Abstract
The performance of 10 clones of the grain aphid Sitobion avenae Fabricius was measured over three generations on three host plants (wheat, bluegrass and ryegrass). The tested clones belonged to six microsatellite genotypes; two genotypes were represented by three clonal lines each that had been collected from different host plants. The clones varied in body color and in the secondary endosymbionts they possessed (Hamiltonella, Regiella or none). The performance of aphids on host plants declined in the order wheat > bluegrass > ryegrass independently of the plant from which they were collected. We also found differences in performance among genotypes and clones of the aphids and among the generations at which the performance was measured. The performance was not affected by the collection site, clone’s original host plant, body color or the presence of endosymbionts.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The population dynamics and genetic analysis of aphids revealed specialization or adaptation in many economically important aphid species (Wilson et al. 2003). For example, the pea aphid (Acyrthosiphon pisum Harris) consists of three specialized clones in Chile (Peccoud et al. 2008) and two specialized clones in North America (Via 1999), while the black bean aphid (Aphis fabae Scopoli) consists of a complex of closely related host plant-associated genotypes (Tosh et al. 2004).
Aphids have a remarkable life cycle which frequently includes a parthenogenetic and a sexual generation, an obligate shift between unrelated host plant taxa, a short generation time, and telescoping of generations, where granddaughters begin to develop directly within the daughters which are themselves not yet born (Dixon 1998). Moreover, it is characteristic for aphids that one parthenogenetic female may give rise to up to eight discrete phenotypes differing in their physiology, morphology, developmental time, longevity, number of progeny, timing of reproduction and fitness, host preferences and ability to use alternative host-plant taxa (Moran 1992). In addition, aphids harbor secondary bacterial endosymbionts, which may contribute to their specialization in respect to host plants and to the way host plants use them (Chen et al. 2000; Tsuchida et al. 2004).
The English grain aphid, Sitobion avenae (F.) is an important pest of cereals worldwide (Wangai et al. 2000). S. avenae is an autoecious aphid restricted to Poaceae and possesses all the life cycles known in aphids (Helden and Dixon 2002). Clones that produce both sexual and asexual morphs are holocylic; they reproduce parthenogenetically for many generations and have just one sexual generation in a year. When a clone produces only asexual morphs, it is anholocyclic; and in this case, they only reproduce parthenogenetically. There are also androcyclic clones, which are the same as anholocyclic, but they produce males in the autumn. There are also intermediate clones, which can be holo- or anholocyclic.
Studies on the genetic structure of S. avenae have found a high genetic variability with spatial and temporal variation and the existence of predominant cyclic parthenogenesis clones (common clones) and host plant adapted clones (De Barro et al. 1995; Haack et al. 2000; Papura et al. 2003; Reimer 2005; Simon et al. 1996; Sunnucks et al. 1997). Also, recent studies in China have indicated the existence of specialized clones (Dai et al. 2014; Gao et al. 2014). S. avenae can harbor secondary bacterial endosymbionts (Alkhedir et al. 2013; Lukasik et al. 2013), but their effects on the clones’ specialization in respect to host plants, and on host plant use, have not yet been adequately investigated.
Previous studies (Dai et al. 2014; De Barro et al. 1995; Gao et al. 2014; Haack et al. 2000; Papura et al. 2003; Reimer 2005; Simon et al. 1996; Sunnucks et al. 1997) considered the original host of the clone and the collection site as the criteria to determine the degree of adaptation and the specialization of S. avenae clones. However, the factors that determine the specialization and fitness of S. avenae clones or other aphid species in respect to their host plants are not understood.
In this study, we investigated the factors that might affect the specialization of the common S. avenae clones in Germany as well as their use of host plants. Such factors include the original host of the clone, collecting site, and body color, in addition to clonal and bacterial diversity, taking into account the life cycle of S. avenae and the fact that even nymphs born from a parthenogenetic mother can be polymorphic. We evaluated the fitness parameters of ten S. avenae clones that were related to most common genotypes in Germany in respect to three host plants in order to assess the factors that influenced the host plant use, specialization and temporal abundance of these clones.
