Abstract
Tomato spotted wilt orthotospovirus (TSWV) is one of the most widespread and damaging pepper viruses worldwide. Until recently, disease control has been achieved by breeding pepper cultivars with the resistance gene Tsw. However, the breakdown of Tsw-resistance gene has been increasingly reported from many locations where resistant cultivars have been grown. This has been caused by the emergence of resistance-breaking isolates because of the high evolutionary and adaptative capacity of TSWV. The resurgence of RB TSWV-strains causing heavy production losses in pepper (Capsicum annuum) plants in Turkey has directed the attention to the factors contributing to the outbreak of these serious epidemics. In this paper, we report the first detailed molecular and biological characterization of TSWV-isolates in the field and greenhouse-grown pepper plants. Biological clones from TSWV field samples segregated into three different types of symptoms on pepper plants carrying the Tsw-resistance gene; (i) only local lesions as a result of hypersensitive reaction, (ii) chlorotic rings, chlorosis, vein clearing, downward leaf curling, and stunting, (iii) severe systemic necrosis and wilting leading to the death of resistant plants. The phylogenetic analysis of the NSs gene has shown that the isolates were clustered regardless of the phenotype RB or non-resistance breaking (NRB). Our study has also supported the hypothesis that developing and expanding the RB isolates has occurred locally. The phylogenetic analyses on TSWV-pepper isolates have revealed the fact for the presence of reassortment in TSWV population which might be caused by a mixed infection of TSWV variants.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Tomato spotted wilt orthotospovirus (TSWV) in the genus Orthotospovirus (Tospoviridae) ranks second among the ten most scientifically or economically crucial plant viruses (Scholthof et al. 2011). It affects more than 1090 different plant species belonging to over 90 monocotyledonous and dicotyledonous plant families (Parrella et al. 2003; Pappu et al. 2009). The virus induces various symptoms in its hosts such as chlorosis, necrosis, ringspot, stunting, streak, ring spot on leaves, stems, and fruits in some cases may lead to the death of plants depending on the growth stage of plants, environmental conditions, and virus strain (Francki and Hatta 1981; Soler et al. 1998; Adkins 2000; Mandal et al. 2006). TSWV infection leads to reduce in annual yield, and symptoms drastically reduce the marketing value of fruits (Griep et al. 2000; McPherson et al. 2003; Riley et al. 2011). Due to having wide host ranges, and the ability to evolve in such a way that it is troublesome to control virus infectious in the fields (Whitfield et al. 2005; Persley et al. 2006; Moury and Verdin 2012).
TSWV has a complex structure compared to most other plant viruses. Like other members of the Tospoviridae family, TSWV are spherical, enveloped viruses, with three single-stranded genomic RNA molecules associated with a nucleocapsid protein (Goldbach and Peters 1996; Cortez et al. 2001). The L (large) RNA segment is negative-sense and encodes RNA-dependent RNA polymerase (RdRp) protein (de Haan et al. 1991). The M (medium) RNA segment is ambisense and has two open reading frames encoding proteins: the non-structural protein NSm in the viral sense, and the Gn/Gc precursor glycoprotein in the viral complementary (vc) sense, play important roles in cell-to-cell movement in the host and in vector transmission, respectively (Sin et al. 2005; Naidu et al. 2008). The S (small) RNA segment is also ambisense and encoding proteins in opposite orientation: in the viral sense for the non-structural protein NSs and the nucleocapsid (N) protein in the viral complementary sense (Pappu et al. 2020). The NSs protein has been identified as an avirulence (avr) and RNA silencing suppressor (RSS) protein (Takeda et al. 2002; de Ronde et al. 2014).
Pepper (Capsicum annuum L.) is one of the most important vegetables for Turkey due to having immense export potential. The first reports for the TSWV infections on pepper were made from the eastern Mediterranean in 1998 (Yurtmen et al. 1998) later from the Black Sea (Arlı-Sökmen et al. 2005) and western Mediterranean (Güneş and Gümüş 2019) as well as on other crops from various regions (Tekinel et al. 1969; Tekinel 1973; Azeri 1981; Arlı-Sökmen et al. 2005; Kamberoğlu et al. 2009; Sertkaya 2015; Fidan et al. 2016). The most effective control against this pathogen seemed to be the production of resistant cultivars. Concerning pepper, Tsw from C. chinense is the single dominant resistance gene that provides hypersensitivity (HR) resistance to TSWV in peppers and is introgressed to commercial cultivars (Black et al. 1991; Boiteux 1995; Moury et al. 1997; Pappu et al. 2009). However, Tsw resistance-breaking isolates (TRB) have been reported in Brazil (Boiteux and Nagata 1993), the USA (Hobbs et al. 1994; Black et al. 1996; Macedo et al. 2019), Italy (Roggero et al. 2002), Australia (Thomas-Carrol and Jones 2003; Sharman and Persley 2006), Spain (Margaria et al. 2004), Korea (Chung et al. 2012; Hoang et al. 2013), and Turkey (Deligöz et al. 2014). The tremendous ability of the virus to generate new virulent isolates has imposed the need to seek and evaluate new sources of resistance or tolerance to TSWV. Some studies in Turkey had focused on using the Tsw gene to develop TSWV resistant pepper lines (Şimşek et al. 2015; Çelik et al. 2018). Developing efficient management strategies against TSWV requires firstly determination of the isolates which present in the critical growing regions and their relation with the Tsw gene. In this study, the biological characterization of TSWV-pepper isolates was done, and four protein-coding regions (RdRp, NSm, NSs, and N) were sequenced. A phylogenetic study was performed to compare Turkish TSWV-pepper isolates with TSWV isolates from different geographical regions to understand sequence variability and population diversity.
