Abstract
The identification of new biomarkers for the early detection of hepatocellular carcinoma is critical in the development of tumor-targeted therapy, which is possibly advantageous on the prognosis of this disease. Results from our previous study indicated that CCL15 can be a specific proteomic biomarker of hepatocellular carcinoma, which plays an important role in tumorigenesis and tumor invasion. In this study, we found that CCL15 can induce hepatocellular carcinoma cell migration and invasion. Furthermore, CCR1, the receptor of CCL15, was demonstrated to play a critical role in metastatic hepatocellular carcinoma. CCR1 short hairpin RNA significantly inhibited CCL15-induced chemotaxis and invasion of HepG2 cells. Moreover, CCR1 knockdown significantly limited the activity and expression of matrix metalloproteinase-2 (MMP-2) and MMP-9. These findings suggest that CCR1 plays critical roles in hepatocellular carcinoma metastasis, which indicates that CCR1 may be a potential molecular target in hepatocellular carcinoma therapy.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Hepatocellular carcinoma (HCC) is one of the most common and aggressive malignancies worldwide [1]. The poor prognosis of HCC is mainly attributed to recurrence and metastasis [2]. Recent studies have shown that chemokines and their receptors play important roles in the metastatic potential of cancer cells [3]. For example, the CXCL12/CXCR4 pair has been identified to mediate metastasis of breast cancer cells [4].
CCL15/leukotactin-1 (Lkn-1), a member of the CC chemokine family and expressed only in the gut and the liver, has been recently cloned and partially characterized [5–7]. CCL15 was demonstrated to have a chemoattractant role for monocytes, lymphocytes, neutrophils, eosinophils, and dendritic cells [7]. In addition, this molecule plays an important role in the development of inflammation and allergic inflammatory diseases. CCL15 has in vitro and in vivo angiogenic activity [8]. Moreover, desensitization studies have indicated that CCL15 acts mainly via the CC chemokine receptor, CCR1 [6, 9]. Accumulating evidence has suggested that CCL15 may have a crucial role in the progression of tumor cells by promoting leukocyte infiltration and modulating tumor cell motility [9–13].
CCR1, the main and specific receptor for CCL15, is a G protein-coupled receptor that is expressed by a variety of cells, such as monocytes, lymphocytes, neutrophils, and eosinophils [14]. Recent studies have verified that CCR1 is also highly expressed in HCC cells and tissues [15], and its downregulation significantly inhibits HCC cell migration and invasion [16]. However, the function and mechanism of CCL15/CCR1 pair in HCC cell migration and invasion are not well understood.
Results from our previous study showed that CCL15 was uniquely expressed in serum samples from HCC patients using SELDI-TOF technique. Biological analysis suggested that CCL15 promoted HCC migration and invasion (unpublished data). Hence, in this study, we investigated our hypothesis that the interaction of CCL15 with CCR1 may have some essential roles in the HCC cell migration and metastasis.
Materials and methods
Cells and materials
Hepatocellular carcinoma cell lines HepG2, HLE, and HuH7 were obtained from American Type Culture Collection (Manassas, VA), and cultured in DMEM 1640 supplemented with 10 % (v/v) fetal bovine serum (FBS). Chemotaxis chambers and membranes were from Neuroprobe (Gaithersburg, MD, USA). Recombinant Human CCL15/68aa was from RD Systems (USA). Antibody to CCR1 was from Abcam Inc. (CA). Antibody to β-actin was from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA, USA). Goat anti-mouse IgG-FITC and Donkey anti-goat IgG-FITC were from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA).
Gene transfection
2 × 105 cells were cultured in 12-well plates 1 day before transfection in DMEM containing 10 % FBS, 1 % non-essential amino acids, and 1 % sodium pyruvate. After starved for 3 h, transfection was performed by using Lipofectamine2000 (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions. To establish stable cell lines, the cells were enriched using 0.6 mg/ml G418 sulfate (Gibco/Invitrogen, USA) or 0.4 mg/ml hygromycin B from BD Biosciences. After the cells grow up, single cell-expressing GFP was picked out and cultured into 96-well plates, and then each clone was validated by RT-PCR and Western blotting analysis.