Materials and methods
Aphid and plant cultures
The ten clones (Table 1) used in this study were collected in 2004 (Alkhedir et al. 2010) and they were genotyped and selected based on five microsatellite loci (Alkhedir et al. 2013). The clones were abundant on wheat in central Germany and represented the six most commonly occurring genotypes in central Germany (Reimer 2005). The clones were collected from different fields to ensure variability.
The aphid cultures were kept on the winter wheat cultivar ‘Bussard’ in rearing cabinets at 20 °C, 16:8 h light/dark conditions and 60–80 % humidity, with water being applied twice a week. The cultures were maintained by transferring new-born aphids of each tested clone to new seedlings. We set the light intensity in the rearing cabinets to 200 µE at which the aphids separated into green color morphs (clones 7 and 8) and brown morphs (the rest of the tested clones). In addition, under this light intensity, the clones have their highest fitness and produce less alatae (Alkhedir et al. 2010). All the clones reproduced parthenogenetically under these conditions. The clones were used in the experiments 6 months after collection.
The following cultivated host plant species were used in the experimental setup: winter wheat cultivar “Bussard”, ryegrass (Lolium perenne L. cv “Herault”) and bluegrass (Poa annua L., unknown cultivar). Ryegrass and bluegrass were selected because they are grown as forage in the collecting area. Seven-day-old seedlings were used.
Clonal performance of S. avenae on the tested host plants
Before the start of the experiments, we performed a set of primary experiments (not shown) under the tested conditions (see preceding paragraph). It was found that all the clones needed between 5 and 7 days to develop from first nymph stage to adults. Having tried different rearing systems, it was decided to use a system based on caging the aphid and plants with transparent ventilated 10 cm × 30 cm cylindrical tubes, as described by Alkhedir et al. (2013), since using small cages or petri-dishes resulted in significant mortality among the nymphs and also the production of more alatae.
In order to exclude the effect of any reduction in host quality and short photoperiods (Dixon 1998) as well as the effect of the crowding of both adults and offspring (Watt and Dixon 1981) on the fitness parameters of S. avenae, host plants of the same age were used and the aphid cultures were maintained every 2 weeks.
The experiments were started with ten nymphs and replicated 10 times for each clone on each of the tested host plants in order to simulate natural conditions and to minimize the effect of polymorphism. The fitness parameters were estimated at intervals of 2 weeks, which is equal to one generation, in order to investigate the reasons behind the temporal abundance of the tested clones in central Germany as reported by Reimer (2005). Two weeks was also the interval between collection times of the clones (Reimer 2005).
The fitness parameters of the ten aphid clones on the tested host plants used in this study were assessed by quantifying the population size (produced offspring), fresh weight, number of alatae, and survival of aphids after 2 weeks following the introduction of ten synchronized first stage nymphs on the respective host plants. This experiment was continued over three consecutive generations. Rearing of the nymphs followed the protocol described above.
Ten aphids from each clone on each host plant at each generation were genotyped to check the genetic identity of the clones during the course of the experiments. These were tested first at the time of collection and then at six-month intervals on all host plants over three successive generations.
Statistical analysis
Analysis of variance was used to analyze the performance of the clones, with population size, number of alatae, surviving aphids and the fresh weight of the population being considered as the dependent variables; and the clone, host plants, genotype, the original host of the clone, collection site, body color and symbionts being considered as the independent variables. Fisher’s LSD adjustment was used to compare the clonal performance on each host plant species. Systat for Windows, version 11.00.01 (SYSTAT 2004) was used to perform these analyses.
