Abstract
Ring finger protein 1 (Ring1) have recently been reported to be closely related to aggressive tumor features in multiple cancer types, including prostate cancer, non-small-cell lung cancer, and bladder cancer. However, the role of Ring1 in human hepatocarcinogenesis remains unclear. In this study, we aimed at investigating the latent role of Ring1 in hepatocellular carcinoma (HCC) development. The expression of Ring1 was evaluated using Western blot analysis in 8 paired fresh HCC tissues and immunohistochemistry on 98 paraffin-embedded sections from 2005 to 2008. Moreover, RNA interference, CCK-8, colony formation, and flow-cytometry analyses were performed to investigate the role of Ring1 in the regulation of HCC cell proliferation. Compared with adjacent normal tissues, the level of Ring1 was significantly increased in HCC specimens. High expression of Ring1 was associated with histological grade (P = 0.011) and tumor size (P = 0.004), and Ring1 expression was positively related with the proliferation marker Ki-67 (P < 0.001). Moreover, knocking down Ring1 induced growth impairment and G1/S cell cycle arrest in HCC cells. Kaplan–Meier survival curves showed that high expression of Ring1 indicated poor prognosis of HCC (P = 0.03). On the basis of these results, we proposed that the expression of Ring1 protein may be a novel indicator of HCC prognosis.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Human hepatocellular carcinoma (HCC) ranks the fifth leading cause of cancer mortality worldwide. Because of the high prevalence of invasion and metastasis, the 5-year postoperative survival rate is 30–40 % [1–3]. The exact molecular mechanisms underlying the occurrence and development of HCC remain to be elucidated, though various risk factors for HCC, such as hepatitis B virus (HBV), hepatitis C virus (HCV), and aflatoxin B1, have been well defined [4]. Moreover, hepatocarcinogenesis is a very complex process associated with complicated changes in host gene expression patterns including oncogenes, tumor suppressor genes, and some cell cycle regulators during the processes of HCC initiation and progression [5]. Therefore, the identification of potential molecule underlying HCC development may provide new strategies for the diagnosis and treatment of this dismal disease.
Ring1 is a member of polycomb-group (PcG) protein that contains a RING finger motif, a specific zinc-binding domain that exists in many regulatory proteins [6]. Ring finger protein 1 (Ring1) was first reported as a transcriptional repressor interacting with the PcG protein complex and exerting tumorigenic activity [7]. Ring1 interacts with polycomb-group proteins to mediate histone ubiquitylation and Hox gene silencing [8]. It is supposed that Ring1 is mainly confined to polycomb repressive complex 1 (PRC1). Ring1-containing PRC1 plays an important role in the regulation of cell proliferation and transformation [9, 10]. For example, PRC1 binds directly to INK4A-ARF locus and repress the expression of p16INK4A and p14ARF, leading to resultant CDK4/CDK6 activation and cell proliferation [11]. Importantly, it has been manifest that polycomb group proteins exert critical roles in the maintenance of somatic and cancer stem cells [12]. Accordingly, substantial evidence has indicated that Ring1 is highly expressed in assorted cancer types, including lung cancer [13, 14] and bladder cancer [15]. However, it remains unclear whether Ring1 exerts a function in hepatocarcinogenesis.
In the current study, we investigated the clinical significance of Ring1 expression in HCC patients and the correlation between Ring1 expression and clinicopathological parameters by immunohistochemistry in HCC specimens. We found that the expression of Ring1 was closely correlated with histological grade (P = 0.011), tumor size (P = 0. 004), and the overall survival of HCC patients. In addition, Kaplan–Meier survival curve analysis indicated the usefulness of Ring1 as a prognostic factor of HCC. All these data implied that Ring1 might be a HCC-associated molecule with a prognostic value.