Reverse transcription-PCR (RT-PCR) analysis
Total RNA was extracted from cells using Trizol (Invitrogen) and then reverse-transcribed and amplified using the One Step RNA PCR Kit (AMV) (TaKaRa, Shiga, Japan). The primers sequences, the predicted amplicon sizes, and the annealing temperatures are depicted in Table 1. The conditions for PCR were as follows: 95 °C for 5 min followed by annealing temperature for 1 min and 72 °C for 1 min in a 25 mL reaction buffer containing cDNA generated from 100 ng of total RNA, 0.2 mM each dNTP, 0.2 mM each primer, and 2.5 U of Taq polymerase (TaKaRa Biotechnology, Dalian, People’s Republic of China). Quantitation of the amount of PCR product after 35 cycles was performed after electrophoresis on 1 % agarose gels and ethidium bromide staining.
Western blotting
Western blotting assay was performed as described by Sun R.H. et al. [17]. Cells and clones were washed twice with ice cold PBS pH 6.8, and then lysed by 1 × SDS sample buffer (Tris–HCl, pH 6.8, 62.5 mM, 2 % SDS, 10 % glycerol.). Equal amounts of cell lysates (20 μg total protein per lane) were loaded onto 10 % or 14 % SDS-PAGE gels, transferred onto PVDF membranes (Millipore, USA), probed with anti-CCR1 primary antibody, followed by HRP-conjugated secondary antibody and visualized by enhanced chemiluminescence reagents ECL (Pierce, Rockford, IL). Western blotting analysis of β-actin was used as loading control.
Chemotaxis assay
Chemotaxis assays were performed essentially as previously described using Boyden chambers equipped with 8-μm porosity polyvinylpyrrolidone-free polycarbonate filters [18]. Briefly, polycarbonate filters were coated on the lower surface with 20 μg/ml human type I collagen (Collaborative Biomedical Products; Bedford, MA) for 30 min at 37 °C and placed between the lower chamber and upper chamber. The lower chamber was filled with DMEM media containing CCL15 as chemoattractants. Serum-deprived HepG2, Huh-7, and HLE cells were separately washed, trypsinized, resuspended in serum-free medium containing 1 % albumin at a concentration of 3 × 105 cells/ml, and placed in the upper chamber. The chambers were incubated at 37 °C in a cell-culture incubator for 6 h. Then the filter membrane was washed, and cells attached were fixed and stained. The number of migrating cells was counted in six randomly chosen fields (×400) by light microscope, and the counts were averaged (means ± SD). Chemotaxis index = the migrating cell number in a chemoattractant gradient/the migrating cell number in a medium control.
Scratch assay
Cells were seeded in 35-mm dishes for 2 days, starved in serum-free medium overnight, after which, scrape wounds were created with a 10-μl sterile plastic pipette tip in the confluent cell monolayers. After washing with PBS, serum-free medium (to prevent cell proliferation) was added. The widths of the wounds were recorded at different time points (0, 3, 6, 9, 12, 24 h) using the number of grids in the ocular of our microscope. Ten grids represented 1 mm in length, and we could therefore calculate the migration distances by the changes of the grid number accordingly. Three measurements at different positions along each scratching line were recorded. And in order to test the effect of CCL15 on the motility of hepatocarcinoma cells, the serum-free medium containing CCL15 (1 μM) was added to the scraped cells and cultured for 24 h.
Matrigel invasion assay
For invasion assays, 105 cells in 400 μL serum-free DMEM medium were plated in the top chamber of Transwells with a Matrigel-coated membrane (Corning Incorporated, USA) containing pores 8 μm diameter. The inserts were placed into the bottom chamber wells of a 24-well plate containing MEM with CCL15 as a chemoattractant. After 36 h of incubation, cells remaining on the insert top layers were removed by cotton swab scrubbing, as recommended by the manufacturer. Cells on the lower surface of the membrane were fixed and stained. Inserts were washed for several times in distilled water before air drying. The membranes were photographed, and cell counts were determined after totaling five random fields.
