation of three independent triplicate determinations. a 17+ after siRNAPCNA using TK promoter primers. b 17+ after siRNAPCNA using VP16 promoter primers. c 17+ after araC using TK promoter primers. d 17+ after araC using VP16 promoter primers
It will be noted that at 8 hpi, DNA replication is in full swing and much newly replicated viral DNA is present in the cell. We have previously shown that the bulk of this newly replicated DNA is not associated with nucleosomes (Oh et al. 2012); consequentially, the small value of nucleosome-protected DNA at 8 hpi, compared to 4 hpi, is due to quantitation of the protected fragment to the total HSV DNA at this time, whereas in the Southern blot analysis (Fig. 3), the total amount of nucleosomal DNA is shown uncorrected for the total amount of viral DNA present.
Although HSV interacts with PCNA, it does not appear to interact with cell DNA polymerases
In order to determine whether HSV-1 uses cellular DNA polymerase to interact with PCNA, we performed immunoprecipitation experiments using antibodies to PCNA and cellular DNA polymerases. The HSV-1 DNA polymerase (UL42) is an early gene and thus is not expressed immediately after entry of the viral genome into the cell nucleus. However, HSV-1 DNA requires repair following its entrance of the nucleus and has been shown to accumulate cellular by at least 1 h PI. Also, it is known that HSV-1 DNA polymerase does not interact with PCNA (Gottlieb et al. 1990). Thus, the possibility that HSV DNA uses cellular polymerases to deposit nucleosomes after infection by recruitment of PCNA and DNA polymerases is possible. A chromatin immunoprecipitation (ChIP) approach using antibodies against DNA polymerase and PCNA was used to test this hypothesis.
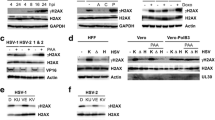
First, we verified that HSV-1 DNA binds to PCNA. Figure 5 shows that PCNA binds to HSV-1 DNA early at 1 hpi using TK promoter primers (Fig. 5a) and at 4 hpi using VP16 promoter primers (Fig. 5b), but that later this complex dissociates.
Chromatin immunoprecipitation (ChIP) using PCNA antibody. Vero cells infected with 17+ HSV virus at a MOI of 5 were harvested at 1, 4, and 8 hpi. Cells were cross-linked using 1 % HCHO for 15 min, and the reaction was stopped by adding glycine at the final concentration of 0.125 M. The chromatin and bound factors were purified and sonicated as described in the “Materials and methods”. Ten percent of the material was retained as the input sample. The rest of the chromatin sample was equally divided and incubated with anti-PCNA antibodies, or IgG antibodies, and precipitated using protein A agarose beads. The DNA-protein complexes were eluted from the beads and cross-linked by heat. The proteins were then digested with proteinase K and DNA purified by phenol-chloroform extraction. These DNA samples were used for quantitative RT-PCR. Quantities of the ChIPed PCNA, IgG, and PCNA were calculated by the formula (quantity sample/quantity input) × 10. This experiment was repeated three times with essentially similar patterns. a PCNA immunoprecipitated and quantitated using TK promoter primers. b PCNA immunoprecipitated and quantitated using VP16 promoter primers. c Polymerase delta precipitated and quantitated using TK promoter primers. d Polymerase epsilon precipitated and quantitated using TK promoter primers. e Polymerase lambda precipitated and quantitated using TK promoter primers. f Polymerase eta precipitated and quantitated using TK promoter primers
Next, we determined if any cell DNA polymerases bind to PCNA and HSV-1 DNA. PCNA is a processivity factor for cell DNA polymerases delta and epsilon, which are mainly responsible for DNA replication, but also play a role in DNA repair and recombination (Oku et al. 1998). However, it also binds to the Y-family of polymerases (Lehmann 2006) and the X-family of polymerases (Shimazaki et al. 2005). The Y-family of polymerases (polymerase eta, iota) are responsible for translesion synthesis past damaged sites (Lehmann 2006), and the X-family of polymerases (polymerase lambda, mu) are involved in non-homologous rejoining of DNA double-strand breaks (Shimazaki et al. 2005). In addition, Muylaert and Elias (2010) suggested that DNA polymerase eta was required by HSV-1 for replication at low MOI, and that it may act as a gap filler after HSV-1 replication. Thus, we performed ChIP assay for polymerases delta, epsilon, eta, and lambda.
