Abstract
We have recently developed a multiplex mass spectrometry imaging (MSI) method which incorporates high mass resolution imaging and MS/MS and MS3 imaging of several compounds in a single data acquisition utilizing a hybrid linear ion trap-Orbitrap mass spectrometer (Perdian and Lee, Anal. Chem. 82, 9393–9400, 2010). Here we extend this capability to obtain positive and negative ion MS and MS/MS spectra in a single MS imaging experiment through polarity switching within spiral steps of each raster step. This methodology was demonstrated for the analysis of various lipid class compounds in a section of mouse brain. This allows for simultaneous imaging of compounds that are readily ionized in positive mode (e.g., phosphatidylcholines and sphingomyelins) and those that are readily ionized in negative mode (e.g., sulfatides, phosphatidylinositols and phosphatidylserines). MS/MS imaging was also performed for a few compounds in both positive and negative ion mode within the same experimental set-up. Insufficient stabilization time for the Orbitrap high voltage leads to slight deviations in observed masses, but these deviations are systematic and were easily corrected with a two-point calibration to background ions.

ᅟ
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Matrix-assisted laser desorption/ionization mass spectrometry imaging (MALDI-MSI) is a rapidly developing and increasingly widespread tool for mapping metabolites and other biomolecules in tissue [1–3]. The development of high-resolution mass spectrometers capable of highly accurate mass measurements has allowed for the analysis of complex tissue samples and the assignment of molecular formulas directly from imaging experiments [4, 5]. MS/MS imaging has also been performed for structural analysis of detected analytes, as well as discrimination of isomers with different on-tissue distributions [6–8]. We have recently developed a multiplex MS imaging methodology to incorporate high mass resolution Orbitrap scans with faster but lower mass resolution ion trap scans in a single imaging experiment. This method allows for the reduction of data acquisition time while achieving MS imaging in both high mass resolution and high spatial resolution. Tandem MS scans can also be included to provide structural information and to reinforce analyte identifications made from accurate mass [6].
Lipids are one commonly studied class of compounds in MSI [9]. The high abundance of lipids in many tissues, established procedures for sample preparation, and minimal delocalization in typical preparation techniques make them good targets for the development and demonstration of MSI techniques. Several recent reviews are available describing lipid analysis and imaging by different ionization techniques, including matrix-assisted laser desorption/ionization (MALDI) [10], desorption electrospray ionization (DESI) [11], and secondary ion mass spectrometry (SIMS) [12, 13]. Lipid classes investigated by MSI include sterols, fatty acids, phosphatidylcholines (PCs), phosphatidylinositols (PIs), phosphatidylserines (PSs), sphingomyelins (SMs), and sulfatides (STs). With the exception of sterols, these lipids consist of a polar headgroup region and a non-polar hydrocarbon tail region. Differences in headgroup composition lead to significant differences in ionization yield between classes. Generally, those lipids that are detected in high abundance as positive ions are poorly detected or not detected at all as negative ions, and vice-versa [10]. Therefore, more comprehensive lipid data can be obtained by detecting in both positive and negative ionization modes than in a single polarity alone.
One method for acquiring both positive and negative ion mode images in MALDI-MSI is to image two separate tissue samples (e.g., sequential tissue sections), one in positive mode and one in negative mode. This allows for the selection of the best matrix for each polarity. In experiments using two tissue sections, however, comparison of features on positive and negative images requires overlapping the two images by correlating structural features and is not always simple or precise. Depending on the sample and preparation methods, there may be variations in sample composition between adjacent sections. Additionally, acquiring opposite polarity images from separate sections requires the preparation of twice as many samples. For these reasons, it is beneficial to obtain both ion mode images on a single tissue. Thomas and coworkers demonstrated that certain matrixes can be optimized for efficient profiling of lipids for both positive and negative ion mode [14]. Using 1,5-diaminonapthalene as a matrix, they successfully obtained both positive and negative MS images on a single tissue section by collecting consecutive positive and negative images with a slight position offset. The same method can be also applied by TOF-SIMS for lipid imaging in both polarities on a single section [15]. TOF-SIMS is very attractive for certain applications [15–17] because of its extremely high spatial resolution (down to nanometer size), no need of optimizing matrix for both polarities, and fast data acquisition time; however, MSI of high molecular weight lipids (m/z > 700) is limited due to its low sensitivity.