Gelatin zymography
Gelatin zymography assay was performed essentially as previously described [19]. Cells transfected with control or CCR1 small interfering RNA (siRNA) were treated with 1 μM CCL15 for 12 h in serum-free medium. The conditioned medium was collected and analyzed on 10 % SDS–polyacrylamide gel incorporated with 0.1 % gelatin.
Statistical analysis
All statistical analyses were carried out with the use of SPSS software Version 13.0. Data were expressed as the mean ± standard error from at least three separate experiments. Statistical significance of the results was analyzed by two-way ANOVA, and p < 0.05 was considered to be statistically significant.
Results
CCL15 induced HCC cells chemotaxis and invasion
CCL15 binds with CCR1 and CCR3 and has a chemoattractant role for neutrophils, monocytes, and lymphocytes [7]. First, we detected the messenger RNA (mRNA) and protein expression of CCR1 in HuH7, HLE, and HepG2 cells (Fig. 1a). The CCR1 expression could be detected in three different HCC cell lines.
Effects of CCL15 on hepatocarcinoma cells migration and invasion. a RT-PCR and Western blotting analysis of CCR1 in Huh-7, HLE, and HepG2 cells. b Chemotaxis assay of HepG2 cells with different concentrations of CCL15 treatment. c Wound healing assay of HepG2 cells and HLE cells with or without CCL15 (1 nM) treatment. d Invasion assays of HepG2 cells and HLE cells under with or without CCL15 treatment, with invasion pictures shown (×200) (* p < 0.05)
To determine the function of CCL15 in HCC cell migration, we performed chemotaxis assay. Chemotaxis refers to the directional cell movement that is dependent on the concentration gradient. A 48-well chemotaxis model was applied to perform CCL15-induced chemotaxis. The chemotaxis activity of HepG2 cells depended on the concentrations of CCL15 used in the induction. Moreover, the highest chemotaxis activity was highest observed at 1-nM recombinant human CCL15 (Fig. 1b). This result demonstrates that the HepG2 cell migration in response to CCL15 is dosage-dependent.
Then, we performed a scratch assay in a medium supplemented with 0.5 % FBS to examine the effects of CCL15 in wound healing and cancer cell directional migration. As shown in Fig. 1c, treatment with recombinant human CCL15 could promote HepG2 cell migration compared with cells without CCL15 HepG2 cells.
Cell migration is essential in tumor invasion and metastasis. To measure the effects of CCL15 on HCC cell invasiveness, we employed a Transwell invasion system. The system consisted of two fluid-filled stacked compartments separated by a porous membrane filter coated with Matrigel. The quantitative analysis of cell numbers revealed that the invaded cells induced by CCL15 in the HepG2 and HLE cells were higher than in the medium alone (Fig. 1d). These results strongly suggest that CCL15 can promote hepatocellular cell migration and invasion.
Knockdown of CCR1 suppresses HCC cells chemotaxis and invasion
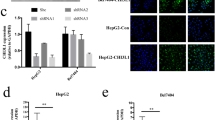
The physiological role of CCR1 in HCC can be elucidated by inhibiting the expression of endogenous CCR1 in HCC cell lines. Efficient knockdown of CCR1 expression was achieved by the successful transfection of three stealth siRNA duplexes (si#1, si#2, and si#3 siRNA) targeting CCR1 in HepG2 cells. Transient transfection of si#1 and si#3 siRNA specifically downregulated CCR1 protein expression (Fig. 2a).
