Abstract
Cytogenetically normal acute myeloid leukemia (CN-AML) constitutes the largest subgroup of AML patients that is associated with molecular alteration. MiRNAs have been shown to be aberrantly expressed in CN-AML. In addition, specific miRNA (miR) expression patterns were found to be associated with certain genetic alterations in these patients. This study investigated the expression level of miR-1, miR-486, and let-7a in 45 CN-AML patients well characterized for FLT3 and/or NPM1 mutations using real-time quantitative RT-PCR and evaluated the association between candidate miRs expression and clinical features. Our data revealed that miR-1 was significantly overexpressed in CN-AML patients, and increasing expression of miR-1 correlated with NPM1 mutation (P < 0.05) and lower hemoglobin level was also observed in patients with miR-1 overexpression (P < 0.05). The expression of miR-1 was much higher in AML-M2 compared with other subtypes. Further, we found significantly increasing miR-486 expression in 40 of 45 (89 %) CN-AML patients. There was no significant association of upregulation of miR-486 with clinical parameters. The expression level of miR-486 was increased in AML-M2 subtype. The levels of let-7a were significantly increased in CN-AML patients compared to the healthy control and significantly higher in the NPM1 ± CN-AML patients. There was no correlation detected between the level of let-7a and FLT3+. An increasing expression level of let-7a was demonstrated in M2 subtype. In addition, our data showed no significant association between increasing let-7a and clinical characteristic. Comparison of peripheral blood and bone marrow results in 30 CN-AML patients showed that there is a considerable concordance between PB and BM in the results of candidate miR levels (P < 0.001). In conclusion, further studies should also be performed to detect functional mechanism of these miRs.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Acute myeloid leukemia (AML) is a group of hematopoietic stem cell disorders that are characterized by the autonomous proliferation of myeloid progenitors and is also associated with genetic instability and molecular changes. Cytogenetic aberrations have classified AML patients into favorable, intermediate, and adverse prognostic groups [1, 2]. AML patients with cytogenetic normal (CN) who comprise the largest cytogenetic group of AML (approximately 45 %) fall into the intermediate category [3, 4]. This heterogeneous AML subgroup shows molecular alterations and gene mutation. Among these genetic abnormalities, Fms-like tyrosine kinase 3 (FLT3) lesions have been reported in about 25 % of AML cases, and patients with this abnormality are associated with a poor prognosis [5–7]. Nucleophosmin (NPM1) abnormalities were predominantly observed in 45 to 60 % of patients with CN-AML, accounting for the most frequent genetic change in this subset [8, 9]. Four main categories may be identified by alterations of these genes including FLT3wt/NPM1wt, FLT3wt/NPM1+, FLT3+/NPM1wt, FLT3+/NPM1+ [10].
MicroRNAs (miRNAs) are small, noncoding RNA which typically regulate the levels of mRNA stability and translation by binding the 3′untraslated region (UTR) of mRNAs. MiRNA plays an important role in hematopoietic stem cells, proliferation, survival, and differentiation [11, 12]. Dysregulation of miRNAs in all of these pathways are observed in AML patients [13, 14]. Aberrant miRNA expressions have been variable in different cytogenetic and molecular subtypes in AML cases [15, 16]. For example, several miRNAs such as rmiR-10a, miR-10b, miR-155, let-7a, miR-125b, and miR-196a act as oncomiRs in hematological malignancies especially in CN-AML patients. A large number of genes in AML patients can be targeted and repressed by these miRs [17–21]. The role of miR-130a, miR-424 as tumor suppressor was reported in CN-AML patients. The expression of these miRNAs is downregulated in these patients. Despite several miRNA profiling studies on AML patients, there are only few reports of miRNA deregulated [10, 19, 21].