An alternative way of acquiring opposite polarity MS images on a single tissue would be through repeated polarity switching in a single experiment. It has been recently demonstrated that polarity switching could be used to detect multiple lipid classes in a shotgun lipidomics experiment with electrospray ionization [18]. However, there has been no such experiment in MS imaging on a single tissue. Here we extend our multiplex MS imaging methodology to include polarity switching for imaging of a wide range of lipid classes in a single acquisition. This greatly expands the flexibility of multiplex MS imaging and allows accurate mass imaging of both positive and negative ions along with MS/MS imaging of a few targeted compounds in both polarities.
2 Experimental
2.1 Materials and Sample Preparation
Coronal sections of mouse brain (striatum, catalog no. MF-201-03) were purchased from Zyagen (San Diego, CA, USA). According to the provider’s description, brains were obtained from CD1 mice, flash frozen, and sectioned in OCT at 7–10 μm thickness. 2,5-dihydroxybenzoic acid (DHB) was purchased from Thermo Fisher (Waltham, MA, USA). Vacuum-sealed tissue sections were allowed to come to room temperature before opening. The tissue was subjected to DHB sublimation without any pretreatment using a procedure described previously [19].
2.2 MSI Data Acquisition
Data was acquired on a linear ion trap-orbitrap mass spectrometer with a MALDI source (MALDI LTQ-Orbitrap Discovery; Thermo Scientific, San Jose, CA, USA) modified to use an external Nd:YAG laser (UVFQ; Elforlight Ltd., Daventry, UK). The laser pump current was set to 80 % demand (~1 μJ pulse energy) and the repetition rate was 60 Hz. The laser spot size was estimated to be ~20 μm by observation of burn marks on a thin film of α-cyanohydroxycinnamic acid. Spectra were collected from 20 shots per scan and one scan per spiral step.
The Thermo TunePlus software was used to define the raster pattern: 9 spiral steps per raster step, 25 μm spiral step size, and 100 μm raster step size. MS and MS/MS scans were defined in the Thermo Xcalibur software. A software patch received from the manufacturer was used to reduce the delay time for polarity switching from 7 to 5 s. Nine scan events were defined, the same as the number of spiral steps, such that one spectrum was collected from each spiral step. Orbitrap scans were acquired from m/z 150–1000 and CID MS/MS scans were acquired in the linear ion trap with normalized collision energies of 40–60 (arbitrary units) and 1.8 Da isolation widths. m/z of 739.47, 772.53, 782.57, and 826.57 were selected for MS/MS in positive ion mode and 834.52, 885.54, and 906.63 were selected for MS/MS in negative ion mode.
2.3 Data Analysis
After data acquisition, mass spectra were post-calibrated using the Recalibrate Offline tool (Thermo Fisher Scientific). Positive and negative Orbitrap spectra were treated independently. All spectra in a given polarity were averaged, omitting the first 90 min of acquisition time. This ensures that masses used for calibration are stable after the initial mass accuracy deviation caused by polarity switching (discussed below). From these averaged spectra, a ‘one for all’ external recalibration was applied to each individual spectrum. Two background peaks were chosen as calibrant peaks for each polarity. In positive mode the peaks used were m/z 215.1543 (protonated 1,8-bis(dimethylamino)naphthalene (DMAN), a persistent instrumental contaminant from a previous experiment with DMAN as a matrix) and m/z 332.3312 (C23H42N+, an ammonium ion from the tissue embedding medium). Negative mode spectra were calibrated using two matrix peaks, m/z 329.0279 and m/z 681.0450, corresponding to [2DHB – 2H + Na]− and [4DHB – 4H + 3Na]−, respectively.
ImageQuest software (Thermo) was used to generate MS images. Mass tolerances used for ion image production were ±0.01 Da for Orbitrap scans and ±0.5 Da for MS/MS scans. MS images were normalized against total ion count, and MS/MS images were generated without normalization. QualBrowser (Thermo) was used to generate averaged mass spectra.