Materials and methods
Virus isolates and DAS-ELISA
Leaf and fruit samples from pepper (C. annuum) plants with or without the Tsw gene showing TSWV-like symptoms (Fig. 1) were collected from commercial greenhouses and open fields in the Eastern Mediterranean region of Turkey (Mersin, Adana, Kahramanmaraş) during 2017–2019. In total, 300 samples were tested by DAS-ELISA (Clark and Adams 1977) for TSWV infection, using a specific polyclonal antiserum (Loewe Biochemica GmbH, Germany) to select virus isolates. The samples were also checked for Cucumber mosaic virus (CMV), Tobacco mosaic virus (TMV) and Potato virus Y (PVY), using specific antisera to avoid the risk of mixed infection with the most frequent viruses found in pepper crops of the surveyed area.
Biological characterization
The 14 field samples resulting single TSWV infection were mechanically inoculated on Nicotiana benthamiana to increase the virus titer, then transferred into Vigna unguiculata to obtain a homogenous intra-isolate population of genetic variants (Moury et al. 1997). Thirty biological clones were obtained after three serial passages of a single local lesions with subsequent subculture. These TSWV clones were subsequently amplified in N. benthamiana plants, later mechanically inoculated onto N. tabacum cv. Samsun NN as well as susceptible C. annuum cv. ‘Naz F1’(Eastern seeds, Turkey), TSWV resistant C. chinense accession ‘PI 152225’ (Black et al. 1991), and C. annuum cv. ‘Mertcan F1’ (Yüksel seeds, Turkey) to evaluate the ability to overcome the resistance provided by the Tsw gene. Mechanical inoculation was performed on seedlings at the 2–4 leaf stage by rubbing infected leaf extracts prepared in four volumes of 0.03 M phosphate buffer (pH 7.0) containing 2% (wt/vol) diethyldithiocarbamate, active charcoal at 20 mg/ml, and carborundum. Negative control plants were prepared with mock inoculation and carborundum. Each of plant species had three replicates. Inoculated plants were kept in a growth chamber with a 16 h photoperiod and constant temperature of 25 °C, and the symptom expression was monitored for four weeks after inoculation. ELISA test was run with inoculated and uninoculated upper leaves up to 15 and 30 days post-inoculation (dpi).
Biological clones amplification and sequencing
N. benthamiana plants with increased TSWV viral titers, after local lesion purification on V. unguiculata plants, used to extract total RNA using TRIzol-based method described by Chomczynski and Sacchi (1987) with slight modifications. Furthermore, RNAs were reverse-transcribed using M-MLV reverse transcriptase (Promega) with the specific reverse primer for the synthesis of the first-strand cDNA. PCR step took place with the primer sets to amplify part of RdRp, NSm, N, and NSs genes (Debreczeni et al. 2015; Tentchev et al. 2011) and Taq DNA polymerase (Thermo Fisher Scientific). PCR products were separated by electrophoresis in agarose gels, stained with ethidium bromide, and the DNAs was visualized under UV lighting. PCR amplicons were directly sequenced by the Sanger method with both primers in two directions (Medsantek, Turkey), and obtained a consensus sequence for each isolate. The nucleotide sequences of 950 nt (position 5443–6392), 771 nt (position 101–871), 846 nt (position 559–1404), 777 nt (1–777) in size representing RdRp, NSm, NSs, and N proteins, respectively were deposited in GenBank database (Supplementary Table 1).