Results
Clonal performance of S. avenae
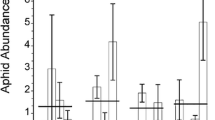
The clonal performance (fitness parameters) was significantly affected by the following factors: the host plant (Fig. 1) (F 2,897 = 327,601, p < 0.001), the genotypes (F 5,894 = 7.635, p < 0.001), the clones (F 9,890 = 4.322, p < 0.001), and the number of generations (F 2,897 = 33.546, p < 0.001) (Figs. 2, 3, 4, 5). However, neither the site at which the clones were collected nor the clone-origin’s host affected the performance of the clones (F 2,897 = 1,782, p > 0.05; F 2,897 = 2,313, p > 0.05, respectively). In addition, neither body color (F 1,898 = 10.879, p > 0.05) nor the performance of the tested genotypes were affected by the symbiotic bacteria (F 2,890 = 0.207, p > 0.05).
The average number of offspring produced by all the Sitobion avenae clones tested was higher on wheat compared to bluegrass and ryegrass (Fig. 1). Moreover, all aphids had a higher weight on wheat compared to bluegrass and ryegrass (Fig. 1). In contrast, all clones produced more alatae on bluegrass compared to ryegrass and wheat (Fig. 1), while more individuals survived on wheat compared to bluegrass and ryegrass (Fig. 1).
The tested S. avenae clones varied in the number of offspring (Fig. 2) and in their fresh weight (Fig. 3) over three successive generations. The number of offspring was highly correlated with fresh weight (p < 0.001; R 2 = 0.965). S. avenae clones showed large variation in alatae production (Fig. 4) but less variation in survival rates over three generations (Fig. 5).
Discussion
Polymorphism of S. avenae clones
Sitobion avenae clones were reared for 6 months on wheat prior to the experiments, thus the variance in fitness parameters was not caused by a sudden change of host plants or by environmental conditions. Rather, it can be ascribed to the genetic make-up and intrinsic polymorphism of the aphids. Reimer (2005) evaluated the temporal abundance of the clones in the field at three collection times with an interval of 2 weeks. In this study, we estimated the population size at the same intervals under constant conditions in the laboratory. The population size of the clones varied over three consecutive generations (Fig. 2). For example, clones 4, 5 and 6 on wheat produced their largest populations in their third generations, while clone 9 had the largest population in the second generation. The experimental conditions were controlled and kept constant; furthermore, the way the performance changed in consecutive generations differed among clones, showing that a systematic effect of experimental conditions can be excluded. We conclude that estimation of fitness parameters for the assessment of adaptation or specialization in aphids over a single generation is insufficient. Furthermore, our results show that variation of environmental conditions alone cannot fully explain temporal variations in the abundance of aphids in the field.
Host specialization in S. avenae
Traditionally, DNA microsatellites are used to assign S. avenae samples to clones (Dai et al. 2014; De Barro et al. 1995; Figueroa et al. 2005; Gao et al. 2014; Haack et al. 2000; Vialatte et al. 2005; Weber 1985; Wilson et al. 1997). Adaptation of the clones of S. avenae to host plants is defined by the host from which they were collected, which are mainly maize, wheat, barley, cocksfoot and other graminoides (Dai et al. 2014; De Barro et al. 1995; Figueroa et al. 2005; Gao et al. 2014; Haack et al. 2000; Vialatte et al. 2005; Weber 1985; Wilson et al. 1997). Microsatellite analysis and the plant from which an aphid was collected, however, do not reveal whether a clone is common and to which host it is specialized. Especially if the interaction between aphids and their host plants is determined by genes responsible for host plant resistance, as in the case of Schizaphis graminum (Ono et al. 1999), clonal performance on resistant and susceptible varieties of the host plants should be studied. In our study, we did not find any impact of the clone’s original host on its adaptation to tested host plants.
Clones of S. avenae that are restricted to one host plant were only found on the cocksfoot cultivar “Prairial” (De Barro et al. 1995; Sunnucks et al. 1997). Recent studies showed that the specialization on cocksfoot depends on the content of water-soluble carbohydrates (Alkhedir et al. 2013). Other cases of restriction of S. avenae to host plants are not known. The term “specialized clones” should be used with caution in respect to aphids; the term “adapted clones” might be a better way of expressing the fact that the clones have a better performance on certain host plants than on others.