In this study, we focused on CN-AML patients that are categorized according to the NPM1 and FLT3 expression, and then three miRNAs (miR-1, miR-486, and let-7a) were selected among them; miR-486 and let-7a are involved in erythroid differentiation, and deregulation of miR-1 plays an important role in many types of cancers [22–25]. In addition, we investigated whether these miR expressions associate with clinical features such as age, gender, white blood cell (WBC), Hb count, and French-American-British (FAB) subtype of CN-AML patients. Here, we evaluated the concordance between peripheral blood (PB) and bone marrow (BM) in detecting expression levels of candidate miRs.
Materials and methods
Patients and samples
In this study, BM and PB samples were obtained from AML adult patients with newly diagnosed AML admitted at Hematology, Oncology and Stem Cell Transplantation Research Center, Shariati Hospital, Tehran, Iran, and this project was approved by the ethics committee board of this research center. The BM and PB samples were collected from 15 healthy donors (9 men and 6 women) with a median age of 46 (range, 29–65), and they did not have anemia. Populations of the patients were collected from all French-American-British (FAB) types except M3, and the patients had a median age of 57 years. Cytogenetic analyses of the samples were performed after short-term culture. Metaphase chromosomes were banded using trypsin-giemsa banding technique and analyzed according to the International System for Human Cytogenetic Nomenclature. The type of this research is experimental study. In this study, 45 patients with normal karyotype were selected. Details of the patients are presented in Table 1.
Gene mutation detection
Identifying the presence or absence of FLT3-internal tandem repeat (ITD) of FLT3 exon 14 was carried out by capillary electrophoresis as reported previously. Mutation in NPM1 was assessed by polymerase chain reaction (PCR) amplification and fragment analysis of NPM1 exon 12. In order to decrease the level of heterogeneity, CN-AML patients only have ITD mutations for FLT3, and type A mutations for NPM1 were selected. Herein, FLT3-ITD and type A NPM1 mutation are showed in FLT3+ and NPM1+, respectively.
qRT-PCR for miRNA
Total RNA was extracted from the samples by using the miRNeasy Mini Kit following the manufacturer’s instructions (Qiagen, Miami, FL, USA). The concentration and purity of the RNA samples were detected spectroscopically. For miRNA quantification, 1 μg of total RNA were transcribed into cDNA via miRNA reverse transcription reaction using stem loop-RT primer and reverse transcriptase kit (Invitrogen). Quantitative real-time PT-PCR (qRT-PCR) was performed using Excite Real Time Mastermix with SYBR green (Biogene, Cambridge, UK) and using ABI prism 7700 (Applied Biosystems). Thermal cycling condition for miRNA was performed as follows: step 1—at 95 °C for 15 min; step 2—at 95 °C for 15 s; step 3—at 60 °C for 30 s, step 4—at 72 °C for 1 min. Steps 2–4 were repeated for 40 cycles. U6 small nuclear RNA was used as an internal normalization. The relative expression levels of miRNAs were calculated using the 2−ΔΔCt method. The primers used for stem-loop RT-PCR for miR-1, miR486, and let-7a are presented in Table 2.
Statistical analysis
The difference in miRNA expression levels between CN-AML patients and control groups was estimated using Student’s t test (for independent samples). Results were analyzed using the GraphPad software (GraphPad PRISM V 5.04 analytical software).
To further investigate, the correlation between the prevalence of deregulation levels of each miRNA and the clinical and hematologic parameters were analyzed with Spearman’s rank correlation. Paired sample t test was utilized to investigate equivalence between paired PB and BM readouts for candidate miRs in this study. All P values were two-tailed, and P < 0.05 was considered statistically significant.
Result
MiR-1 expression in CN-AML
Significant increase in miR-1expression in CN-AML patients compared to the healthy donors was observed (Fig. 1a). We also analyzed the relationship between miR-1 and FLT3-ITD and NPM1 mutation. The upregulation of miR-1 was observed in CN-AML carrying the NPM1+. However, overexpression of miR-1 is not statistically different between FLT3+ and FLT3-wt/NPM1-wt in CN-AML patients (Fig. 1b).