Phylogenetic analysis and detection of recombination events
Alignments of the obtained nucleotide and deduced amino acid sequences of TSWV-pepper isolates are from Turkey with additional homologous sequences of other comparative TSWV isolates retrieved from GenBank. MEGA 7 software (Kumar et al. 2016) was used for multiple alignment of the obtained nucleotide and deduced amino acid sequences of Turkish TSWV-pepper isolates with homologous sequences of other TSWV isolates retrieved from GenBank, and phylogenetic inference with the neighbor-joining method and Kimura 2-parameters as nucleotide substitution model. Bootstrap analysis with 1000 replicates was performed to evaluate the significance of the interior branches. Also, the possible recombination and/or reassortment events were investigated using the RDP4 (Martin et al. 2015) software with default settings. For recombination (and reassortment) analyses were conducted in the four regions (RdRp, NSm, NSs, N) separately, and concatenating the sequences of the four genomic regions into one sequence file of 3344 nt in length. The suite of recombination detection programs used for the detection of recombination events and the corresponding average P-values for each event were: RDP, GeneConv, Bootscan, MaxChi, Chimaera, SiScan. Bootscan, RDP and SiScan are phylogeny-based methods, GeneConv, MaxChi, and Chimaera are substitution-based methods. Putative recombination events were identified with a Bonferroni corrected P-value cut-off of 0.05. Later, nucleotide sequences of potential reassortants were compared with the parental sequences by SimPlot Version 3.5 (Lole et al. 1999) using a 200 nt window and step size of 20 nt.
Results
Biological characterization
A total of fourteen pepper samples with single TSWV infection and free for CMV, TMV, PVY in ELISA were used to have biological clones from local lesions in V. unguiculata plants. Isolates TRpep 37, 47, 59, 67, 125, 146, 150, 154, 160, 171, 175 were obtained from greenhouse-infected C. annuum plants, while TRpep 209, 219 and 272 were obtained from field-infected C. annuum plants, all carrying the Tsw gene. Transmission of 30 TSWV isolates (obtained by biological cloning from 14 TSWV-infected pepper plants from field) to pepper plants with the Tsw gene (C. chinense accession 'PI 152225', C. annuum cv. 'Mertcan F1'), showed three different types of symptoms (Table 1): (i) only local lesions as a result of HR, and neither symptoms were observed and nor virus detected by ELISA at 15 and 30 dpi, (ii) chlorotic rings, chlorosis, vein clearing, downward leaf curling and stunting on inoculated and newly emerged leaves, (iii) severe systemic necrosis and wilting that lead to death resistant pepper plants (Fig. 2). Systemic symptoms (chlorotic rings, chlorosis, downward leaf curling, stunting) occured in susceptible C. annuum cv. ‘Naz F1’ plants at 6–10 dpi. The virus was detected in ELISA both in inoculated and newly emerging leaves in the pepper plants carrying the gene Tsw and the other hosts used for the biological characterization. Some biological clones from the same field-isolates have different biotypes: RB and NRB, ex. TRpep-47/3 (RB) and TRpep-47/5 (NRB) mentioned in Supplementary Table 1.
Symptoms of TSWV-pepper isolates on N. benthamiana (a: wilting; b, c: chlorotic rings, downward leaf curling, yellow leaf), N. tabacum cv. Samsun NN (d: vein clearing; e, f: necrotic and chlorotic rings), susceptiple C. annuum cv. ‘Naz F1’ (g: yellow leaf, chlorotic rings; h: chlorotic rings, stunting, leaf deformation, i: chlorotic spots, yellow leaf) and resistant C. chinense ‘PI1522255’ (j: hypersensitive reaction; k: systemic necrosis; l: chlorotic rings) pepper genotypes
Phylogenetic relationships of TSWV isolates and recombination events
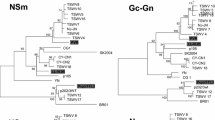