Our results indicate that both polymerase delta and epsilon, involved mainly in replication, are not used by HSV-1 (Fig. 5c and d, respectively). Our results indicate that X-family polymerases are also unlikely to bind to HSV-1 to aid replication, repair, or histone deposition (Fig. 5e). Figure 5f shows that the Y-family polymerases do not bind to HSV-1 either. These results suggest that, under the conditions tested, HSV-1 DNA binds to PCNA but does not employ the PCNA-DNA polymerase relationship for this binding.
Does PCNA interact with the HSV genome through ICP34.5?
Previous work by Harland et al. (2003) showed that HSV ICP34.5 can interact with PCNA. In order to determine if ICP34.5, through its interaction with PCNA, has a role in HSV DNA replication, we compared the growth of a mutant lacking ICP34.5 with wild-type virus, in the presence of siRNAPCNA knockdown or araC inhibition. We performed RT-PCR for nuclear HSV DNA purified from untreated, siRNAPCNA-treated, and araC-treated cells, at various times post-infection with HSV-1 strain 1716, using TK promoter primers (Fig. 6). The results indicate that both PCNA-specific siRNA and araC treatments lead to almost complete inhibition of 1716 replication (Fig. 6a, b). Additionally, after negative control siRNA treatment, we observed the same pattern of 1716 DNA replication as in untreated cells (Fig. 6a, b).
AraC or SiRNAPCNA treatment affects HSV-1 mutant 1716 similar to parental HSV-1 strain 17. Vero cells were treated with or without araC and incubated for 72 h followed by infection with 1716 virus at a MOI of 5. Cells were harvested at 0 (mock infection), 1, 4, and 8 h post-infection and the genomic DNA extracted. Quantitative real-time PCR was used to determine the level of HSV and cell DNA. Each data point represents the mean ± standard deviation of three independent triplicate determinations. a 1716 virus before and after araC. b 1716 virus before and after siRNAPCNA. c Vero cells were incubated with and without araC or transfected with siGENOME SMART pool Human PCNA and incubated for 72 h followed by infection with 1716 virus at MOI 5. Cells were harvested at 0 (mock infection), 1, 4, 8, and 20 h post-infection and each sample was titered in triplicate on Vero cells using immunoperoxidase-based assay. Each data point represents the mean ± standard deviation of three independent triplicate determinations
To confirm the inhibition of HSV DNA synthesis after PCNA inhibition, as determined by PCR, we examined the replication of HSV-1 strain 1716 before and after PCNA inhibition using siRNAPCNA or araC treatment (Fig. 6c). Measurement of plaque formation, using an immunoperoxidase-based assay, showed that the titer of HSV-1 strain 1716 was decreased by almost 2 logs at 20 h post-infection, a result similar to that seen for the parental HSV-1 strain 17 (Fig. 2). Thus, we conclude that PCNA is required for efficient HSV replication, but that its involvement is independent of its ability to complex with ICP34.5.
Discussion
The fact that HSV-1 uses cellular proteins for its propagation is well established and numerous papers have shown the interaction of cellular genes with HSV. For example, HSV-1 gene ICP8, which binds to viral DNA, has been co-localized with cellular gene products SSB, PCNA, Rb, p53, DNA ligase 1, and pol α (Wilcock and Lane 1991). Furthermore, ICP0 has been shown to interact with cyclin D3, elongation factor EF-1, transcription factor BMAL1, and the ubiquitin-specific protease USP7 (Boutell and Everett 2003). HSV-1 has been shown to associate with nuclear domain-10 (ND10) in order to gain access to certain ND10-associated proteins involved in recombination and repair (Barr et al. 2003; Everett 2001; Maul et al. 1993). Components of the homologous recombination pathway (RPA, RAD51, and NBS1) have also been shown to be recruited to some types of pre-replicative sites (Wilkinson and Weller 2004). HSV replication is known to be regulated by ND10 components ATRX and hDaxx (Lukashchuk and Everett 2010). Our data from Figs. 1 and 2 support a role for the cell DNA polymerase-associated protein PCNA playing a role in HSV replication.