Materials and methods
Patients
Hepatocellular carcinoma tissues were obtained from 98 patients who underwent hepatic resection without preoperative systemic chemotherapy at the Surgery Department of the Affiliated Hospital of Nantong University from 2005 to 2008. All HCC tissues were collected using protocols approved by the Ethics Committee of Cancer Hospital of Nantong University, and written informed consent was obtained from every patient. The main clinical and pathologic variables are shown in Table 1. Their ages ranged from 25 to 77 years, with an average age of 53.04 years. Most of the patients were male and the male to female ratio was 65:33. Histological grades were classified to well (grade I-II; n = 54) and poorly differentiated (grade III-IV; n = 44). The follow-up time was 5 years, with a range of 2 to 70 months. Formalin-fixed, paraffin-embedded sections were prepared for all tissues and reviewed by 3 pathologists. Clinical data (patient history, diagnosis, staging, and survival) were obtained from the National Cancer Institute “Regina Elena” databases. Survival data were integrated by periodic interviews with their relatives. Tissue specimens were immediately processed after surgical removal. For histological examination, all tumorous and surrounding non-tumorous tissue portions were processed into 10 % buffered formalin-fixed, paraffin-embedded blocks. Protein was analyzed in 8 snap-frozen tumorous and adjacent non-tumorous tissue samples that were stored at −80 °C.
Immunohistochemistry
The sections were deparaffinized using a graded ethanol series, and endogenous peroxidase activity was blocked by soaking in 0.3 % hydrogen peroxide. Thereafter, the sections were processed in 10 mM citrate buffer (pH 6.0) and heated to 121 °C in an autoclave for 20 min to retrieve the antigen. After rinsing in phosphate-buffered saline (PBS; pH 7.2), 10 % goat serum was applied for 1 h at room temperature to block any non-specific reactions. The sections were then incubated 4 h with anti-human Ring1 monoclonal antibody (diluted 1:100; Santa Cruz Biotechnology, Santa Cruz, CA) and anti-Ki-67 mouse monoclonal antibody (diluted 1:100; Santa Cruz Biotechnology). Negative control slides were also processed in parallel using a non-specific immunoglobulin IgG (diluted 1:100; Santa Cruz Biotechnology) at the same concentration as the primary antibody. All slides were processed using the peroxidase-anti-peroxidase complex method (Dako, Hamburg, Germany). After rinsing in water, the peroxidase sections were counterstained with hematoxylin, dehydrated, and cover-slipped [16].
Immunohistochemical evaluation
Stained sections were observed under a microscope. All of the immunostained sections were evaluated in a blinded manner without knowledge of the clinical and pathological parameters of the patients. For assessment of Ring1 and Ki67, 5 high-power fields in each specimen were selected randomly, and cell staining was examined under high-power magnification. More than 500 cells were counted to determine the mean percent, which represented the percentage of immunostained cells related to the total number of cells. We defined that: (+): ≥ mean percent; (−): < mean percent. In half of the samples, staining was repeated twice to avoid possible technical errors, but similar results were obtained in these samples [17].
Western blot analysis
Before Western blotting, cells were washed three times with ice-cold PBS, re-suspended in lysis buffer (50 mM Tris–HCl, 120 mM NaCl, 0.5 % Nonidet P-40, 100 mM NaF, 200 lMNa3VO4, and protease inhibitor mixture) or frozen tissues were homogenized in lysis buffer (1 % NP-40, 50 mM/l Tris, pH 7.5, 5 mM/l EDTA, 1 % SDS, 1% sodium deoxycholate, 1 % Triton X-100, 1 mM/l PMSF, 10 mg/ml aprotinin, and 1 mg/ml leupeptin), and then incubated for 20 min at 4 °C while rocking. Lysates were cleared by centrifugation (10 min, 12,000 rpm, 4 °C). Fifty micrograms of total protein was resolved by SDS-PAGE and transferred onto polyvinylidene difluoride membranes (Immbilon, Millipore). The membranes were firstly blocked with 5 % nonfat dry milk and then incubated with the primary antibody described above for 2 h at room temperature. After three times of washes, filters were incubated with horseradish peroxidase-conjugated secondary human anti-mouse or anti-rabbit antibodies (1:5000; pierce) for 1 h at room temperature according to the manufacturer’s instructions. Detection of immunocomplexes was performed with an enhanced chemiluminescence system (NEN Life Science Products, Boston, MA) [18].