Phylogenetic trees were drawn to characterize genetic variability of the Turkish TSWV-pepper isolates and their phylogenetic relationship among other TSWV isolates with different geographical origins based on the nucleotide sequences of RdRp, NSm, NSs, and N genes. The identity of the nucleotide sequence of the RdRp gene among the 30 Turkish TSWV-pepper isolates ranged between 96.2 and 100% while the amino acid sequence identity ranging between 97.4 and 100%. Four clades could be clearly observed in the phylogenetic tree of the isolates based on the RdRp gene. The first clade contained two sub-clades (corresponding to sub-clades Ia, Ib) comprising 26 Turkish TSWV-pepper isolates (Fig. 3a). The second clade contained isolates from Italy (KJ575619.1, MH763621.1, MH763622.1, MH763623.1), France (MK792774.1), South Korea (KC261971.1), and TswvTRpep isolate (MK682812.1) previously reported from Turkey whereas four-isolates (TRpep-154/5, TRpep-154/8, TRpep-171/3, TRpep-160/4) formed distinct clade from others (Fig. 3a). The nucleotide sequence identity of the NSm gene among the 30 Turkish TSWV-pepper isolates ranged between 97.2 and 100%, while the percentage identity at amino acid level ranged between 97.2 and 100%. The phylogenetic tree based on the NSm gene also had two main clades. The first clade was subdivided into two sub-clades (corresponding to sub-clades a1, a2): the first sub-clade contained the 23 Turkish TSWV-pepper isolates as well as isolates from Australia (KT717692.1, MG025803.1), USA (AY744486.1), South Korea (KC261966.1, KC261972.1), Italy (HQ830185.1), Spain (HM015511.1, HM015519.1), and Turkey (MK367503.1) (Fig. 3b). And, the remaining Turkish TSWV isolates formed a separate sub-clade. The nucleotide sequence identity of the NSs gene among the 30 Turkish TSWV-pepper isolates ranged between 96.8 and 100%, while the amino acid sequence identity was between 96.4 and 100%. The nucleotide and amino acid sequence identity of NSs gene for 5 TSWV isolates (TRpep-47/3, TRpep-47/5, TRpep-47/9, TRpep-47/12, TRpep-47/17) originated from one plant was 98.3–100% and 98.2–100%, respectively. Similarly, the identity scores of NSs gene among the other 4 TSWV isolates (TRpep175/3, TRpep175/5, TRpep175/8, TRpep175/11) originated from one plant was 97.7–100% on nucleotide level and 97.1–100% on amino acid level. The phylogenetic tree based on the NSs gene isolates was grouped into two main clades. The first clade further fell into six sub-clades (corresponding to sub-clades w1, w2, w3, w4, w5, w6), the first one of which included the 20 present isolates with the isolates from France (FR692822.1, FR692835.1), and Italy (HQ830187.1, HQ830186.1) (Fig. 3c). The second sub-clade contained two Turkish TSWV-pepper isolates (TRpep-47/17 RB and TRpep-125/13); the third sub-clade included isolates from Hungary (KJ649612.1), Spain (MK922149.1), and Turkey (MK922150.1). The fourth sub-clade contained a single Turkish-pepper isolate (TRpep-175/3). The fifth sub-clade contained six Turkish TSWV-pepper isolates (TRpep-175/5 RB, TRpep-59/2, TRpep-67/2 RB, TRpep-59/7, TRpep-125/6, TRpep-150/2). The sixth sub-clade consisted of South Korea (MF159063.1, KC261958.1), and France (FR692829.1, FR693031.1) isolates. TRpep-160/4 as being the most divergent isolate among all the others was clustered together in a second clade with Tsw-resistance breaking isolates from Brazil (DQ915947.1), Italy (DQ376179.1, DQ376183.1), Spain (DQ376185.1), and Turkey (MK922155.1, MK922154.1, MK922152.1).
The nucleotide sequence identity of the N gene among the 30 Turkish TSWV-pepper isolates was from 97.1 to 100%, while the amino acid sequence identity ranging between 97.2 and 100%. The phylogenetic tree based on the N gene isolates was clustered in two main clades. The first clade included three sub-clades (corresponding to sub-clades x1, x2, x3), one of which contained the 13 isolates of Turkish TSWV-pepper isolates, and isolates from France (FR693046.1, FR693058.1, FR693060.1), Italy (DQ431237.1, HQ830187.1), Hungary (KJ649612.1), and South Korea (HQ267713.1, MF467373.1) (Fig. 3d). Both the second and third sub-clades included Turkish TSWV-pepper isolates. The second clade was divided into three sub-clades (corresponding to sub-clades y1, y2, y3). The first sub-clade contained TRpep-150/7, TRpep-47/12, TRpep-67/1, 272/4 isolates, while only TRpep-219/9 isolate was in the second sub-clade. Three Turkish TSWV-pepper isolates (TRpep-146/8, TRpep-47/9, TRpep-47/5) were clustered in the third sub-clade with isolates from Argentina (MK524183.1), South Korea (DQ453158.1, MG001348.1, MF159070.1, MF159071.1) (Fig. 3d). The phylogenetic trees of L, M, and S segments had multiple clades. In any case, some of the Turkish TSWV-pepper isolates have clustered with previously reported TSWV isolates in GenBank; however, the isolates from Turkey seemed to be very diverse from the other TSWV isolates.