Although HSV has its own DNA polymerase, cellular DNA polymerases are known to influence HSV replication, repair, and nucleosome deposition. Muylaert and Elias have shown that HSV-1 plaque efficiency was reduced 104-fold in DNA polymerase η mutant cells (Muylaert and Elias 2010). They showed that DNA polymerase η was strictly required for virus replication at low, but was not important at high, multiplicities of infection. Cavanaugh and Kuchta showed that DNA polymerase α can elongate herpes primase-synthesized RNA primers much more efficiently than herpes polymerase (Cavanaugh and Kuchta 2009). They hypothesize that if indeed HSV-1 replication requires polymerase α, it is likely to form a complex involving herpes polymerase, polymerase α, and herpes primase-helicase. No in vivo data is available to support this hypothesis. Although it has been shown that cellular DNA polymerase β is involved in HSV-2 repair, neither HSV DNA polymerase nor cellular DNA polymerase α appear to be involved in this process (Nishiyama et al. 1983).
It is known that HSV-1 uses the cell DNA repair machinery from the comparison of HSV replication in DNA-repair-competent and DNA-repair-deficient cell cultures after exposure to DNA-damaging agents (Miller and Smith 1991). The authors found that DNA damage stimulates DNA-repair-competent cells to amplify HSV replication, the extent of cellular DNA damage influenced the level of virus replication, and the effect of the DNA damage on HSV replication was time dependent. DNA repair mechanisms that act on a variety of chromosomal lesions may be involved in the repair and biological activation. Host recombination and repair proteins are also involved in viral DNA replication (Wilkinson and Weller 2004). They observed that RPA, RAD51, and NBS1 are present in the pre-replicative sites but require viral polymerase and other replication proteins within these sites.
Another conclusion that can be made from our data is that PCNA plays a role in histone deposition on HSV-1 genome. We digested the linkers between nucleosomes using Micrococcal nuclease (MNase) and purified 150-bp DNA fragments protected by histones. This DNA was used for RT-PCR analysis using TK and VP16 promoter primers and Southern blot analysis using the radioactively labeled probes covering the entire HSV-1 genome. Our data shows that PCNA inhibition leads to the failure in histone assembly on the HSV-1 genome (Fig. 4). In the cell, PCNA does not deposit histone by itself but serves as a link between Asf1 and CAF-1 proteins, which are critical for managing the flow of histones, and is thought to tether them to DNA polymerase (Peng et al. 2010). It is possible that PCNA is involved in HSV-1 replication and histone deposition via Asf1 and CAF-1 proteins, as PCNA and Asf1 are responsible for the progression of the replication fork via coordinated disassembly and reassembly of nucleosomes. Asf1 interacts with cellular transcriptional co-activator HCF-1 and it is thought that this complex regulates nucleosome assembly and disassembly, and viral replication and repair (Peng et al. 2010). Although this mechanism could account for nucleosome deposition during viral DNA replication, it would not account for nucleosome deposition prior to DNA replication. Nucleosome deposition during this time would have to rely on another mechanism, such as DNA repair or transcription linked deposition. Our data support the hypothesis that PCNA may play a role in early nucleosome deposition via the DNA repair processes. Previously, our work showed that H3.3 was incorporated into HSV chromatin at the earliest infection times, supporting this DNA repair–transcription hypothesis (Placek et al. 2009).