Cell culture and cell cycle analysis
The SMMC-7721, HepG2, SK-Hep1, and 97H human HCC lines along with LO2—a normal human liver cell line—were obtained from the Institute of Cell Biology, Academic Sinica and cultured in DMEM supplemented with 10 % fetal bovine serum, 100 units/ml penicillin, and 100 μg/ml streptomycin in 5 % CO2 at 37 °C. For cell cycle analysis, cells were fixed in 70 % ethanol for 1 h at 4 °C, and then incubated with 1 mg/ml RNase A for 30 min at 37 °C. Subsequently, cells were stained with propidiumiodide (50 μg/ml PI) (Becton Dickinson, San Jose, CA) in PBS, 0.5 % triton- × 100, and analyzed using a Becton Dickinson flow cytometer BD FACSCAN (San Jose, CA) and Cell Quest acquisition and analysis programs [19].
SiRNA and transfection
Small interference RNAs (siRNA) were chemically synthesized (GenePharma Co. Ltd). The synthesized oligonucleotides for RNA interference (RNAi) Ring1 targeted the sequences: siRNA#1: 5′-AGAUCUUAGAGAUCAGGGCTT-3′; siRNA#2: 5′-AAACAGCAAUCUCUGUGCCTT-3′. A nonspecific scrambled siRNA with a sequence of 5′-UUCUCCGAACGUGUCACGUTT-3′ was used as a negative control. Huh7 cells were seeded the day before transfection using the Dulbecco-modified Eagle medium with 10 % FBS but without antibiotics. For transient transfection, the Ring1 siRNA vector and the negative control vector were carried out by adding the mixture of siRNA and the siRNA transfection reagent (Santa Cruz Biotechnology) and plus siRNA dilution buffer as suggested by the manufacturer. Transfected cells were used for the subsequent experiments 48 h after transfection.
Cell proliferation assay
Cell viability was measured using the Cell Counting Kit-8(CCK-8) assay following the manufacturer’s instructions. Briefly, cells were plated at a density of 2 × 104 cells/well in a 96-well plate in a volume of 100 μL and grown overnight. After adding Cell Counting Kit-8 (Dojindo, Kumamoto, Japan) reagents to a subset of wells under different treatments and the cells were incubated for 2 h at 37 °C, we quantified the absorbance on an automated plate reader.
Colony formation assay
Huh7 cells were seeded at 200 cells/well in 6-well culture plates after transfected according to the manufacturer’s instructions. After cultured for 10 days, the colonies (≥50 cells/colony) were counted after 0.5 % crystal violet staining.
Statistical analysis
Statistical analysis was performed using the Stat View 5.0 software package. The association between Ring1 and Ki-67 expression and clinicopathological variables were computed using the χ2 test. Ki-67 and Ring1 expression in HCC was studied using the Spearman rank correlation test because the data were not normally distributed. For analysis of survival data, Kaplan–Meier curves were constructed, and the log-rank test was performed. Multivariate analysis was performed using Cox’s proportional hazards model. P < 0.05 was considered statistically significant in the statistical analysis. The results are expressed as mean ± SE [20].
Results
Increased expression of Ring1 in HCC tissues and cell lines
To determine whether the level of Ring1 is associated with the progression of HCC, comparative analysis of Ring1 expression was conducted on eight pairs of matched HCC tissues and adjacent normal tissues and five HCC cell lines using Western blotting analysis. As shown in Fig. 1, the expression of Ring1 was dramatically increased in HCC tissues compared with the adjacent normal tissues (Fig.1a, b). In addition, Huh7 cells displayed the highest abundance of Ring1 whereas LO2 hepatocytes exhibited the lowest level of Ring1 expression among these available HCC cells and hepatocytes (Fig.1c, d).