The recombination analyses were performed based on each coding region using sequences obtained in this study, and no intra-gene recombination events were identified for particular coding regions. However, the predicted breakpoints in the concatenated alignments of the four genomic regions (3344 nt in length) indicated that TRpep-171/3 and TRpep-175/11 were predicted to be products of reassortments. The isolate TRpep-171/3 grouped with TRpep-154/5 based on the phylogenetic analysis of RdRp, NSs, and N sequences showed 99.1%, 99.6%, 100% nucleotide sequence identity respectively; while the same isolate grouped with TRpep-150/2 had 99.7% nt identity for the NSm sequences. Likewise, the isolate TRpep-175/11 together with TRpep-175/8 showed 100% nt identity based on phylogenetic analysis of RdRp, NSs, and N sequences, however, the nucleotide identity for the NSm sequences was 100% when it was with TRpep-175/3.
The results of analysis of concatenated genome sequences of TSWV isolates in RDP4, showed two recombination events identified in TRpep-171/3, with as TRpep-154/5 major parents and TRpep-150/2 as a minor parent, and in TRpep-175/11, with as TRpep-175/8 major parents and TRpep-175/3 as a minor parent at stringent P-values (Table 2).
Discussion
Turkey is one of the foremost countries concerning pepper production in open field and greenhouse in the Mediterranean basin. The outbreak of TSWV has been reported as the major concern in the pepper cultivating areas. The most efficient way to control the virus spread is the use the pepper cultivars with the resistance gene Tsw because all the other control methods have been found insufficient to reduce virus epidemics up to now. Therefore, molecular and biological characterizations of the Turkish TSWW-pepper isolates from the greenhouse and field-grown plants in small, but economically very important locations were investigated to gather preliminary information. Symptom variation has shown the presence of mixed infection of various TSWV isolates in the fields causing biodiversity in the TSWV population. Mixed infection either with different variants of viruses or with viruses may expose a range of consequences (Garnsey and Randles 1987). The genetic exchange by segment reassortment or recombination event in TSWV population was stated in previous studies (Tentchev et al. 2011; Lian et al. 2013; Margaria et al. 2014). In RNA viruses with the segmented genome, reassortment may offer an important role in evolution (Thekke-Veetil et al. 2015). Lian et al. (2013) mentioned that the rate of TSWV recombination in nature is significantly lower than that of reassortment. The results of all analyses did not show any evidence for recombination events between TSWV isolates. It should be not having full-length genomic sequences. According to the indication of recombination signals in the RDP program and phylogenetic tree analyses, two pepper isolates could be originated through reassortment. One possible reassortant isolate was found in the same pepper plant with its major and minor parents.
NSs protein has a role in suppressing the gene silencing mechanism of the host plant and is also responsible for breaking the Tsw-resistance (Takeda et al. 2002; de Ronde et al. 2013, 2014). It has been stated that there is no specific point mutation in all the resistance breaking phenotypes, and they show differences in various amino acid changes in the NSs protein (Tsompana et al. 2005; Margaria et al. 2007; Almási et al. 2020). For this reason, we compared the amino acid sequences of the NSs gene of the Turkish TSWV-pepper isolates, it was expressed that a common specific mutation in the NSs gene has not been detected. The phylogenetic analysis of the NSs gene has also shown that the isolates were clustered regardless of the phenotype (RB or NRB). Our study supported the hypothesis that resistance breakdown could have evolved independently several times in Turkey.
Here, we described the relationships of Turkish TSWV-pepper isolates with previously reported TSWV isolates from other geographical regions. Analysis of the molecular variation of the four genes revealed low genetic diversity in Turkish TSWV-pepper isolates. The close phylogenetic relationship between TSWV isolates from Turkey and other countries suggests that some isolates might have been introduced in Turkey with intercontinental shipping of virus-infected plant materials and/or virus vectors.