It is possible that PCNA may be involved in HSV-1 replication, repair, and histone deposition via cell DNA polymerases. PCNA is a processivity factor for DNA polymerases delta and epsilon (Maul et al. 1993; Oku et al. 1998) and binds to X- and Y-families of DNA polymerases (Lehmann 2006; Shimazaki et al. 2005). Initially, we confirmed that PCNA binds to HSV-1 DNA by performing ChIP experiments using anti-PCNA antibodies to immunoprecipitate PCNA in samples infected with HSV-1 virus. Using two different promoter primers, TK and VP16, we were able to detect the complex of PCNA and HSV-1 at 1 and 4 hpi, respectively (Fig. 5a, b). We also performed ChIP experiments to immuno-precipitate four different types of cell DNA polymerase binding PCNA in samples infected with HSV-1 virus. However, our ChIP data demonstrated that although HSV-1 binds to PCNA early in infection (Fig. 5a, b), at the sensitivity of our assay, it does not form any complexes with cell DNA polymerases that normally bind to PCNA at our level of detection (Fig. 5c–f). Thus, we conclude that cell DNA polymerases are not involved in HSV-1 histone deposition and PCNA may act through some other protein interaction with viral DNA. It is also possible that other polymerases (alpha or beta) could bind to viral DNA—at least it was demonstrated that polymerase alpha extends herpes primase-synthesized RNA primers more efficiently than the viral polymerase (Cavanaugh and Kuchta 2009).
The finding that ICP34.5 forms a DNA-binding complex with PCNA and HSV replication proteins suggests a role for PCNA in HSV replication (Harland et al. 2003) even though HSV has its own processivity protein (UL42) (Zuccola et al. 2000). Our data suggest that HSV-1 replication is PCNA dependent, in contrast to conclusions presented by Muylaert and Elias (2010). We compared the replication of a wild type HSV-1 virus and a mutant virus lacking the ICP34.5 gene, with and without PCNA inhibition, and demonstrated that PCNA inhibition reduces replication and plaque forming abilities similarly for both viruses (Figs. 2 and 6). Thus, we concluded that PCNA is not involved in HSV replication via interaction with ICP34.5.
Although nucleosome assembly occurs during cell DNA replication (Burgess and Zhang 2010), replication-coupled nucleosome assembly does not appear to be the primary mechanism used to deposit nucleosomes during HSV lytic infection for two reasons. First, nucleosomes are placed on the HSV genome within 1 h post-infection (Kent et al. 2004), a time long before HSV DNA polymerase is expressed and an increase in viral DNA is measurable. Second, the level of nucleosomes on viral DNA peaks before “late” gene expression, and they do not appear to accumulate on newly replicated DNA (Oh and Fraser 2008). Thus, a more likely process leading to nucleosome deposition on HSV is DNA repair and end joining of the linear genome after entry into the cell nucleus. During cellular DNA repair, chromatin structure is re-established.
As HSV virion DNA entering the cell nucleus has single-stranded breaks in it (Wilkie 1973) and is linear, it is probable that the cell perceives the HSV DNA as damaged and requiring repair of the breaks, joining of the ends, and restoration of the chromatin. Both CAF-1 and Asf1 are involved in chromatin reassembly after repair of DNA lesions (Linger and Tyler 2007). It is known that interaction between chromatin assembly factor 1 (CAF-1) and PCNA is important in recruiting CAF1 to sites of UV damage (Polo et al. 2006). Most chromatin repair mechanisms are believed to involve complete removal of nucleosomes prior to lesion repair and chromatin reassembly. In the case of HSV DNA, there is no chromatin on the virion DNA; thus, if the cell DNA repair machinery is utilized to repair HSV DNA, this initial step of nucleosome removal is not required, and post-repair step nucleosome deposition may occur because it is part of the repair machinery rather than because there is a requirement for the HSV lytic cycle. Alternatively, HSV entering a cell may be mimicking sperm fertilization of an egg, as sperm has little histone (4 % nucleosomes; Hammoud et al. 2009) and much protamine, which is replaced by histone on cell entry (Yang et al. 2014). It is clear that nucleosomes on HSV DNA contain epigenetic marks (acetylation, methylation, etc.) in a pattern consistent with cellular genes (Amelio et al. 2006; Kent et al. 2004), yet it is unclear how these marks arise through a repair process (Groth et al. 2007). The need for epigenetic marks during acute infection is unclear and may simply be a consequence of using the cellular transcription machinery.