Expression of Ring1 in human hepatocellular carcinoma tissues and cell lines. Eight representative paired samples of HCC tissue (T) and adjacent normal tissues (N) were used for Western blot analysis. The results of Western blot analysis (a) and the quantification graph (b) shows that, in all samples except one tested, Ring1 expression levels were significantly higher in HCC than in paired adjacent normal tissues. Western blot analysis shows that Ring1 protein expression is increased in HCC cells compared with the normal liver cell line (L02) (c). The bar chart demonstrates the ratio of Ring1 protein to GAPDH by densitometry (d). The data are mean ± SEM. GAPDH was used as a control for protein load and integrity. The same experiment was repeated at least 3 times
Ring1 expression correlates consistently with Ki-67 expression in HCC
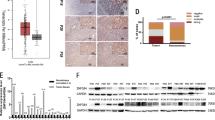
Next, we determined the association between Ring1 expression and the histological characteristics of HCC. Ninety-eight paraffin-embedded HCC clinical specimens, which included 54 cases of well-differentiated carcinoma (stage I-II) and 44 cases of poor-differentiated carcinoma (stage III-IV), were examined using immunohistochemical staining. We observed a positive correlation between the expression of Ring1 and Ki-67 and identified that high expression of Ring1 (A) correlated with high Ki-67 expression (B) in HCC specimens, whereas Ring1 and Ki-67 expression were barely identified in adjacent normal liver tissues (Fig. 2). The percentage of Ring1-positive tumor cells ranged from 2.32 to 98.21. The mean percentage of positive cells was 57.22. The percentage of Ki-67-positive tumor cells ranged from 3.11 to 98.42. The mean percentage of positive cells was 59.33. Moreover, there was a positive correlation between Ring1 and Ki-67 expression, with a correlation coefficient of 0.581 (P < 0.001, Fig. 3). These data implied that upregulated expression of Ring1 potentially contributed to HCC progression.
Ring1 expression in HCC and adjacent non-cancerous tissues by immunohistochemical staining paraffin-embedded HCC tissue sections were stained with antibodies against Ring1, Ki-67, and counterstained with hematoxylin. High Ring1 (a) and Ki-67 (b) expression was detected in HCC specimen (SP × 400). Meanwhile, low levels of Ring1 expression (c) were seen in the adjacent non-cancerous tissues, whereas low concentrations of Ki-67 (d) were observed in the same adjacent non-cancerous tissues (SP × 400). Details of the experiments are given in “Materials and methods”
Correlation between Ring1 expression and clinicopathological features in HCC
We further evaluated the association between Ring1 expression and the clinicopathological parameters of HCC patients. The patients’ clinicopathological data are summarized in Table 1. For statistical analysis of the expression of Ring1 and Ki-67, the HCC specimens were separated into two groups, high expressers and low expressers, according to the percentage of Ring1 and Ki-67-positive cells using the cutoff values of 57.22 and 59.33 % representing the mean value of Ring1 and Ki-67 expression, respectively. Ring1 expression was significantly correlated with histological grade (P = 0.011) and tumor size (P = 0.004). However, there was no significant correlation between Ring1 expression and the prognostic factors such as patients’ age, gender, serum AFP level, liver cirrhosis status, No. of tumor nodes, tumor metastasis status, and microvascular invasion. Therefore, we speculated that upregulation of Ring1 might specifically facilitate HCC growth.
The prognostic significance of Ring1 expression
Thereafter, we investigated whether Ring1 expression could predict the prognosis of HCC patients. The follow-up data of 98 patients were totally collected and subjected to survival analysis. As shown in Table 2, we evaluated the association between patients’ survival status and clinicopathological parameters. Through the analysis, we found that there was significant correlation between patients’ survival status and clinicopathological parameters such as Ring1 expression (P < 0.001) and Ki-67 expression (P < 0.001). So we further carried out Kaplan–Meier analysis to study the correlation between Ring1 and patients’ survival. In this way, it was found that patients with high Ring1 expression predicted significantly worsened overall survival (P < 0.001, Fig. 4). Multivariate analysis using the Cox proportional hazards model showed that the levels of Ring1 (P = 0.014) and Ki-67 (P = 0.008) were independent prognostic indicators of overall survival (Table 3).