CCR1 silencing impaired the migration and invasion of HepG2 cells. (a Western blotting analysis of HepG2 cells transfected with CCR1 siRNA plasmid. Overexpression of CCR1 and β-actin was used as loading control. b CCR1 downregulation inhibits the migration of HepG2 cells by wound healing scratch assay. The migrating distances of monolayer cells were measured at 0, 3, 6, 9, 12, and 24 h after scratches created under microscope. c Chemotaxis assay of parental cells and siCCR1 transfected cells. d Invasion assay of parental cells and siCCR1 transfected cells. e Cell migration and invasion were not significantly changed in siCCR1 cells with or without CCL15 treatment
The migratory ability of CCR1 knockdown cells was assessed by performing wound-healing, chemotaxis, and Matrigel invasion assays. Differences in wound-closure ability were more evident in CCR1 knockdown cells compared with the control cells. As shown in Fig. 2b, the cells with downregulated CCR1 expression took a longer time to fill the wound gap compared with the control cells. Consistently, the chemotaxis assay revealed that HepG2 cells exhibited significant responses to 1 nM CCL15, but the downregulation of CCR1 dramatically inhibited CCL15-induced chemotaxis (Fig. 2c). In addition, Matrigel-based invasion assay was applied to explore the effect of CCR1 silencing on the invasive capability of HepG2 cells. As shown in Fig. 2d, the number of migrated cells in the control HepG2 cells was significantly higher than that in the CCR1 knockdown cells. However, no significant difference was observed on cell migration and invasion with or without CCL15 after the downregulation of CCR1 expression in the HepG2 cells (Fig. 2e). Hence, these results demonstrate that the knockdown of CCR1 reduced the migratory and invasive potentials of HCC cells.
Overexpression of CCR1 promotes HCC cells chemotaxis and invasion
Overexpression of CCR1 in HCC cells was also adopted as a complementary method to characterize the role of CCR1 in HCC. Thus, we generated stable cell lines expressing the CCR1 vector or the control vector (HepG2/CCR1 and HepG2/control, respectively) by selecting transfected cells in G418. CCR1 stable cell lines were confirmed by Western blot and RT-PCR (Fig. 3a). Cells that overexpressed CCR1 were subjected to wound-healing, chemotaxis, and invasion assays. Results revealed that cells that overexpressed CCR1 displayed significantly enhanced cell migration and invasion than the vector control cells (Fig. 3b–d).
The migration and invasion assays of overexpressed CCR1 clones of HepG2 cells. a RT-PCR and Western blotting analysis of HepG2 clones with overexpression of CCR1. b Wound healing assay of HepG2 cells and HepG2 /Clone1 at indicated time points. c Wound healing assay of CCR1 overexpressed HepG2 cells with or without CCL15 treatment. c Chemotaxis assay of HepG2 cells and HepG2/Clone1 cells with indicated amount of CCL15 as chemoattractant for 6 h. e Invasion assay of HepG2 cells and HepG2/Clone1 cells with CCL15 as chemoattractant for 36 h, with pictures shown (×200) (* p < 0.05, ** p < 0.005)
Furthermore, we explored the functions of CCL15 in the migration of cells that overexpress CCR1. The wound-healing assay showed that recombinant human CCL15-treated cells took a much longer time to fill the wound gap compared with the untreated cells (Fig. 3e).
CCR1 silencing inhibits hepatocellular carcinoma cells migration and invasion by regulating MMP-2 and MMP-9 expression and activity
The MMP overexpression plays an important role in cancer metastasis [20]. Thus, we also investigated the role of CCL15/CCR1 axis on MMP expression and activity in HepG2 cells. The expression levels of MMP-1, MMP-2, MMP-3, MMP-9, and MMP-10 between control and siCCR1 cells were compared using qPCR. The results showed that mRNA levels of MMP-2 and MMP-9 were downregulated in the siCCR1 cells, whereas the expression of MMP-1, MMP-3, and MMP-10 did not change significantly (Fig. 4a). However, no difference was observed between the MMP-2 and MMP-9 expression of the CCL15-treated and untreated siCCR1 cell (Fig. 4b).
Disruption of CCR1 by siRNA impairs MMP-2 and MMP-9 expression. a Expressions of MMPs were compared by qPCR between scr cells and siCCR1 cells, *p < 0.05. b The expression of MMP-2 and MMP-9 were not significantly changed in siCCR1 cells with or without CCL15 treatment. c Activated form of MMP-2 and MMP-9 in indicated cells was assessed using gelatin zymography assays
The active forms of MMP-2 and MMP-9 were detected in the culture medium of both cells using gelatin zymography. The amounts of active MMP-2 and MMP-9 in the culture medium were downregulated when CCR1 was reduced in siCCR1/HepG2 cells. However, the treatment of recombinant human CCL15 in siCCR1/HepG2 cells cannot alter the activity of MMP-2 and MMP-9.