This study shows that PCNA plays a role in nucleosome deposition and DNA replication of HSV in tissue culture cells. As with many other studies of acute HSV infection, it utilizes non-neuronal cells. However, the results are an important first step in the study of the early stages of reactivation of latent viral genomes from peripheral neuronal cells.
References
Amelio AL, Giordani NV, Kubat NJ, O’Neil JE, Bloom DC (2006) Deacetylation of the herpes simplex virus type 1 latency-associated transcript (LAT) enhancer and a decrease in LAT abundance precede an increase in ICP0 transcriptional permissiveness at early times postexplant. J Virol 80:2063–8
Barr SM, Leung CG, Chang EE, Cimprich KA (2003) ATR kinase activity regulates the intranuclear translocation of ATR and RPA following ionizing radiation. Curr Biol 13:1047–51
Boutell C, Everett RD (2003) The herpes simplex virus type 1 (HSV-1) regulatory protein ICP0 interacts with and ubiquitinates p53. J Biol Chem 278:36596–602
Burgess RJ, Zhang Z (2010) Histones, histone chaperones and nucleosome assembly. Protein Cell 1:607–12
Cavanaugh NA, Kuchta RD (2009) Initiation of new DNA strands by the herpes simplex virus-1 primase-helicase complex and either herpes DNA polymerase or human DNA polymerase alpha. J Biol Chem 284:1523–32
Chou J, Kern ER, Whitley RJ, Roizman B (1990) Mapping of herpes simplex virus-1 neurovirulence to gamma34.5, a gene nonessential for growth in culture. Science 250:1262–1266
Cunningham C, Davison AJ (1993) A cosmid-based system for constructing mutants of herpes simplex virus type 1. Virology 197:116–24
Everett RD (2001) DNA viruses and viral proteins that interact with PML nuclear bodies. Oncogene 20:7266–73
Garber DA, Beverley SM, Coen DM (1993) Demonstration of circularization of herpes simplex virus DNA following infection using pulsed field gel electrophoresis. Virology 197:459–462
Gehen SC, Vitiello PF, Bambara RA, Keng PC, O’Reilly MA (2007) Downregulation of PCNA potentiates p21-mediated growth inhibition in response to hyperoxia. Am J Physiol Lung Cell Mol Physiol 292:L716–24
Gibson W, Roizman B (1971) Compartmentalization of spermine and spermidine in the herpes simplex virion. Proc Natl Acad Sci U S A 68:2818–21
Gottlieb J, Marcy AI, Coen DM, Challberg MD (1990) The herpes simplex virus type 1 UL42 gene product: a subunit of DNA polymerase that functions to increase processivity. J Virol 64:5976–87
Groth A, Rocha W, Verreault A, Almouzni G (2007) Chromatin challenges during DNA replication and repair. Cell 128:721–33
Hammoud SS, Nix DA, Zhang H, Purwar J, Carrell DT, Cairns BR (2009) Distinctive chromatin in human sperm packages genes for embryo development. Nature 460:473–8
Harland J, Dunn P, Cameron E, Conner J, Brown SM (2003) The herpes simplex virus (HSV) protein ICP34.5 is a virion component that forms a DNA-binding complex with proliferating cell nuclear antigen and HSV replication proteins. J Neurovirol 9:477–88
Huertas D, Sendra R, Munoz P (2009) Chromatin dynamics coupled to DNA repair. Epigenetics 4:31–42
Kent JR, Zeng PY, Atanasiu D, Gardner J, Fraser NW, Berger SL (2004) During lytic infection herpes simplex virus type 1 is associated with histones bearing modifications that correlate with active transcription. J Virol 78:10178–10186
Knipe DM, Cliffe A (2008) Chromatin control of herpes simplex virus lytic and latent infection. Nat Rev Microbiol 6:211–21