Kaplan–Meier survival curves for low versus high Ring1 expression in 98 patients with HCC. Based on the median Ring1 percentages, patients were divided into two groups: high Ring1 expressers (57 %) and low Ring1 expressers (43 %). Low versus high Ring1 expression in 98 patients of HCC showed a highly significant separation (P < 0.001, log-rank test)
Ring1 was high expressed in proliferating HCC cell lines
Based on the above results, we considered that Ring1 might promote HCC development through modulating HCC growth. To verify this piece of hypothesis, we further analyzed the expression of Ring1 during cell cycle progression in HCC cells. Huh7 cells were arrested in G1 phase by serum deprivation for 72 h. After serum-refeeding, the cells were released from G1 phase and entered into S phase (Fig. 5a). Western blot result showed that the expression of Ring1 was increased as early as 4 h after serum-stimulation. Meanwhile, the expression of Cyclin D1 and PCNA was correlatively upregulated (Fig.5b, c). These results suggested Ring1 might work as a cell cycle promoter in regulating HCC cell proliferation.
Expression of Ring1 in proliferating HCC cells. Flow-cytometry quantitation of cell cycle progress in Huh7 cells (a). Cells were synchronized at G1 after serum starvation for 72 h, then progressed into cell cycle by adding medium containing 10 % FBS for the indicated times (R4 h, R8 h, R12 h, R24 h). Mean ± SD of 3 independent experiments. Huh7 cells were synchronized by serum starvation for 72 h and upon serum releasing, and cell lysates were prepared and analyzed by Western blot using antibodies directed against Ring1, PCNA, and cyclin D1 (b). GAPDH was used as a control for protein load and integrity. The bar chart below demonstrates the ratio of Ring1, cyclin D1, and PCNA protein to GAPDH by densitometry (c). S serum starvation, R serum release
Ring1 knockdown suppress cellular proliferation and inhibit cell cycle progress
To further determine the regulatory role of Ring1 on HCC growth, we used chemically synthesized siRNA to knock down endogenous Ring1 in Huh7 cells. The interference efficiencies of the Ring1-targeting siRNAs were assessed 48 h post-transfection using Western blot analysis. As predicted, Ring1 siRNA oligos, especially siRing1#2, markedly attenuated the cellular level of Ring1 in Huh7 cells, compared with cells transfected with control siRNA (Fig.6a). It was also found that, after siRing1#2 transfection, the levels of cyclin D1 and PCNA significantly declined in Huh7 cells (Fig. 6b, c). Moreover, we also showed that Ring1-depleted Huh7 cells exhibited significantly decreased proliferation, compared with negative control siRNA- or mock-transfected cells (Fig.6d) using CCK-8 assay. Similar results were obtained using colony formation assay (Fig.6e). To determine whether the cell cycle distribution was altered after depletion of Ring1 in Huh7 cells, flow cytometry was performed 48 h after transient transfection of the Ring1 siRNA#2. Cell cycle analysis showed a significantly decreased population in the S phase, whereas an increased population in the G1 phase after transfection of Ring1 siRNA#2 in Huh7 cells, when compared with negative control cells (Fig. 6f). These results indicated that Ring1 interference retarded cell cycle progression, leading to impaired proliferation of HCC cells.
Ring1 knockdown suppress cellular proliferation and inhibit cell cycle progress. Western blot analysis showed that siRNA treatment of Ring1 in Huh7 cells (a). The expression level of the cells transfected by siRNA#2 displayed a more significant decrease. After siRNA transfection, accompanied with the decrease of Ring1 expression in Huh7 cells, the level of cyclin D1 and PCNA diminished (b). The bar chart below demonstrates the ratio of Ring1, PCNA, and cyclin D1 protein to GAPDH by densitometry (c). The data are mean ± SEM. CCK-8 assay showed that Ring1 knockdown inhibited proliferation progress of Huh 7 cells (d). Cell Counting Kit-8 reagents were added to the medium and incubated for additional 2 h. Absorbance was measured at each indicated time (0, 1, 2, and 3 days). Each time point was derived from three independent experiments. Silencing endogenous Ring1 inhibits cell growth as determined by colony formation assay (e). Ring1 knockdown resulted in a decrease of S transition of Huh7 cells, whereas an increased population in the G1 phase (f). The data are showed as mean ± SD for three experiments
Discussion
Hepatocarcinogenesis is associated with complex molecular events, and the prognosis for HCC patients remains poor because of the limitation of conventional treatment strategies [21]. Hence, for clinicians and basic scientists, it is a big challenge to discover novel molecular markers correlated with the prognosis of HCC and develop more effective anti-neoplastic therapies, such as molecular-targeted drugs and cancer vaccines or antibodies [22, 23]. In the current study, we detected the expression of Ring1 protein in HCC tissue specimens and found differential expression of this protein in HCC and normal liver tissues. We also found that high expression of Ring1 protein was significantly associated with poor patient survival compared to that of the patients with low expression. We investigated Ring1 expression and its role in HCC, and significant correlations were observed between Ring1 expression and HCC specimen’s histological grade and tumor size. These findings validated that Ring1 was a novel PcG protein involved in HCC development.