Expression level of miR-1 in CN-AML and healthy control. a Relative expression of miR-1 in AML patients was quantified by real-time PCR, normalized to the expression of U6 and calculated according to the 2−ΔΔCt method. b CN-AML patients were divided in four groups: NPM1-wt/FLT3-wt; NPM1+ = FLT3-wt and NPM1mutA; FLT3+ = FLT3-ITD and NPM1-wt; NPM1+/FLT3+ = FLT3-ITD and NPM1mutA. c Relative expression of miR-1 in these patients demonstrated in all of subtype except M3.
Associations between the miR-1 and clinical characteristics (sex, age, WBC count, Hb count, and platelet count) have been detected in the current study; no significant difference in age, gender and other clinical parameters except Hb levels was found in patients with miR-1 overexpression compared to the patients without any increased miR-1 expression. Patients with higher miR-1 expression had lower Hb level than those without miR-1 overexpression (P < 0.05) (Table 2). Overexpression of miR-1 was also observed in almost all of the subtypes except M5, and the expression level of miR-1 was much higher in AML-M2 than in other subtypes of these patients (Fig. 1c).
Overexpression of miR-486-5p in CN-AML
Analysis of miR-486-5p expression level in CN-AML cases showed higher levels of miR-486-5p expression in 40 of the 45 patients (89 %) (Fig. 2a). The status of miR-486 expression was not correlated with NPM1+ and FLT3+ (Fig. 2b). Our data showed no significant difference in sex, age, WBC, and Hb and platelet levels between patients with and without miR-486-5p overexpression. Upregulated miR-486-5p expression was observed in almost all of the subtypes, and the highest expression of miR-486 was detected in patients with M2 (P < 0.001) (Fig. 2c).
Expression level of miR-486 in CN-AML and healthy control. a Relative expression of miR-486 was obtained by qRT-PCR and normalizing with U6. b Expression value of miR-486 in subgroups of CN-AML patients(NPM1-wt/FLT3-wt,NPM1+, FLT3+, NPM1+/FLT3+) were compared. There is no correlation between miR-486 level and NPM1+ and FLT3+ c Relative expression of miR-486 was compared in different FAB subtypes of AML except M3. High expression level of miR-486 was detected in almost all of the subtypes
Let-7a expression in CN-AML
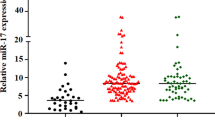
When expression levels of let-7a in CN-AML cases were compared with normal samples, we detected upregulation of let-7a in these patients (Fig. 3a). The expression of let-7a was significantly higher in the NPM1 ± CN-AML group compared to the FLT3-wt/NPM1-wt group (P < 0.01), and the statue of let-7a was also not correlated to the FLT3+ group (Fig. 3b). Overexpression of let-7a was detected in each AML subtype. However, the prevalence of let-7a upregulation in M2 subtype was higher than in other subtypes (Fig. 3c). We also examined whether the overexpression of let-7a could be associated with clinical factors. This result showed that expression levels of let-7a were not significantly associated with clinical parameters.
Expression level of let-7a in CN-AML and healthy control. a Let-7a expression level at CN-AML patients was presented. b Comparison in subgroups of CN-AML patients (NPM1-wt/FLT3-wt, NPM1+, FLT3+, NPM1+/FLT3+) was demonstrated. c Comparison of let-7a expression level in different FAB subtype of AML except M3
Analysis of PB and BM paired samples
Thirty PB and BM from the same patients were compared at the time of diagnosis to detect whether PB could serve as well as BM and provide similar information in candidate miRs. The expression level comparison of candidate miRs in paired PB and BM samples demonstrated that there is a considerable concordance between the expression levels of miRs for PB and BM samples (P < 0.001) (Fig. 4).
Discussion
MiRNAs as a noncoding RNA regulate several biological processes such as cell cycle regulation, differentiation, proliferation, and apoptosis of cells in human body. Some miRs may act as tumor suppressors and others as oncogenes in many cancers. Despite the presence of miRNAs in biological functions, pathway regulations based on miRNA expression remain unknown.
In the present study, we assessed the expression level of miR1, let-7a, and miR486 in CN-AML, and association of the deregulation of these miRs with clinical parameters and FLT3+ and NPM1+ were detected.