3 Results and Discussion
As explained in the Experimental section, polarity switching between positive and negative ion mode was adapted by modifying the multiplex MS imaging method previously developed [6]. In short, each traditional ‘raster step’ is now broken down to nine spiral steps of smaller MALDI plate movement. The spiral step function is intended to enable spectral averaging within the raster step, but we are utilizing this functionality to acquire multiple data sets within each raster step. Figure 1 illustrates the MALDI plate spiral movement utilized in the current study for multiplex MS imaging with polarity switching. Each raster step is composed of nine spiral steps. Orbitrap scans are acquired in spiral steps 1 (positive) and 6 (negative). MS/MS scans are collected in steps 2–5 (positive) and 7–9 (negative). The polarity is automatically switched from positive to negative between step 5 and 6, and from negative to positive between step 9 and step 1 of the next raster step. It should be noted that we are incorporating not only high-resolution MS spectra in both polarities but also multiple MS/MS spectra in positive and negative ion modes. Hence, our experimental design corresponds to nine separate MS imaging experiments performed on a single tissue. This represents the first acquisition of MS and MS/MS images in both polarities on a single tissue. Compared to a simple consecutive MS imaging experiment in two polarities [14], the inclusion of multiple MS/MS scans here allows for much more chemical information to be extracted from a single tissue sample.
Illustration of spiral movement of MALDI plate and spectral acquisition method at each spiral step. The numbers indicate the order of spiral movement. A spiral step size of 25 μm (e.g., 1 → 2) and a raster step size of 100 μm (e.g., 1 → 1) were used in this experiment. FT indicates Orbitrap data acquisition. MS2 indicates ion trap MS/MS scans. MS/MS spectra were obtained for four positive ions and three negative ions using the linear ion trap
Figure 2 shows the phospholipid m/z range of averaged positive and negative mode spectra for the entire tissue surface after post-calibration with known background peaks. Peaks were assigned based on accurate mass data and comparison to known lipid compounds in mouse brain [9]. Comparison of the two spectra shows completely different spectral patterns, as expected. In positive ion mode, most lipid peaks are assigned to PC and SM species in the form of [M + H]+, [M + Na]+, and [M + K]+. Several fragment peaks are also observed, especially –N(CH3)3 loss from PC and/or SM, [M + H/K/Na – 59]+. Lipid species assigned in negative mode include [M − H]− species from PS, ST, PG, and PI. Observation of these lipid compounds is consistent with previous findings [14] and suggests both positive and negative ion spectra are necessary to reveal the distribution of various lipid compounds through MS imaging. Mass errors in the recalibrated averaged spectra are below 3 ppm for both positive ions and negative ions.
Phospholipid m/z range of averaged positive (above axis) and negative (below axis) spectra obtained from coronal mouse brain section showing assignments for selected lipid ions and deviations from calculated values. Spectra were recalibrated against background ions after acquisition. X indicates fragments such as the loss of N(CH3)3 and * represents matrix ion peaks
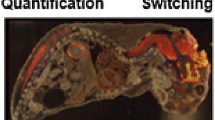
Figure 3 shows images for selected positive and negative ions. Distinct localizations are observed for a number of lipids across a range of classes, including cholesterol, PC, SM, PI, ST, and PS. The distributions of several ions correlate with anatomical structures. As an example, PI (18:0/20:4) is seen in higher abundance in gray matter tissue, while cholesterol is seen in higher abundance in white matter. These distributions are similar to those seen in SIMS imaging performed on similar mouse brain tissue sections by Sjovall et al. [20]. It should be noted that the analysis was performed without tissue washing to remove endogenous salts, and may result in slight differences in apparent distributions of some lipid signals [21]. MS/MS spectra support the assignments of peaks observed in the Orbitrap spectra (Supplementary Figure S1). Figure 3 shows MS/MS images for several structurally informative transitions, including neutral loss (NL) of 183 (phosphocholine headgroup) from PC (32:0), NL of 284 (18:0 fatty acid) from PC (36:1) + K, NL of 304 (20:4 fatty acid) from PI (18:0/20:4), and NL of 338 (C23H46O fragment of the h24:0 chain). Such fragmentation patterns can be helpful in conjunction with accurate mass data to identify unknowns and to distinguish potentially interfering lipids from different classes.
This experiment incorporated MS/MS scans at fixed m/z at seven of the nine spiral steps included in the method. This allows for the generation of images for specific CID fragmentation patterns (Figure 3). The method can be easily modified to provide different information. MS/MS scans can be replaced by MSn scans if further structural discrimination is necessary or ion trap MS scans for improved spatial resolution [6]. More importantly, we can perform MS/MS analysis of unknown compounds on-the-fly. This could be accomplished by utilizing the data-dependent scan function of Xcalibur, which selects a few of the most intense ions for MS/MS based on the initial FT scan. Combining this technique with the dynamic exclusion function, one could acquire thousands of MS/MS spectra in a single MS imaging experiment [6]. Namely, as MS imaging is being performed in high resolution MS, MS/MS spectra of thousands of the most intense ions at various locations can be acquired in-tandem for structural analysis later on. While data-dependent MS/MS is commonly adapted to identify peptides using MALDI-tandem mass spectrometry [22], its application to MS imaging is unique in our multiplex-MSI approach [6] and cannot be achieved by a simple back-to-back experiment.