These results suggest that the metastasis-promoting function of CCL15/CCR1 axis may occur through extracellular matrix remodeling.
Discussion
Metastasis is one of the main obstacles in the improvement of the survival rate and long-term prognosis of patients with HCC. The final site of metastasis of tumor cells is organ-specific, in which chemokine receptors play important roles in the chemotaxis of tumor cells to target organs [21, 22]. Identification of these metastasis-related chemokine receptors may provide potential targets in tumor therapy. CCR1 was the first identified CC chemokine receptor that binds with CC chemokines, including MIP-1a/CCL3, RANTES, CCL8, CCL7, Lkn-1 (CCL15)/MIP-5, CCL23, and HCC-1 [23–29]. CCR1 is expressed in neutrophils, monocytes, eosinophils, dendritic cells, activated T cells, and B lymphocytes [30–33]. CCR1 expression was detected in ovarian cancer tissues [34, 35] and malignant plasma cells [36]. Mukaida et al. demonstrated that CCR1 was the only chemokine receptor that was expressed consistently in all hepatoma cell lines, suggesting that CCR1 may induce survival signals in hepatoma cells [15]. In this study, HepG2 clone cells with CCR1 overexpression have stronger migration abilities under CCL15 stimulation, whereas the disruption of CCR1 yielded contrasting results, suggesting that CCL15 acts mainly via its receptor CCR1 to induce hepatoma cell migration and chemotaxis.
MMPs are part of a large family of proteolytic enzymes, which plays a key role in cancer invasion and metastasis because of their ability to degrade the extracellular matrix and basement membrane in various cancers [37]. Among these enzymes, MMP-9 and MMP-2 have been found to be highly associated with metastatic spread in various cancers [38]. MMP-2 activity is necessary for Matrigel invasion of HCC cells [39]. In this study, we found that CCR1 gene silencing inhibited the CCL15-induced MMP-2 and MMP-9 upregulation, which suggests that CCL15/CCR1 axis might regulate MMP-2 and MMP-9 production in human HCC. The functions of CCL15/CCR1 axis on tumor cell migration and invasion may be through induced MMP-2 and MMP-9 production, thereby degrading the extracellular matrix.
In summary, we have shown for the first time that CCL15/CCR1 axis has an important role in HCC cell migration and invasion, which may be mediated by MMP-2 and MMP-9.
References
Parkin DM, Bray F, Ferlay J, Pisani P. Global cancer statistics, 2002. CA Cancer J Clin. 2005;55:74–108.
Thorgeirsson SS, Grisham JW. Molecular pathogenesis of human hepatocellular carcinoma. Nat Genet. 2002;31:339–46.
Singh S, Sadanandam A, Singh RK. Chemokines in tumor angiogenesis and metastasis. Cancer Metastasis Rev. 2007;26:453–67.
Muller A, Homey B, Soto H, Ge N, Catron D, Buchanan ME, et al. Involvement of chemokine receptors in breast cancer metastasis. Nature. 2001;410:50–6.
Zhang S, Youn BS, Gao JL, Murphy PM, Kwon BS. Differential effects of leukotactin-1 and macrophage inflammatory protein-1 alpha on neutrophils mediated by CCR1. J Immunol. 1999;162:4938–42.
Pardigol A, Forssmann U, Zucht HD, Loetscher P, Schulz-Knappe P, Baggiolini M, et al. HCC-2, a human chemokine: gene structure, expression pattern, and biological activity. Proc Natl Acad Sci U S A. 1998;95:6308–13.
Youn BS, Zhang SM, Lee EK, Park DH, Broxmeyer HE, Murphy PM, et al. Molecular cloning of leukotactin-1: a novel human beta-chemokine, a chemoattractant for neutrophils, monocytes, and lymphocytes, and a potent agonist at CC chemokine receptors 1 and 3. J Immunol. 1997;159:5201–5.
Hwang J, Kim CW, Son KN, Han KY, Lee KH, Kleinman HK, et al. Angiogenic activity of human CC chemokine CCL15 in vitro and in vivo. FEBS Lett. 2004;570:47–51.