Kutluay SB, Triezenberg SJ (2009a) Regulation of histone deposition on the herpes simplex virus type 1 genome during lytic infection. J Virol 83:5835–45
Kutluay SB, Triezenberg SJ (2009b) Role of chromatin during herpesvirus infections. Biochim Biophys Acta 1790:456–66
Lehmann AR (2006) New functions for Y family polymerases. Mol Cell 24:493–5
Leinbach SS, Summers WC (1980) The structure of herpes simplex virus type 1 DNA as probed by micrococcal nuclease digestion. J Gen Virol 51:45–59
Linger JG, Tyler JK (2007) Chromatin disassembly and reassembly during DNA repair. Mutat Res 618:52–64
Lu X, Triezenberg SJ (2010) Chromatin assembly on herpes simplex virus genomes during lytic infection. Biochim Biophys Acta 1799:217–22
Lukashchuk V, Everett RD (2010) Regulation of ICP0-null mutant herpes simplex virus type 1 infection by ND10 components ATRX and hDaxx. J Virol 84:4026–40
Marchion DC, Pfau JC, Weber PA, Grobe AC, Duran CM, Cheung DT (2003) An in vitro model of pericardial tissue healing. Biomaterials 24:89–95
Maul GG, Guldner HH, Spivack JG (1993) Modification of discrete nuclear domains induced by herpes simplex virus type 1 immediate early gene 1 product (ICP0). J Gen Virol 74(Pt 12):2679–90
Miller CS, Smith KO (1991) Enhanced replication of herpes simplex virus type 1 in human cells. J Dent Res 70:111–7
Muggeridge MI, Fraser NW (1986) Chromosomal organization of the herpes simplex virus genome during acute infection of the mouse central nervous system. J Virol 59:764–767
Muylaert I, Elias P (2010) Contributions of nucleotide excision repair, DNA polymerase eta, and homologous recombination to replication of UV-irradiated herpes simplex virus type 1. J Biol Chem 285:13761–8
Nishiyama Y, Tsurumi T, Aoki H, Maeno K (1983) Identification of DNA polymerase(s) involved in the repair of viral and cellular DNA in herpes simplex virus type 2-infected cells. Virology 129:524–8
Oh J, Fraser NW (2008) Temporal association of the herpes simplex virus genome with histone proteins during a lytic infection. J Virol 82:3530–7
Oh J, Ruskoski N, Fraser NW (2012) Chromatin assembly on herpes simplex virus 1 DNA early during a lytic infection is Asf1a dependent. J Virol 86:12313–21
Oku T, Ikeda S, Sasaki H, Fukuda K, Morioka H, Ohtsuka E, Yoshikawa H, Tsurimoto T (1998) Functional sites of human PCNA which interact with p21 (Cip1/Waf1), DNA polymerase delta and replication factor C. Genes Cells 3:357–69
Olivo JF, Guille F, Lobel B (1989) Microscopic hematuria. Semiologic value in urology. Management of microscopic hematuria. J Urol 95:453–8
Paunesku T, Mittal S, Protic M, Oryhon J, Korolev SV, Joachimiak A, Woloschak GE (2001) Proliferating cell nuclear antigen (PCNA): ringmaster of the genome. Int J Radiat Biol 77:1007–21
Peng H, Nogueira ML, Vogel JL, Kristie TM (2010) Transcriptional coactivator HCF-1 couples the histone chaperone Asf1b to HSV-1 DNA replication components. Proc Natl Acad Sci U S A 107:2461–6
Pignatti PF, Cassai E (1980) Analysis of herpes simplex virus nucleoprotein complexes extracted from infected cells. J Virol 36:816–28
Placek BJ, Huang J, Kent JR, Dorsey J, Rice L, Fraser NW, Berger SL (2009) The histone variant H3.3 regulates gene expression during lytic infection with herpes simplex virus type 1. J Virol 83:1416–21