Since their discoveries, the significance of polycomb-group proteins in cancer biology has attracted intensive research concern [24]. Mounting evidence indicated that PcG proteins, such as Bmi1, EZH2, and EED, played integral roles in the development of multiple cancer types through epigenetic modification of target genes [25]. Recent studies have demonstrated that PcG proteins function in multiprotein complexes. Two of the best characterized PcG complexes are PRC1 and PRC2 [26]. PRC1 is a very large complex of more than 10 subunits, including the oncoprotein BMI as well as the other PcG proteins, such as Ring1, HPC, HPH, and SCML [27]. PRC2 is a smaller complex containing the PcG proteins EZH2, EED, SUZ12, and the histone binding protein RbAp46. The PRC2 complex possesses histone methytransferase (HMT) activity, specific for lysine (K) residues K27 of histone H3, which maybe related with the development of carcinomas [28, 29]. Watanabe reported that deregulation of histone lysine methyltransferases contributed to oncogenic transformation in human bronchoepithelial cells [30]. Kondo Y revealed that alterations in histone modifications were related to gene silencing in hepatocellular carcinomas [31]. Recently, member of PRC2 complex, especially of EZH2, is often reported in various cancers [32]. However, in spite of the fact that PRC1 subunit Ring1 interacts with multiple polycomb-group proteins and displays tumorigenic activity, little has been done with regard to the role of Ring1 in HCC development. Our study conceivably demonstrated that Ring1 was an oncoprotein implicated in the development of HCC. In addition, upregulated expression of Ring1 contributed to aberrant growth of HCC. These data are consistent with several previous reports indicating that Bmi1 played a crucial role in HCC progression. Given the fact that PRC1 complex plays a facilitating role in transformation and tumorigenesis in multiple cancers, we speculated that increased expression of PRC1 subunits might promote hyperactivation of PRC1 complex, leading to consequent cell transformation and HCC development.
Recently, Ring1 expression had been reportedly implicated in the development of various cancer types, including prostate cancer, breast cancer non-small-cell lung cancer, and bladder cancer [33]. However, the molecular mechanism underlying the oncogenic property of Ring1 remains largely elusive. In this regard, early studies suggested that overexpression of Ring1 led to elevated expression of immediate early genes, including c-jun and c-fos, which is accompanied with cell transformation and in vivo tumorigenesis of NIH 3 T3 cells [9]. Recently, through gene-targeting approaches, it was found that Ring1 was essential for the maintenance of stem cell identity, and knockout of Ring1 significantly reduced the proliferation of mesenchymal stem cells [25, 34]. Our study suggested that Ring1 promoted the proliferation of HCC cells, whereas interference of Ring1 led to significant reduction of Cyclin D1 and PCNA expression, as well as proliferative impairment in HCC cells. The findings implicated that Ring1 may regulate the expression of multiple cell cycle regulators and resultantly play a key role in the regulation of cell proliferation.
In summary, we reported for the first time that high expression of PcG protein Ring1 correlates with unfavorable prognosis and shorter survival rate in HCC. Elevated Ring1 expression may be associated with aberrant proliferation of HCC cells. Ring1 can be used potentially as a therapeutic target, which would represent a major target point in the treatment of HCC.
References
Malek NP, Schmidt S, Huber P, Manns MP, Greten TF. The diagnosis and treatment of hepatocellular carcinoma. Dtsch Arztebl Int. 2014;111:101–6.