References
Adkins S (2000) Tomato spotted wilt virus-positive steps towards negative success. Mol Plant Pathol 1(3):151–157
Almási A, Nemes K, Salánki K (2020) Increasing diversity of resistance breaking pepper strains of Tomato spotted wilt virus in the Mediterranean region. Phytopathol Mediterr 59(2):385–391. https://doi.org/10.14601/Phyto-11346
Arlı-Sökmen M, Mennan H, Sevik MA, Ecevit O (2005) Occurrence of viruses in field-grown pepper crops and some of their reservoir weed hosts in Samsun, Turkey. Phytoparastica 33(4):347–358. https://doi.org/10.1007/BF02981301
Azeri T (1981) Preliminary report of tomato spotted wilt virus and its epidemy on tobacco in the Çanakkale region of Turkey. J Turk Phytopathol 10(2–3):79–87
Black LL, Hobbs HA, Gatti JM Jr (1991) Tomato spotted wilt virus resistance in Capsicum chinense PI 152225 and 159236. Plant Dis 75:863. https://doi.org/10.1094/PD-75-0863A
Black LL, Hobbs HA, Kammerlohr DS (1996) Resistance of Capsicum chinense lines to tomato spotted wilt virus isolates from Louisiana, USA, and inheritance of resistance. Acta Hortic 431:393–401. https://doi.org/10.17660/ActaHortic.1996.431.34
Boiteux LS (1995) Allelic relationships between genes for resistance to tomato spotted wilt tospovirus in Capsicum chinense. Theor Appl Genet 90:146–149
Boiteux LS, Nagata T (1993) Susceptibility of Capsicum chinense PI 159236 to tomato spotted wilt virus isolates in Brazil. Plant Dis 77:210. https://doi.org/10.1094/PD-77-0210F
Çelik İ, Özalp R, Çelik N, Polat İ, Sülü G (2018) Improvement of long type pepper lines resistant to tomato spotted wilt virus (TSWV). Derim 35(1):27–36. https://doi.org/10.16882/derim.2018.325765
Chomczynski P, Sacchi N (1987) Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem 162(1):156–159. https://doi.org/10.1006/abio.1987.9999 (PMID: 2440339)
Chung BN, Choi HS, Yang EY, Cho JD, Cho IS, Choi GS, Choi SK (2012) Tomato spotted wilt virus isolates giving different infection in Commercial Capsicum annuum cultivars. Plant Pathol J 28(1):87–92. https://doi.org/10.5423/PPJ.NT.09.2011.0169
Clark MF, Adams AN (1977) Characteristic of the microplate method of enzyme-linked immunosorbent assay for the detection of plant viruses. J Gen Virol 34:475–483. https://doi.org/10.1099/0022-1317-34-3-475
Cortez I, Saaijer J, Wongjkaew KS, Pereira AM, Goldbach R, Peters D, Kormelink R (2001) Identification and characterization of a novel tospovirus species using a new RT-PCR approach. Adv Virol 146:265–278. https://doi.org/10.1007/s007050170174
de Haan P, Kormelink R, de Oliveira RR, van Poelwijk F, Peters D, Goldbach R (1991) Tomato spotted wilt virus L RNA encodes a putative RNA polymerase. J Gen Virol 72:2207–2216. https://doi.org/10.1099/0022-1317-72-9-2207
de Ronde D, Butterbach P, Lohuis D, Heild M, van Lent JWM, Kormelink R (2013) Tsw gene-based resistance is triggered by a functional RNA silencing suppressor protein of the Tomato spotted wilt virus. Mol Plant Pathol 14:405–415
de Ronde D, Pasquier A, Ying S, Butterbach P, Lohuis D, Kormelink R (2014) Analysis of tomato spotted wilt virus NSs protein indicates the importance of the N-terminal domain for avirulence and RNA silencing suppression. Mol Plant Pathol 15:185–195
Debreczeni DE, López C, Aramburu J, Darós JA, Soler S, Galipienso L, Falk BW, Rubio L (2015) Complete sequence of three different biotypes of tomato spotted wilt virus (wild type, tomato Sw-5 resistance-breaking and pepper Tsw resistance-breaking) from Spain. Adv Virol 160(8):2117–2123. https://doi.org/10.1007/s00705-015-2453-8
Deligöz I, Sökmen MA, Sarı S (2014) First report of resistance breaking strain of tomato spotted wilt virus (Tospovirus; Bunyaviridae) on resistant sweet pepper cultivars in Turkey. New Dis Rep 30:26
Fidan H, Koç G, Topçu T (2016) Infection and molecular characterization of tomato spotted wilt virus (TSWV) on Anthurium sp. Alatarım 15(2):28–36
Francki RIB, Hatta T (1981) Tomato spotted wilt virus. In: Kurstak E (ed) Handbook of Plant Virus Infections and Comparative Diagnosis. Elsevier/North- Holland Biomedical Press, Amsterdam, pp 491–512
Garnsey SM, Randles JW (1987) Biological interactions and agricultural implications of viroids. In: Semancik JS (ed) Viroids and viroid-like pathogens. CRC Press, Boca Raton, FL, pp 127–160
Goldbach R, Peters D (1996) Molecular and biological aspects of tospoviruses. In: Elliott RM (ed) The Bunyaviridae. Plenum Press, New York
Griep RA, Prins M, van Twisk C, Keller HJHG, Kerschbaumer RJ, Kormelink R, Golbach RW, Schots A (2000) Application of Phage display in selecting tomato spotted wilt virus –Specific single -Chain antibodies (scFvs) for sensetive diagnosis in ELISA. Phytopathology 90:183–190
Güneş N, Gümüş M (2019) Detection and characterization of tomato spotted wilt virus and cucumber mosaic virus on pepper growing areas in Antalya. J Agric Sci 25:259–271
Hoang NH, Yang HB, Kang BC (2013) Identification and inheritance of a new source of resistance against tomato spotted wilt virus (TSWV) in Capsicum. Sci Hortic 161:8–14
Hobbs HA, Black LL, Johnson RR, Valverde RA (1994) Differences in reactions among tomato spotted wilt virus isolates to three resistant Capsicum chinense lines. Plant Dis 78:1220 (Abstr.)