Ko J, Kim IS, Jang SW, Lee YH, Shin SY, Min DS, et al. Leukotactin-1/CCL15-induced chemotaxis signaling through CCR1 in HOS cells. FEBS Lett. 2002;515:159–64.
Gerard C, Rollins BJ. Chemokines and disease. Nat Immunol. 2001;2:108–15.
Mukaida N. Interleukin-8: an expanding universe beyond neutrophil chemotaxis and activation. Int J Hematol. 2000;72:391–8.
Kunz M, Hartmann A, Flory E, Toksoy A, Koczan D, Thiesen HJ, et al. Anoxia-induced up-regulation of interleukin-8 in human malignant melanoma. A potential mechanism for high tumor aggressiveness. Am J Pathol. 1999;155:753–63.
Desbaillets I, Diserens AC, Tribolet N, Hamou MF, Van Meir EG. Upregulation of interleukin 8 by oxygen-deprived cells in glioblastoma suggests a role in leukocyte activation, chemotaxis, and angiogenesis. J Exp Med. 1997;186:1201–12.
Gao JL, Wynn TA, Chang Y, Lee EJ, Broxmeyer HE, Cooper S, et al. Impaired host defense, hematopoiesis, granulomatous inflammation and type 1–type 2 cytokine balance in mice lacking CC chemokine receptor 1. J Exp Med. 1997;185:1959–68.
Lu P, Nakamoto Y, Nemoto-Sasaki Y, Fujii C, Wang H, Hashii M, et al. Potential interaction between CCR1 and its ligand, CCL3, induced by endogenously produced interleukin-1 in human hepatomas. Am J Pathol. 2003;162:1249–58.
Wu X, Fan J, Wang X, Zhou J, Qiu S, Yu Y, et al. Downregulation of CCR1 inhibits human hepatocellular carcinoma cell invasion. Biochem Biophys Res Commun. 2007;355:866–71.
Sun R, Gao P, Chen L, Ma D, Wang J, Oppenheim JJ, et al. Protein kinase C zeta is required for epidermal growth factor-induced chemotaxis of human breast cancer cells. Cancer Res. 2005;65:1433–41.
Carloni V, Romanelli RG, Mercurio AM, Pinzani M, Laffi G, Cotrozzi G, et al. Knockout of alpha6 beta1-integrin expression reverses the transformed phenotype of hepatocarcinoma cells. Gastroenterology. 1998;115:433–42.
Das G, Shiras A, Shanmuganandam K, Shastry P. Rictor regulates MMP-9 activity and invasion through Raf-1-MEK-ERK signaling pathway in glioma cells. Mol Carcinog. 2011;50:412–23.
Gialeli C, Theocharis AD, Karamanos NK. Roles of matrix metalloproteinases in cancer progression and their pharmacological targeting. FEBS J. 2011;278:16–27.
Luker KE, Lewin SA, Mihalko LA, Schmidt BT, Winkler JS, Coggins NL, Thomas DG, Luker GD. Scavenging of CXCL12 by CXCR7 promotes tumor growth and metastasis of CXCR4-positive breast cancer cells. Oncogene. 2012.
Li J, Sun R, Tao K, Wang G. The CCL21/CCR7 pathway plays a key role in human colon cancer metastasis through regulation of matrix metalloproteinase-9. Dig Liver Dis. 2011;43:40–7.
Yang X, Walton W, Cook DN, Hua X, Tilley S, Haskell CA, et al. The chemokine, CCL3, and its receptor, CCR1, mediate thoracic radiation-induced pulmonary fibrosis. Am J Respir Cell Mol Biol. 2011;45:127–35.
Suffee N, Hlawaty H, Meddahi-Pelle A, Maillard L, Louedec L, Haddad O, Martin L, Laguillier C, Richard B, Oudar O, Letourneur D, Charnaux N, Sutton A. RANTES/CCL5-induced pro-angiogenic effects depend on CCR1, CCR5 and glycosaminoglycans. Angiogenesis. (2012).