Tsochatzis EA, Meyer T, Burroughs AK. Hepatocellular carcinoma. N Engl J Med. 2012;366:92. author reply 92–93.
Yang JD, Roberts LR. Hepatocellular carcinoma: a global view. Nat Rev Gastroenterol Hepatol. 2010;7:448–58.
Hwang S, Ahn CS, Kim KH, Moon DB, Ha TY, Song GW. Survival rates among patients awaiting deceased donor liver transplants at a single high-volume Korean center. Transplant Proc. 2013;45:2995–6.
Hwang S, Moon DB, Ahn CS, Kim KH, Ha TY, Song GW, et al. Risk-based long-term screening for hepatocellular carcinoma recurrence after living donor liver transplantation. Transplant Proc. 2013;45:3076–84.
Niwa J, Ishigaki S, Doyu M, Suzuki T, Tanaka K, Sobue G. A novel centrosomal ring-finger protein, dorfin, mediates ubiquitin ligase activity. Biochem Biophys Res Commun. 2001;281:706–13.
Hinz S, Kempkensteffen C, Christoph F, Krause H, Schrader M, Schostak M, et al. Expression parameters of the polycomb group proteins bmi1, suz12, ring1 and cbx7 in urothelial carcinoma of the bladder and their prognostic relevance. Tumour Biol. 2008;29:323–9.
Cao R, Tsukada Y, Zhang Y. Role of bmi-1 and ring1a in h2a ubiquitylation and hox gene silencing. Mol Cell. 2005;20:845–54.
Satijn DP, Otte AP. Ring1 interacts with multiple polycomb-group proteins and displays tumorigenic activity. Mol Cell Biol. 1999;19:57–68.
Boukarabila H, Saurin AJ, Batsche E, Mossadegh N, van Lohuizen M, Otte AP, et al. The prc1 polycomb group complex interacts with plzf/rara to mediate leukemic transformation. Genes Dev. 2009;23:1195–206.
Kia SK, Gorski MM, Giannakopoulos S, Verrijzer CP. Swi/snf mediates polycomb eviction and epigenetic reprogramming of the ink4b-arf-ink4a locus. Mol Cell Biol. 2008;28:3457–64.
Gil J, Bernard D, Peters G. Role of polycomb group proteins in stem cell self-renewal and cancer. DNA Cell Biol. 2005;24:117–25.
Oyanagi H, Takenaka K, Ishikawa S, Kawano Y, Adachi Y, Ueda K, et al. Expression of lun gene that encodes a novel ring finger protein is correlated with development and progression of non-small cell lung cancer. Lung Cancer. 2004;46:21–8.
Senthi S, Lagerwaard FJ, Haasbeek CJ, Slotman BJ, Senan S. Patterns of disease recurrence after stereotactic ablative radiotherapy for early stage non-small-cell lung cancer: a retrospective analysis. Lancet Oncol. 2012;13:802–9.
Syrios J, Logothetis M, Grivas A, Lianos E, Athanasiou AE. Primary signet-ring adenocarcinoma of the urinary bladder with gastric metastasis or a second primary cancer? J BUON. 2012;17:396.
Lu C, Liu G, Cui X, Zhang J, Wei L, Wang Y, et al. Expression of sgta correlates with prognosis and tumor cell proliferation in human hepatocellular carcinoma. Pathol Oncol Res. 2014;20:51–60.
Ni S, Zhu J, Zhang J, Zhang S, Li M, Ni R, et al.: Expression and clinical role of nf45 as a novel cell cycle protein in esophageal squamous cell carcinoma (escc). Tumour biology : the journal of the International Society for Oncodevelopmental Biology and Medicine 2014
Wang Y, Fei M, Cheng C, Zhang D, Lu J, He S, et al. Jun activation domain-binding protein 1 negatively regulate p27 kip1 in non-Hodgkin’s lymphomas. Cancer Biol Ther. 2008;7:460–7.
Wang Y, Yang S, Ni Q, He S, Zhao Y, Yuan Q, et al. Overexpression of forkhead box j2 can decrease the migration of breast cancer cells. J Cell Biochem. 2012;113:2729–37.