Kamberoğlu MA, Calışkan AF, Alan B (2009) First Report of tomato spotted wilt virus on eggplant in Turkey. J Plant Pathol 91(1):231–231
Kumar S, Stecher G, Tamura K (2016) Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33(7):1870–1874. https://doi.org/10.1093/molbev/msw054
Lian S, Lee JS, Cho WK, Yu J, Kim MK, Choi HS, Kim KH (2013) Phylogenetic and recombination analysis of tomato spotted wilt virus. PLoS ONE 8(5):e63380. https://doi.org/10.1371/journal.pone.0063380
Lole KS, Bollinger RC, Paranjape RS, Gadkari D, Kulkarni SS, Novak NG, Inger-soll R, Sheppard HW, Ray SC (1999) Full-length human immuno deficiency virus type 1 genomes from subtype C-infected seroconverters in India, with evidence of inter subtype recombination. J Virol 73:152–160. https://doi.org/10.1128/jvi.73.1.152-160.1999
Macedo MA, Rojas MR, Gilbertson RL (2019) First report of a resistance-breaking strain of Tomato spotted wilt orthotospovirus infecting sweet pepper with the Tsw resistance gene in California, USA. Plant Dis 103(5):1048. https://doi.org/10.1094/PDIS-07-18-1239-PDN
Mandal B, Pappu HR, Csinos AS, Culbreath AK (2006) Response of peanut, pepper, tobacco, and tomato cultivars to two biologically distinct isolates of tomato spotted wilt virus. Plant Dis 90:1150–1155. https://doi.org/10.1094/PD-90-1150
Margaria P, Ciuffo M, Pacifico D, Turina M (2007) Evidence that the nonstructural protein of tomato spotted wilt virus is the avirulence determinant in the interaction with resistant pepper carrying the Tsw gene. Mol Plant Microbe Interact 20:547–558
Margaria P, Ciuffo M, Turina M (2004) Resistance breaking strain of tomato spotted wilt virus (Tospovirus; Bunyaviridae) on resistant pepper cultivars in Almeria, Spain. Plant Pathol 53:795–795. https://doi.org/10.1111/j.1365-3059.2004.01082.x
Margaria P, Miozzi L, Ciuffo M, Pappu H, Turina M (2014) The first complete genome sequences of two distinct European tomato spotted wilt virus isolates. Arch Virol 160:591–595. https://doi.org/10.1007/s00705-014-2256-3
Martin DP, Murrell B, Golden M, Khoosal A, Muhire B (2015) RDP4: Detection and analysis of recombination patterns in virus genomes. Virus Evol 1:vev003. https://doi.org/10.1093/ve/vev003
McPherson RM, Jones DC, Pappu HR, Moore JM (2003) Reducing the risks of spotted wilt virus in tobacco with selected Thrips (Thysanoptera: Thripidae) control practices. J Agric Urban Entomol 20(1):11–23
Moury B, Palloix A, Gebre-Selassie K, Marchoux G (1997) Hypersensitive resistance to tomato spotted wilt virus in three Capsicum chinense accessions is controlled by a single gene and is overcome by virulent strains. Euphytica 94:45–52
Moury B, Verdin E (2012) Viruses of pepper crops in the Mediterranean basin: a remarkable stasis. Adv Virus Res 84:127–162
Naidu RA, Sherwood JL, Deom CM (2008) Characterization of a vector-non-transmissible isolate of tomato spotted wilt virus. Plant Pathol 57:190–200
Pappu HR, Jones RAC, Jain RK (2009) Global status of tospovirus epidemics in diverse cropping systems: Successes achieved and challenges ahead. Virus Res 141:219–236. https://doi.org/10.1016/j.virusres.2009.01.009
Pappu HR, Whitfield AE, de Oliveria AS (2020) Tomato spotted wilt virus (Tospoviridae). Elseveir. https://doi.org/10.1016/B978-0-12-809633-8.21329-0
Parrella G, Gognalons P, Gebre-Selassie K, Vovlas C, Marchoux G (2003) An update of the host range of tomato spotted wilt virus. J Plant Pathol 85:227–264
Persley DM, Thomas JE, Sharman M (2006) Tospoviruses: an Australian perspective. Australas Plant Pathol 35:161–180. https://doi.org/10.1071/AP06015
Riley DG, Joseph SV, Srinivasan R, Diffie S (2011) Thrips vectors of tospoviruses. J Integr Pest Manag 2:1–10. https://doi.org/10.1603/IPM10020
Roggero P, Masenga V, Tavella L (2002) Field Isolates of tomato spotted wilt virus overcoming resistance in pepper and their spread to other hosts in Italy. Plant Dis 86:950–954. https://doi.org/10.1094/PDIS.2002.86.9.950