In this study, we found increased expression of miR-1 in CN-AML patients compared to normal controls which is consistent with observation made by Marcucci et.al. [26], and a study on AML cell lines showed that overexpression of miR-1 promotes cell proliferation [27]. Result of this study showed for the first time that miR-1 upregulation has association with NPM1+. However, there was no significant difference between the increasing miR-1 expression and clinical characteristic. Aberrant expression of miR-1 has been shown to have a role in tumorigenesis, and downmodulation of miR-1 has been identified in many types of cancer, such as bladder, prostate, lung, thyroid, colon, and colorectal cancer [23, 28, 29]. However, there is only one study on AML cell lines that showed overexpression of miR-1 might act as an oncogene in hematological malignancies [27]. Controversial results in miR-1 function depend on the type of malignant cells, and finding functional mechanism in each type of cancer needs more investigation.
We showed for the first time that miR-486-5p was upregulated in CN-AML patients. No significant association was observed between miR-486 overexpression and clinical features. Overexpression of miR-486-5p was also not correlated with FLT3+ and NPM1+ in these patients. Recently, Wang et al. have showed the overexpression of chronic myeloid leukemia (CML) in patients. Their result implicated that expression of miR-486-5p causes FoxO1 and PTEN protein levels downregulated and enhanced phospho-AKT levels in PI3K/AKT signaling in hematopoietic cells. Therefore, increased expression of miR-486 modulates leukemic hematopoietic progenitor growth, survival, and erythroid differentiation [25]. In another study, Shaham et al. showed that expression of miR-486 increased in myeloid leukemia of Down syndrome [30].
The results of our study showed upregulated expression of let-7a in CN-AML patients compared with healthy controls which is in accordance to the reported finding in these patients [20, 31]. Our findings showed higher let-7a expression in nucleophosmin positive CN-AML patients, which is consistent with results that were revealed by Becker et al. [21]. Comparison between the expression status of let-7a and its clinical features in CN-AML was similarly in line with investigation made by Jinlong et al. [31] and also in concurrence with the report by Li et al. [24]. However, in both studies, they reported no significant difference in let-7a-2-3p expression values between AML patients with or without FLT3-ITD and NPM1 mutations [24, 31]
Using PB for various laboratory investigations is more convenient than BM if PB could provide similar information as BM. Research have been shown that patients with newly diagnosis and over 30 % blasts have significant concordance between PB and BM in the results of cytogenetic and flow cytometric analysis [32, 33]. In our study, comparison between the levels of miRs in BM and PB was determined for the first time. Our data revealed that there was strong concordance between the expression levels of miRs for PB and BM samples.
In conclusion, further work is needed to determine the roles of candidate miRs and links between gene mutations and miR deregulation in CN-AML and also large patient number to confirm our data.
References
Seedhouse CH, Pallis M, Grundy M, Shang S, Russell NH. FLT3-ITD expression levels and their effect on STAT5 in AML with and without NPM mutations. Br J Haematol. 2009;147(5):653–61.
Schlenk RF, Dohner K, Krauter J, Frohling S, Corbacioglu A, Bullinger L, et al. Mutations and treatment outcome in cytogenetically normal acute myeloid leukemia. N Engl J Med. 2008;358(18):1909–18.
Lu Y, Chen W, Stein A, Weiss LM, Huang Q. C/EBPA gene mutation and C/EBPA promoter hypermethylation in acute myeloid leukemia with normal cytogenetics. Am J Hematol. 2010;85(6):426–30.
Bacher U, Schnittger S, Haferlach T. Molecular genetics in acute myeloid leukemia. Curr Opin Oncol. 2010;22(6):646–55.
Pietschmann K, Bolck HA, Buchwald M, Spielberg S, Polzer H, Spiekermann K, et al. Breakdown of the FLT3-ITD/STAT5 axis and synergistic apoptosis induction by the histone deacetylase inhibitor panobinostat and FLT3-specific inhibitors. Mol Cancer Ther. 2012;11(11):2373–83.