Effect of the body color of S. avenae
We did not find any difference between green and brown clones in respect to their host plant use, which agrees with our previous results (Alkhedir et al. 2010). An exception is cocksfoot; green phenotypes used in this study were well adapted to cocksfoot varieties with low soluble carbohydrates content (Alkhedir et al. 2013).
Clonal and endosymbiont diversity of S. avenae
Genetic variability of Sitobion avenae was considered as the main source of differences among clones not only in this study but also in all relevant studies that we are aware of. In our study, however, we could not find any effect of clonal identity on host plant use on any tested host plant species. The genetic diversity among endosymbionts of S. avenae clones used in this study was very low (Alkhedir et al. 2015). Although the effect of endosymbionts on host plant use cannot be conclusively excluded, our results support the report by Lukasik et al. (2013) who did not find any effect of secondary endosymbionts on host plant’s use by S. avenae in England.
Conclusions
Host plant origin, clonal identity and color of aphid do not affect the host plant specialization in S. avenae. All clones in this study performed best on wheat.
Estimation of performance of S. avenae requires monitoring several generations.
References
Alkhedir H, Karlovsky P, Vidal S (2010) Effect of light intensity on colour morph formation and performance of the grain aphid Sitobion avenae F (Homoptera: Aphididae). J Insect Physiol 56:1999–2005
Alkhedir H, Karlovsky P, Vidal S (2013) Relationship between water soluble carbohydrate content, aphid endosymbionts and clonal performance of Sitobion avenae on cocksfoot cultivars. PLoS One 8:e54327
Alkhedir H, Karlovsky P, Mashaly AMA, Vidal S (2015) Phylogenetic relationships of the symbiotic bacteria in the aphid Sitobion avenae (Hemiptera: Aphididae). Environ Entomol. doi:10.1093/ee/nvv114
Chen DQ, Montllor CB, Purcell AH (2000) Fitness effects of two facultative endosymbiotic bacteria on the pea aphid Acyrthosiphon pisum and the blue alfalfa aphid A. kondoi. Entomol Exp Appl 95:315–323
Dai X, Gao S, Liu D (2014) Genetic basis and selection for life-history trait plasticity on alternative host plants for the cereal aphid Sitobion avenae. PLoS One 9:e106179
De Barro PJ, Sherratt TN, David O, Maclean N (1995) An investigation of the differential performance of clones of the aphid Sitobion avenae on two host species. Oecologia 104:379–385
Dixon AFG (1998) Aphid ecology. Chapman & Hall, London
Figueroa CC, Simon JC, Le Gallic JF, Prunier-Leterme N, Briones LM, Dedryver CA, Niemeyer HM (2005) Genetic structure and clonal diversity of an introduced pest in Chile, the cereal aphid Sitobion avenae. Heredity 95:24–33
Gao S-X, Liu D-G, Chen H, Meng X-X (2014) Fitness traits and underlying genetic variation related to host plant specialization in the aphid Sitobion avenae. Insect Sci 21:352–362
Haack L, Simon JC, Gauthier JP, Plantegenest M, Dedryver CA (2000) Evidence for predominant clones in a cyclically parthenogenetic organism provided by combined demographic and genetic analyses. Mol Ecol 9:2055–2066
Helden AJ, Dixon AFG (2002) Life-cycle variation in the aphid Sitobion avenae: Costs and benefits of male production. Ecol Entomol 27:692–701
Lukasik P, Dawid MA, Ferrari J, Godfray HC (2013) The diversity and fitness effects of infection with facultative endosymbionts in the grain aphid Sitobion avenae. Oecologia
Moran NA (1992) The evolution of aphid life cycles. Annu Rev Entomol 37:321–348