Scholthof KB, Adkins S, Czosnek H, Palukaitis P, Jacquot E, Hohn T, Hohn B, Saunders K, Candresse T, Ahlquist P, Hemenway C, Foster GD (2011) Top 10 plant viruses in molecular plant pathology. Mol Plant Pathol 12(9):938–954. https://doi.org/10.1111/j.1364-3703.2011.00752.x
Sertkaya G (2015) Investigation of some viruses in lettuce and spinach fields in Hatay province of Turkey. Mustafa Kemal Univ J Agric Sci 20(1):7–12
Sharman M, Persley DM (2006) Field isolates of tomato spotted wilt virus overcoming resistance in capsicum in Australia. Australas Plant Pathol 35:123–128. https://doi.org/10.1071/AP06014
Şimşek D, Pınar H, Mutlu N (2015) Development of new bell pepper (Capsicum annum L.) lines and cultivars resistance to tospovirus and tobamovirus using molecular breeding methods. Alatarım 14(1):1–8
Sin SH, McNulty BC, Kennedy GG, Moyer JW (2005) Viral genetic determinants for thrips transmission of tomato spotted wilt virus. Proc Natl Acad Sci USA 102:5168–5173
Soler S, Diez MJ, Nuez F (1998) Effect of temperature regime and growth stage interaction on pattern of virus presence in TSWV-resistant accessions of Capsicum chinense. Plant Dis 82:1199–1204. https://doi.org/10.1094/PDIS.1998.82.11.1199
Takeda A, Sugiyama K, Nagano H, Mori M, Kaido M, Mise K, Tsuda S, Okuno T (2002) Identification of a novel RNA silencing suppressor, NSs protein of Tomato spotted wilt virus. FEBS Lett 532:75–79. https://doi.org/10.1016/s0014-5793(02)03632-3
Tekinel N (1973) Investigations on the dissemination and rates of tomato virus disease Adana, Antalya, Hatay, and Içel provinces. Plant Prot Bull 13(3):107–141
Tekinel N, Dolar MS, Sağsöz S, Salcan Y (1969) Research on viruses of some economically important vegetables in the Mersin region. Plant Prot Bull 9:37–49
Tentchev D, Verdin E, Marchal C, Jacquet M, Aguilar JM, Moury B (2011) Evolution and structure of tomato spotted wilt virus populations: evidence of extensive reassortment and insights into emergence processes. J Gen Virol 92:961–973. https://doi.org/10.1099/vir.0.029082-0
Thekke-Veetil T, Polashock JJ, Marn MV, Plesko IM, Schilder AC, Keller KE, Martin RR, Tzanetakis IE (2015) Population structure of blueberry mosaic associated virus: Evidence of reassortment in geographically distinct isolates. Virus Res 2(201):79–84. https://doi.org/10.1016/j.virusres.2015.02.022
Thomas-Carrol ML, Jones RAC (2003) Selection, biological properties and fitness of resistance-breaking strains of tomato spotted wilt virus in pepper. Ann Appl Biol 142:235–243
Tsompana M, Abad J, Purugganan M, Moyer JW (2005) The molecular population genetics of the Tomato spotted wilt virus (TSWV) genome. Mol Ecol 14:53–66. https://doi.org/10.1111/j.1365-294X.2004.02392.x
Whitfield AE, Ullman DE, German TL (2005) Tospovirus-thrips interactions. Annu Rev Phytopathol 43:459–489. https://doi.org/10.1146/annurev.phyto.43.040204.140017
Yurtmen M, Güldür ME, Yılmaz MA (1998) Tomato spotted wilt virus on pepper in Içel province of Turkey. Ninth Conference of the I.S.H.S Vegetable Virus Working Group, Recent Advance in Vegetable Virus Research, Torino, Italy, pp 91–92, 22–27 August 1998
Funding
This work was supported by the Research Fund of Kahramanmaraş Sütçü Imam University (Project No: 2019/3–18 M). We are grateful to Dr. Bekir Bülent Arpacı for valuable suggestions.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Ethical approval
This study did not involve human or animal participants.
Conflict of interest
All authors declare that they have no conflict of interest.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Balsak, S.C., Buzkan, N. Biological and molecular characterization of Tomato spotted wilt orthotospovirus isolates from pepper cultivars with the resistance gene Tsw from Turkey. Australasian Plant Pathol. 51, 543–551 (2022). https://doi.org/10.1007/s13313-022-00890-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13313-022-00890-9