Gong X, Gong W, Kuhns DB, Ben-Baruch A, Howard OM, Wang JM. Monocyte chemotactic protein-2 (MCP-2) uses CCR1 and CCR2B as its functional receptors. J Biol Chem. 1997;272:11682–5.
Corbin ME, Pourciau S, Morgan TW, Boudreaux M, Peterson KE. Ligand up-regulation does not correlate with a role for CCR1 in pathogenesis in a mouse model of non-lymphocyte-mediated neurological disease. J Neurovirol. 2006;12:241–50.
Escher SE, Forssmann U, Frimpong-Boateng A, Adermann K, Vakili J, Sticht H, et al. Functional analysis of chemically synthesized derivatives of the human CC chemokine CCL15/HCC-2, a high affinity CCR1 ligand. J Pept Res. 2004;63:36–47.
Hwang J, Son KN, Kim CW, Ko J, Na DS, Kwon BS, et al. Human CC chemokine CCL23, a ligand for CCR1, induces endothelial cell migration and promotes angiogenesis. Cytokine. 2005;30:254–63.
Tsou CL, Gladue RP, Carroll LA, Paradis T, Boyd JG, Nelson RT, et al. Identification of C-C chemokine receptor 1 (CCR1) as the monocyte hemofiltrate C-C chemokine (HCC)-1 receptor. J Exp Med. 1998;188:603–8.
Choi SW, Hildebrandt GC, Olkiewicz KM, Hanauer DA, Chaudhary MN, Silva IA, et al. CCR1/CCL5 (RANTES) receptor-ligand interactions modulate allogeneic T-cell responses and graft-versus-host disease following stem-cell transplantation. Blood. 2007;110:3447–55.
Sebag M. CCR1 blockade and myeloma bone disease. Blood. 2012;120:1351–2.
Semnani RT, Mahapatra L, Dembele B, Konate S, Metenou S, Dolo H, et al. Expanded numbers of circulating myeloid dendritic cells in patent human filarial infection reflect lower CCR1 expression. J Immunol. 2010;185:6364–72.
Furuichi K, Gao JL, Horuk R, Wada T, Kaneko S, Murphy PM. Chemokine receptor CCR1 regulates inflammatory cell infiltration after renal ischemia-reperfusion injury. J Immunol. 2008;181:8670–6.
Scotton C, Milliken D, Wilson J, Raju S, Balkwill F. Analysis of CC chemokine and chemokine receptor expression in solid ovarian tumours. Br J Cancer. 2001;85:891–7.
Long H, Xie R, Xiang T, Zhao Z, Lin S, Liang Z, et al. Autocrine CCL5 signaling promotes invasion and migration of CD133(+) ovarian cancer stem-like cells via NF-kappaB-mediated MMP-9 upregulation. Stem Cells. 2012;30:2309–19.
Trentin L, Miorin M, Facco M, Baesso I, Carraro S, Cabrelle A, et al. Multiple myeloma plasma cells show different chemokine receptor profiles at sites of disease activity. Br J Haematol. 2007;138:594–602.
Kessenbrock K, Plaks V, Werb Z. Matrix metalloproteinases: regulators of the tumor microenvironment. Cell. 2010;141:52–67.
Seo JM, Park S, Kim JH. Leukotriene B4 receptor-2 promotes invasiveness and metastasis of ovarian cancer cells through signal transducer and activator of transcription 3 (STAT3)-dependent up-regulation of matrix metalloproteinase 2. J Biol Chem. 2012;287:13840–9.
Hayashi Y, Osanai M, Lee GH. Fascin-1 expression correlates with repression of E-cadherin expression in hepatocellular carcinoma cells and augments their invasiveness in combination with matrix metalloproteinases. Cancer Sci. 2011;102:1228–35.
Acknowledgments
This work was supported by research grants from the National Scientific Foundation of China (NSFC #81101754)
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflicts of interest
None
Rights and permissions
About this article
Cite this article
Li, Y., Wu, J. & Zhang, P. CCL15/CCR1 axis is involved in hepatocellular carcinoma cells migration and invasion. Tumor Biol. 37, 4501–4507 (2016). https://doi.org/10.1007/s13277-015-4287-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13277-015-4287-0