Ono M, Swanson JJ, Field LM, Devonshire AL, Siegfried BD (1999) Amplification and methylation of an esterase gene associated with insecticide-resistance in greenbugs Schizaphis graminum (Rondani) (Homoptera: Aphididae). Insect Biochem Mol Biol 29:1065–1073
Papura D, Simon JC, Halkett F, Delmotte F, Le Gallic JF, Dedryver CA (2003) Predominance of sexual reproduction in Romanian populations of the aphid Sitobion avenae inferred from phenotypic and genetic structure. Heredity 90:397–404
Peccoud J, Figueroa CC, Silva AX, Ramirez CC, Mieuzet L, Bonhomme J, Stoeckel S, Plantegenest M, Simon JC (2008) Host range expansion of an introduced insect pest through multiple colonizations of specialized clones. Mol Ecol 17:4608–4618
Reimer L (2005) Clonal diversity and population genetic structure of the grain aphid Sitobion avenae (F) in central Europe. PhD thesis, University of Goettingen
Simon JC, Martinez-Torres D, Latorre A, Moya A, Hebert PD (1996) Molecular characterization of cyclic and obligate parthenogens in the aphid Rhopalosiphum padi (L). Proc Biol Sci 263:481–486
Sunnucks P, De Barro PJ, Lushai G, Maclean N, Hales D (1997) Genetic structure of an aphid studied using microsatellites: cyclic parthenogenesis differentiated lineages and host specialization. Mol Ecol 6:1059–1073
Systat (2004) Release 110001 Systat Software Inc 1735 Technology Drive Ste 430 San Jose CA 95110
Tosh CR, Vamvatsikos PG, Hardie J (2004) A highly viable cross between Aphis fabae (Homoptera: aphididae) clones with different plant preference. Environ Entomol 33:1081–1087
Tsuchida T, Koga R, Fukatsu T (2004) Host plant specialization governed by facultative symbiont. Science 303:1989
Via S (1999) Reproductive isolation between sympatric races of pea aphids. I.Gene flow restriction and habitat choice. Evolution 53:1446–1457
Vialatte A, Dedryver CA, Simon JC, Galman M, Plantegenest M (2005) Limited genetic exchanges between populations of an insect pest living on uncultivated and related cultivated host plants. Proc R Soc B Biol Sci 272:1075–1082
Wangai AW, Plumb RT, Van Emden HF (2000) Effects of sowing date and insecticides on cereal aphid populations and barley yellow dwarf virus on barley in Kenya. J Phytopathol 148:33–37
Watt AD, Dixon AF (1981) The role of cereal growth stages and crowding in the induction of alatae in Sitobion avenae and its consequences for population growth. Ecol Entomol 6:441–447
Weber G (1985) On the ecological genetics of Sitobion avenae (F) (Hemiptera Aphididae). J Appl Entomol 100:100–108
Wilson ACC, Sunnucks P, Hales DF (1997) Random loss of X chromosome at male determination in an aphid Sitobion near fragariae detected using an X-linked polymorphic microsatellite marker. Genet Res 69:233–236
Wilson ACC, Sunnucks P, Hales DF (2003) Heritable genetic variation and potential for adaptive evolution in asexual aphids (Aphidoidea). Biol J Linn Soc 79:115–135
Acknowledgments
The authors extend their appreciation to the Deanship of Scientific Research at King Saud University for funding the work through the research group project no. RGPVPP-028. In addition, the authors thank the anonymous reviewer for their reading of our manuscript and their helpful comments and suggestions.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
All authors declare no conflict of interests.
Rights and permissions
About this article
Cite this article
Alkhedir, H., Karlovsky, P., Mashaly, A.M.A. et al. Specialization and host plant use of the common clones of Sitobion avenae (Homoptera: Aphididae). Appl Entomol Zool 51, 289–295 (2016). https://doi.org/10.1007/s13355-016-0400-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13355-016-0400-0