One initial concern with the current approach is that switching the polarity of the Orbitrap high voltage may result in the loss of mass resolving power and/or mass accuracy. However, we found there is virtually no change in mass resolving power and the deviation in measured mass is easily correctable by recalibrating after acquisition. In ESI-Orbitrap, Schuhmann et al. found that mass accuracy is initially degraded with successive polarity switching but stabilizes over time, taking about 30 min to come down below 2 ppm [18]. The degraded mass error is attributed to the temporarily unstable operation of electronics [18]. We found the same behavior with polarity switching in MALDI-Orbitrap. The mass values, however, are stabilized with somewhat shifted mass errors and consistent even after several hours. The amount of mass shift was not consistent depending on the type of experiment, i.e. negative to positive ion mode and vice versa, or the number of MS/MS included. It was as little as a few ppm and as high as 30 ppm, but in any case it is always stabilized in about 30 min in the given experimental condition.
Figure 4 plots the observed mass errors of several ions across the tissue, before and after recalibration. Rastering was performed in the x-direction, so this plot represents mass deviations over the entire scan time, with ~30 to 100 polarity switches between each point, depending upon the raster direction. Recalibration was performed for the entire spectra, with positive and negative ions separately, as described in detail in the Experimental section. After recalibration, average root mean square errors for the plotted ions are 1.6 ppm for positive ions and 1.5 ppm for negative ions. The mass errors following this simple, two-point recalibration are comparable to the instrument’s performance without polarity switching.
Mass errors of selected compounds across the tissue along the arrow shown on the left, before and after a two-point mass recalibration. Orange-red symbols indicate positive ions: (●) m/z 369.351, [cholesterol – H2O + H]+; (▲) m/z 577.519, [34:1 diacylglycerol fragment + H]+; (♦) m/z 772.525, [PC (32:0) + K]+; (×) m/z 798.541, [PC (34:1) + K]+; green-blue symbols indicate negative ions: (+) m/z 465.044, [3DHB – H2O – 2H + Na]−; (♦) m/z 806.548, [ST (18:0) – H]−; (×) m/z 888.624, [ST (24:1) – H]−
The greatest limitation in the current multiplex imaging with polarity switching method is the increase in data acquisition time. The total data acquisition time for a 5 × 8 mm2 rat brain tissue with 100 μm raster steps and nine 25 μm spiral steps is 7.6 h without polarity switching (i.e., spiral steps 6–9 also in positive ion mode); with polarity switching, the same acquisition takes about 23.9 h using the original data acquisition software. The additional 16.3 h are the result of a software-imposed delay of ~7 s for each polarity switch. Some delay time is necessary to turn-off the high voltage on Orbitrap and turn it back on in the opposite polarity. The physical limit of this time delay is about 3 s in the current design (private communication with the manufacturer). A software patch was generously provided by Thermo Fisher Scientific to reduce the delay time to ~5 s. The data presented in this work was obtained with the new software patch and we were able to reduce the dead time to 10.8 h, with the total data acquisition time of 18.4 h. This is a slightly excessive data acquisition time compared to typical imaging experiments, but it is still within the manageable time frame and we could routinely run the polarity switching experiment without any problem.
An ultimate solution to this issue would be the use of dual high voltage supplies. The two high voltage supplies can be maintained at stable voltages in opposite polarities. Polarity switching is then accomplished by simply switching the high voltage supply from one to the other in <0.25 s. This approach has already been adapted in a commercial ESI-Orbitrap mass spectrometer (Orbitrap Exactive Plus), which allows acquisition of a set of orbitrap spectra in both positive and negative ion mode in less than 1 s without compromising mass accuracy (http://www.thermo.com/eThermo/CMA/PDFs/Articles/articlesFile_7841.pdf). Incorporation of this dual supply system would eliminate the need for a delay between polarity switching and acquisition, and significantly shorten the time required for the described experiment.