Poffenberg KL, Roizman B (1985) A non-inverting genome of a viable herpes simplex virus 1: presence of head-to-tail linkage in packaged genomes and requirements for circularization of infection. J Virol 53:587–595
Polo SE, Roche D, Almouzni G (2006) New histone incorporation marks sites of UV repair in human cells. Cell 127:481–93
Roizman B, Knipe D, Whitley R (2013) Herpes simplex viruses. In: Fields virology. Knipe D, Howely, PM (ed) Lippincott, Williams & Wilkins, Philadelphia, PA, pp 1823–1897
Schang LM, Hossain A, Jones C (1996) The latency-related gene of bovine herpesvirus 1 encodes a product which inhibits cell cycle progression. J Virol 70:3807–3814
Shimazaki N, Yazaki T, Kubota T, Sato A, Nakamura A, Kurei S, Toji S, Tamai K, Koiwai O (2005) DNA polymerase lambda directly binds to proliferating cell nuclear antigen through its confined C-terminal region. Genes Cells 10:705–15
Spiker S, Murray MG, Thompson WF (1983) DNase I sensitivity of transcriptionally active genes in intact nuclei and isolated chromatin of plants. Proc Natl Acad Sci U S A 80:815–9
Thompson RL, Wagner EK, Stevens JG (1983) Physical location of a herpes simplex virus type-1 gene function(s) specifically associated with a 10 million-fold increase in HSV neurovirulence. Virology 131:180–192
Ulrich HD, Takahashi T (2013) Readers of PCNA modifications. Chromosoma 122:259–74
Ward SL, Scheuner D, Poppers J, Kaufman RJ, Mohr I, Leib DA (2003) In vivo replication of an ICP34.5 second-site suppressor mutant following corneal infection correlates with in vitro regulation of eIF2 alpha phosphorylation. J Virol 77:4626–34
Wilcock D, Lane DP (1991) Localization of p53, retinoblastoma and host replication proteins at sites of viral replication in herpes-infected cells. Nature 349:429–31
Wilkie NM (1973) The synthesis and substructure of herpesvirus DNA: the distribution of alkali-labile single strand interruptions in HSV-1 DNA. J Gen Virol 21:453–67
Wilkinson DE, Weller SK (2004) Recruitment of cellular recombination and repair proteins to sites of herpes simplex virus type 1 DNA replication is dependent on the composition of viral proteins within prereplicative sites and correlates with the induction of the DNA damage response. J Virol 78:4783–96
Yang P, Wu W, Macfarlan TS (2014) Maternal histone variants and their chaperones promote paternal genome activation and boost somatic cell reprogramming. Bioessays
Zuccola HJ, Filman DJ, Coen DM, Hogle JM (2000) The crystal structure of an unusual processivity factor, herpes simplex virus UL42, bound to the C terminus of its cognate polymerase. Mol Cell 5:267–78
Acknowledgments
We acknowledge the outstanding technical support of Sindura Penubarthi and Nick Ruskoski. We are grateful to Sarah Ratcliffe (Penn CFAR core services; P30-AI-045008) for statistical help, and Erle Robertson and Matt Weitzman for commenting on our manuscript. This work was supported by NIH grant NS33768 and NS 082240. I.S. was supported by NIH training grant T32-CA115299.
Conflict of interest statement
The authors, Iryna Sanders, Mark Boyer, and Nigel W. Fraser, declare that they have no conflicts of interest.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Sanders, I., Boyer, M. & Fraser, N.W. Early nucleosome deposition on, and replication of, HSV DNA requires cell factor PCNA. J. Neurovirol. 21, 358–369 (2015). https://doi.org/10.1007/s13365-015-0321-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13365-015-0321-7