Deeb KK, Smonskey MT, DeFedericis H, Deeb G, Sait SN, Wetzler M, et al. Deletion and deletion/insertion mutations in the juxtamembrane domain of the FLT3 gene in adult acute myeloid leukemia. Leuk Res Rep. 2014;3(2):86–9.
Gale RE, Hills R, Pizzey AR, Kottaridis PD, Swirsky D, Gilkes AF, et al. Relationship between FLT3 mutation status, biologic characteristics, and response to targeted therapy in acute promyelocytic leukemia. Blood. 2005;106(12):3768–76.
Barakat FH, Luthra R, Yin CC, Barkoh BA, Hai S, Jamil W, et al. Detection of nucleophosmin 1 mutations by quantitative real-time polymerase chain reaction versus capillary electrophoresis: a comparative study. Arch Pathol Lab Med. 2011;135(8):994–1000.
Garzon R, Garofalo M, Martelli MP, Briesewitz R, Wang L, Fernandez-Cymering C, et al. Distinctive microRNA signature of acute myeloid leukemia bearing cytoplasmic mutated nucleophosmin. Proc Natl Acad Sci U S A. 2008;105(10):3945–50. doi:10.1073/pnas.0800135105.
Faraoni I, Laterza S, Ardiri D, Ciardi C, Fazi F, Lo-Coco F. MiR-424 and miR-155 deregulated expression in cytogenetically normal acute myeloid leukaemia: correlation with NPM1 and FLT3 mutation status. J Hematol Oncol. 2012;5:26. doi:10.1186/1756-8722-5-26.
Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–97.
Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136(2):215–33.
Marcucci G, Maharry KS, Metzeler KH, Volinia S, Wu YZ, Mrozek K, et al. Clinical role of microRNAs in cytogenetically normal acute myeloid leukemia: miR-155 upregulation independently identifies high-risk patients. J Clin Oncol. 2013;31(17):2086–93.
Bryant A, Palma CA, Jayaswal V, Yang YW, Lutherborrow M, Ma DD. miR-10a is aberrantly overexpressed in Nucleophosmin1 mutated acute myeloid leukaemia and its suppression induces cell death. Mol Cancer. 2012;11(8):1476–4598.
Chen XX, Lin J, Qian J, Qian W, Yang J, Ma JC, et al. Dysregulation of miR-124-1 predicts favorable prognosis in acute myeloid leukemia. Clin Biochem. 2014;47(1–2):63–6.
Wieser R, Scheideler M, Hackl H, Engelmann M, Schneckenleithner C, Hiden K, et al. microRNAs in acute myeloid leukemia: expression patterns, correlations with genetic and clinical parameters, and prognostic significance. Genes Chromosomes Cancer. 2010;49(3):193–203.
Havelange V, Ranganathan P, Geyer S, Nicolet D, Huang X, Yu X, et al. Implications of the miR-10 family in chemotherapy response of NPM1-mutated AML. Blood. 2014;123(15):2412–5.
Shibayama Y, Kondo T, Ohya H, Fujisawa S, Teshima T, Iseki K. Upregulation of microRNA-126-5p is associated with drug resistance to cytarabine and poor prognosis in AML patients. Oncol Rep. 2015;33(5):2176–82.
Schotte D, Pieters R, Den Boer ML. MicroRNAs in acute leukemia: from biological players to clinical contributors. Leukemia. 2012;26(1):1–12.
Chen Y, Jacamo R, Konopleva M, Garzon R, Croce C, Andreeff M. CXCR4 downregulation of let-7a drives chemoresistance in acute myeloid leukemia. J Clin Invest. 2013;123(6):2395–407.
Becker H, Marcucci G, Maharry K, Radmacher MD, Mrozek K, Margeson D, et al. Favorable prognostic impact of NPM1 mutations in older patients with cytogenetically normal de novo acute myeloid leukemia and associated gene- and microRNA-expression signatures: a Cancer and Leukemia Group B study. J Clin Oncol. 2010;28(4):596–604.