4 Conclusion
A multiplex MALDI-MSI method was adapted to allow for collection of positive and negative ion mass spectrometry images from a single tissue section in a single experiment. Complementary lipid classes were observed in positive and negative mode, and MS/MS data acquisition was incorporated in both polarities. Orbitrap mass values were reproducible across the whole imaging experiment, even after over 10,000 polarity changes, with systematic mass shifts for positive and negative ions. A simple two-point post-calibration to background ion signals yielded RMS mass errors that averaged <2 ppm, enabling accurate assignments of most lipid compounds. The current limitation is the long data collection time, as a result of the dead time for high voltage stabilization after each polarity switching. However, the data acquisition could still be performed within manageable time frame, ~15 h, and we could routinely run these experiments without any problem.
The proposed approach has pros and cons compared to acquiring separate MS images for positive and negative ions in a back-to-back experiment. If one simply wants precursor MS data in the two polarities, two separate experiments might be easier and could be done in a shorter time. However, the proposed multiplex MS imaging can easily incorporate MS/MS or MS3 scans in both polarities into a single experimental scheme. For example, the application demonstrated in this work corresponds to nine separate MS imaging experiments, which is not trivial to be performed in a back-to-back experiment on a single tissue because the experimental set-up would need to be made very carefully to avoid any possible overlapping between each imaging acquisition. Additionally, manual intervention between positive and negative data acquisition is essential to allow complete stabilization, typically requiring 2 h, after the polarity switching. In contrast, multiplex MS imaging is rather straightforward to set up and once started runs without further intervention. Furthermore, incorporation of data-dependent MS/MS acquisition during MS imaging is not plausible in a back-to-back experiment. The primary drawback is the longer acquisition time due to the delay between each switching. Considering the facts discussed above, we expect multiplex MS imaging with polarity switching could be quite useful in many applications.
References
Svatos, A.: Mass spectrometric imaging of small molecules. Trends Biotechnol. 28, 425–434 (2010)
Burnum, K.E., Frappier, S.L., Caprioli, R.M.: Matrix-assisted laser desorption/ionization imaging mass spectrometry for the investigation of proteins and peptides. Annu. Rev. Anal. Chem. 1, 689–705 (2008)
Goto-Inoue, N., Hayasaka, T., Zaima, N., Setou, M.: Imaging mass spectrometry for lipidomics. Biochim. Biophys. Acta 1811, 961–969 (2011)
Smith, D.F., Aizikov, K., Duursma, M.C., Giskes, F., Spaanderman, D.-J., McDonnell, L.A., O’Conner, P.B., Heeren, R.M.A.: An external matrix-assisted laser desorption ionization source for flexible FT-ICR mass spectrometry imaging with internal calibration on adjacent samples. J. Am. Soc. Mass Spectrom. 22, 130–137 (2011)
Schober, Y., Schramm, T., Spengler, B., Römpp, A.: Protein identification by accurate mass matrix-assisted laser desorption/ionization imaging of tryptic peptides. Rapid Commun. Mass Spectrom. 25, 2475–2483 (2011)
Perdian, D.C., Lee, Y.J.: Imaging MS methodology for more chemical information in less data acquisition time utilizing a hybrid linear ion trap-Orbitrap mass spectrometer. Anal. Chem. 82, 9393–9400 (2010)
Lunsford, K.A., Peter, G.F., Yost, R.A.: Direct matrix-assisted laser desorption/ionization mass spectrometric imaging of cellulose and hemicellulose in populus tissue. Anal. Chem. 83, 6722–6730 (2011)
Cerruti, C.D., Benabdellah, F., Laprevote, O., Touboul, D., Brunelle, A.: MALDI imaging and structural analysis of rat brain lipid negative ions with 9-aminoacridine matrix. Anal. Chem. 84, 2164–2171 (2012)
Gode, D., Volmer, D.A.: Lipid imaging by mass spectrometry: a review. Analyst 138, 1289–1315 (2013)