Zhang Y, Peng C, Wu G, Wang Y, Liu R, Yang S, et al. Expression of nlk and its potential effect in ovarian cancer chemotherapy. Int J Gynecol Cancer. 2011;21:1380–7.
Buchenau P, Hodgson J, Strutt H, Arndt-Jovin DJ. The distribution of polycomb-group proteins during cell division and development in drosophila embryos: Impact on models for silencing. J Cell Biol. 1998;141:469–81.
Caldas C, Aparicio S. Cell memory and cancer--the story of the trithorax and polycomb group genes. Cancer Metastasis Rev. 1999;18:313–29.
Xing C, Zhou W, Ding S, Xie H, Zhang W, Yang Z, et al. Reversing effect of ring finger protein 43 inhibition on malignant phenotypes of human hepatocellular carcinoma. Mol Cancer Ther. 2013;12:94–103.
Chen Z, Li Z, He Z. Plasticity of male germline stem cells and their applications in reproductive and regenerative medicine. Asian Journal of Andrology 2014
Lapthanasupkul P, Feng J, Mantesso A, Takada-Horisawa Y, Vidal M, Koseki H, et al. Ring1a/b polycomb proteins regulate the mesenchymal stem cell niche in continuously growing incisors. Dev Biol. 2012;367:140–53.
Hallson G, Syrzycka M, Beck SA, Kennison JA, Dorsett D, Page SL, et al. The drosophila cohesin subunit rad21 is a trithorax group (trxg) protein. Proc Natl Acad Sci U S A. 2008;105:12405–10.
Schoeftner S, Sengupta AK, Kubicek S, Mechtler K, Spahn L, Koseki H, et al. Recruitment of prc1 function at the initiation of x inactivation independent of prc2 and silencing. EMBO J. 2006;25:3110–22.
Illingworth RS, Botting CH, Grimes GR, Bickmore WA, Eskeland R. Prc1 and prc2 are not required for targeting of h2a.Z to developmental genes in embryonic stem cells. PLoS One. 2012;7:e34848.
Tolhuis B, de Wit E, Muijrers I, Teunissen H, Talhout W, van Steensel B, et al. Genome-wide profiling of prc1 and prc2 polycomb chromatin binding in drosophila melanogaster. Nat Genet. 2006;38:694–9.
Sasaki A, Masuda Y, Iwai K, Ikeda K, Watanabe K. A ring finger protein praja1 regulates dlx5-dependent transcription through its ubiquitin ligase activity for the dlx/msx-interacting mage/necdin family protein, dlxin-1. J Biol Chem. 2002;277:22541–6.
Cahn YKA. A novel approach to the treatment of diabetic foot abscesses—a case series. J Wound Care. 2014;23(394):396–9.
Abd Al Kader L, Oka T, Takata K, Sun X, Sato H, Murakami I, et al. In aggressive variants of non-hodgkin lymphomas, ezh2 is strongly expressed and polycomb repressive complex prc1.4 dominates over prc1.2. Virchows Arch. 2013;463:697–711.
van Leenders GJ, Dukers D, Hessels D, van den Kieboom SW, Hulsbergen CA, Witjes JA, et al. Polycomb-group oncogenes ezh2, bmi1, and ring1 are overexpressed in prostate cancer with adverse pathologic and clinical features. Eur Urol. 2007;52:455–63.
Endoh M, Endo TA, Endoh T, Fujimura Y, Ohara O, Toyoda T, et al. Polycomb group proteins ring1a/b are functionally linked to the core transcriptional regulatory circuitry to maintain es cell identity. Development. 2008;135:1513–24.
Conflicts of interest
None
Author information
Authors and Affiliations
Corresponding author
Additional information
Yicheng Xiong and Baoying Hu contributed equally to this work.
Rights and permissions
About this article
Cite this article
Xiong, Y., Hu, B., Wei, L. et al. Upregulated expression of polycomb protein Ring1 contributes to poor prognosis and accelerated proliferation in human hepatocellular carcinoma. Tumor Biol. 36, 9579–9588 (2015). https://doi.org/10.1007/s13277-015-3721-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13277-015-3721-7