Gutierrez NC, Sarasquete ME, Misiewicz-Krzeminska I, Delgado M, De Las RJ, Ticona FV, et al. Deregulation of microRNA expression in the different genetic subtypes of multiple myeloma and correlation with gene expression profiling. Leukemia. 2010;24(3):629–37. doi:10.1038/leu.2009.274.
Nohata N, Hanazawa T, Enokida H, Seki N. microRNA-1/133a and microRNA-206/133b clusters: dysregulation and functional roles in human cancers. Oncotarget. 2012;3(1):9–21.
Li Y, Lin J, Yang J, Qian J, Qian W, Yao DM, et al. Overexpressed let-7a-3 is associated with poor outcome in acute myeloid leukemia. Leuk Res. 2013;37(12):1642–7.
Wang LS, Li L, Chu S, Shiang KD, Li M, Sun HY, et al. MicroRNA-486 regulates normal erythropoiesis and enhances growth and modulates drug response in CML progenitors. Blood. 2015;125(8):1302–13.
Marcucci G, Maharry K, Wu YZ, Radmacher MD, Mrozek K, Margeson D, et al. IDH1 and IDH2 gene mutations identify novel molecular subsets within de novo cytogenetically normal acute myeloid leukemia: a Cancer and Leukemia Group B study. J Clin Oncol. 2010;28(14):2348–55. doi:10.1200/jco.2009.27.3730.
Gomez-Benito M, Conchillo A, Garcia MA, Vazquez I, Maicas M, Vicente C, et al. EVI1 controls proliferation in acute myeloid leukaemia through modulation of miR-1-2. Br J Cancer. 2010;103(8):1292–6. doi:10.1038/sj.bjc.6605874.
Wu X, Li S, Xu X, Wu S, Chen R, Jiang Q, et al. The potential value of miR-1 and miR-374b as biomarkers for colorectal cancer. Int J Clin Exp Pathol. 2015;8(3):2840–51.
Xu L, Zhang Y, Wang H, Zhang G, Ding Y, Zhao L. Tumor suppressor miR-1 restrains epithelial-mesenchymal transition and metastasis of colorectal carcinoma via the MAPK and PI3K/AKT pathway. J Transl Med. 2014;12:244. doi:10.1186/s12967-014-0244-8.
Shaham L, Vendramini E, Ge Y, Goren Y, Birger Y, Tijssen MR, et al. MicroRNA-486-5p is an erythroid oncomiR of the myeloid leukemias of Down syndrome. Blood. 2015;125(8):1292–301.
Jinlong S, Lin F, Yonghui L, Li Y, Weidong W. Identification of let-7a-2-3p or/and miR-188-5p as prognostic biomarkers in cytogenetically normal acute myeloid leukemia. PLoS One. 2015;10(2):e0118099.
Tong WG, Sandhu VK, Wood BL, Hendrie PC, Becker PS, Pagel JM, et al. Correlation between peripheral blood and bone marrow regarding FLT3-ITD and NPM1 mutational status in patients with acute myeloid leukemia. Haematologica. 2015;100(3):e97–8. doi:10.3324/haematol.2014.118422.
Weinkauff R, Estey EH, Starostik P, Hayes K, Huh YO, Hirsch-Ginsberg C, et al. Use of peripheral blood blasts vs bone marrow blasts for diagnosis of acute leukemia. Am J Clin Pathol. 1999;111(6):733–40.
Acknowledgments
This work was supported by Tehran University of Medical Sciences (Grant No, 21389) and Hematology, Oncology and Stem Cell Transplantation Research Center, Shariati Hospital, Tehran, Iran.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Seyyedi, S.S., Soleimani, M., Yaghmaie, M. et al. Deregulation of miR-1, miR486, and let-7a in cytogenetically normal acute myeloid leukemia: association with NPM1 and FLT3 mutation and clinical characteristics. Tumor Biol. 37, 4841–4847 (2016). https://doi.org/10.1007/s13277-015-4289-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13277-015-4289-y