Berry, K.A., Hankin, J.A., Barkley, R.M., Spraggins, J.M., Caprioli, R.M., Murphy, R.C.: MALDI imaging of lipid biochemistry in tissues by mass spectrometry. Chem. Rev. 111, 6491–6512 (2011)
Eberlin, L.S., Ferreira, C.R., Dill, A.L., Ifa, D.R., Cooks, R.G.: Desorption electrospray ionization mass spectrometry for lipid characterization and biological tissue imaging. Biochim. Biophys. Acta 1811, 946–950 (2011)
Passarelli, M.K., Winograd, N.: Lipid imaging with time-of-flight secondary ion mass spectrometry (TOF-SIMS). Biochim. Biophys. Acta 1811, 976–990 (2011)
Fletcher, J.S., Vickerman, J.C., Winograd, N.: Label free biochemical 2D and 3D imaging using secondary ion mass spectrometry. Curr. Opin. Chem. Biol. 15, 733–740 (2011)
Thomas, A., Charbonneau, J.L., Fournaise, E., Chaurand, P.: Sublimation of new matrix candidates for high spatial resolution imaging mass spectrometry of lipids: enhanced information in both positive and negative polarities after 1,5-diaminonaphthalene deposition. Anal. Chem. 84, 2048–2054 (2012)
Benabdellah, F., Seyer, A., Quinton, L., Touboul, D., Brunelle, A., Laprevote, O.: Mass spectrometry imaging of rat brain sections: nanomolar sensitivity with MALDI versus nanometer resolution by TOF-SIMS. Anal. Bioanal. Chem. 396, 151–162 (2010)
Leefmann, T., Heim, C., Siljeström, S., Blumenberg, M., Sjövall, P., Thiel, V.: Spectral characterization of 10 cyclic lipids using time-of-flight secondary ion mass spectrometry. Rapid Commun. Mass Spectrom. 27, 565–581 (2013)
Solé-Domènech, S., Sjövall, P., Vukojević, V., Fernando, R., Codita, A., Salve, S., Bogdanović, N., Mohammed, A.H., Hammarström, P., Nilsson, K.P., LaFerla, F.M., Jacob, S., Berggren, P.O., Giménez-Llort, L., Schalling, M., Terenius, L., Johansson, B.: Localization of cholesterol, amyloid and glia in Alzheimer’s disease transgenic mouse brain tissue using time-of-flight secondary ion mass spectrometry (TOF-SIMS) and immunofluorescence imaging. Acta Neuropathol. 125, 145–157 (2013)
Schuhmann, K., Almeida, R., Baumert, M., Herzog, R., Bornstein, S.R., Shevchenko, A.: Shotgun lipidomics on a LTQ Orbitrap mass spectrometer by successive switching between acquisition polarity modes. J. Mass Spectrom. 47, 96–104 (2012)
Hankin, J.A., Barkley, R.M., Murphy, R.C.: Sublimation as a method of matrix application for mass spectrometric imaging. J. Am. Soc. Mass Spectrom. 18, 1646–1652 (2007)
Sjovall, P., Lausmaa, J., Johannson, B.: Mass spectrometric imaging of lipids in brain tissue. Anal. Chem. 76, 4271–4278 (2004)
Hankin, J.A., Farias, S.E., Barkley, R.M., Heidenreich, K., Frey, L.C., Hamazaki, K., Kim, H.-Y., Murphy, R.C.: MALDI mass spectrometric imaging of lipids in rat brain injury models. J. Am. Soc. Mass Spectrom. 22, 1014–1021 (2011)
Martha, D.S., Douglas, F.B.: Complementary use of MALDI and ESI for the HPLC-MS/MS analysis of DNA-binding proteins. Anal. Chem. 76, 5423–5430 (2004)
Acknowledgments
The authors thank Emile de Leeuw at Thermo Fisher Scientific for providing them a software patch and for helpful conversations. This work was supported by the US Department of Energy (DOE), Office of Basic Energy Sciences, Division of Chemical Sciences, Geosciences, and Biosciences. The Ames Laboratory is operated by Iowa State University under DOE Contract DE-AC02-07CH11358.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Figure S1
MS/MS spectra used to generate MS/MS images in Figure 3. MS/MS of [PC (32:0) + K]+ (a), MS/MS of [PC (36:1) + K]+ (b), MS/MS of [PI (18:0/20:4) – H]− (c) and MS/MS of [ST (d18:1/h24:0) – H]− (d). Transitions used to generate images are labeled. m/z 886.7 in (c) is an isotope of precursor (m/z 885.5) that was included as a result of incomplete isolation but did not completely fragment because its secular frequency is slightly off from the activation frequency (PDF 129 kb) (PDF 129 kb)
Rights and permissions
About this article
Cite this article
Korte, A.R., Lee, Y.J. Multiplex Mass Spectrometric Imaging with Polarity Switching for Concurrent Acquisition of Positive and Negative Ion Images. J. Am. Soc. Mass Spectrom. 24, 949–955 (2013). https://doi.org/10.1007/s13361-013-0613-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13361-013-0613-1