Abstract
Aberrant expression of miR-10b has been described in many cancers but remains unexplored in nasopharyngeal carcinoma (NPC). Therefore, we aimed to study the miR-10b expression level in 43 NPC biopsies collected from Tunisian patients and three NPC xenografts. Then, we investigated the correlation between miR-10b expression and its upstream regulators LMP1/Twist1 as well as its adjacent gene HoxD4. We showed that miR-10b was significantly up-regulated in NPC biopsies compared to non-tumor nasopharyngeal tissues (fold change 153; p = 0.004) and associated with advanced clinical stage and young age at diagnosis (p = 0.005 and p = 0.011, respectively). In addition, over-expression of miR-10b was positively associated with the transcription factor Twist1 as well as the EBV oncoprotein LMP1 (fold change 6.32; p = 0.014, fold change 6.58; p = 0.01 respectively). Furthermore, higher level of miR-10b was observed in tumors with simultaneous expression of LMP1 and Twist1, compared to those expressing only Twist1 (fold change 2.49; p = 0.033). Meanwhile, the analysis of the link between miR-10b and its neighbor gene HoxD4 did not show any significant correlation (Fisher test p = 0.205; Mann–Whitney test p = 0.676). This study reports the first evidence of miR-10b over-expression in NPC patients. Furthermore, our findings can support hsa-miR-10b gene regulation through LMP1/Twist1 in NPC malignancy.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Nasopharyngeal carcinoma (NPC) is a malignant tumor that occurs in epithelial cells lining the nasopharynx. In most regions, the age-standardized incidence of NPC is less than 1/100,000 individuals. However, higher rates are observed in the Cantonese population of southern China, and intermediate rates are observed in North Africa [1]. One intriguing characteristic of the North African NPCs is their bimodal age distribution, with a major peak of incidence occurring around 50 and a minor peak between 15 and 25 years old [2]. As a distinct type of head and neck cancers, non-keratinizing NPC is closely associated with Epstein–Barr Virus (EBV) [3, 4]. It is well established that EBV infection contributes to tumorigenesis of NPC, mainly through the expression of the oncoprotein latent membrane protein 1 (LMP1). LMP1 acts as a constitutive active receptor, mimicking the “receptor of tumor necrosis factor (TNF)” super-family to stimulate multiple signalling pathways, including NF-κB, JAK/STAT, p38/MAPK, PI3K/Akt, and ERK-MAPK/JNK [5, 6]. Therefore, LMP1 is able to achieve pleiotropic effects on cell migration, proliferation, apoptosis, and stemness [7, 8].
MicroRNAs (miRNAs) belong to a class of short, highly conserved non-coding RNAs that are known to regulate gene expression at the post-transcriptional level through imperfect base pairing with the 3′ untranslated regions of target mRNAs [9]. Growing evidence shows that miRNAs exhibit a variety of crucial regulatory functions related to cell growth, development, and differentiation [10, 11]. Therefore, alteration of miRNA expression profile could contribute to the pathogenesis of a wide range of human diseases including neoplasia. miRNAs can act as oncogenes by repressing the expression of target tumor suppressor genes or as tumor suppressors by repressing the expression of target oncogenes. Recently, large-scale miRNA screening has been performed for several human cancers, including NPC [12–14]. Recently, it has been reported that LMP1 could modulate miRNA expression pattern, such as the down-regulation of miR-203 and miR-29b and the up-regulation of miR-155, miR-21, and miR-10b [15–19].
The miR-10b, defined as an oncogenic miRNA, has been reported to promote metastatic progression through targeting the transcription factors HoxD10 in breast cancer and KLF4 in esophageal cancer [20, 21]. Similar findings on the migration and invasion processes were also shown for NPC cell lines [19, 22]. In addition, the implication of miR-10b in tumor cell proliferation was also concluded through the repression of BCL2L11, CDKN1A, and NF1 in glioma cells and PTEN in thyroid cancer cells [23–25]. To explore hsa-miR-10b gene regulation, two upstream regions have been studied. In fact, Ma et al. have found that the transcriptional factor Twist1 regulates miR-10b expression through binding to E-box sequence located at the putative promoter region (−460 to −111 bp) of hsa-miR-10b [20]. Moreover, retinoic acid responsive element (RARE) boxes located at −1.5 kb upstream of hsa-miR-10b were involved in the transcriptional regulation of miR-10b as well as its 5′-adjacent gene, HoxD4 [26]. In NPC, the role of LMP1 in the up-regulation of miR-10b has been demonstrated through Twist1 in C666.1 cell line [19].
The present work aimed to study the expression level of miR-10b in NPC primary tumors and then to examine the statistical correlations between miR-10b and its upstream regulators LMP1, Twist1, and its 5′-adjacent gene HoxD4.
Materials and methods
Patients and tissue samples
Primary NPC biopsies were collected with informed consent from 43 patients, prior to any treatment, at the University Hospital of Sfax (Tunisia). Seven cases were of the juvenile form (patients aged less than 30 years). The clinical stage of the disease was determined according to the tumor, node, and metastasis (TNM) classification of the American Joint Committee on Cancer/International Union Against cancer (AJCC/UICC, 1997) [27, 28]. The histological type of NPC was determined on tissue sections according to the World Health Organization (WHO, 2005) [29]. In addition, six biopsy samples totally devoid of malignant cells from nasopharyngeal tissues were used as controls. Three NPC xenografts, C15, C17, and X666, derived from a North African primary NPC, from a cutaneous metastasis of a European NPC, and from a Chinese primary NPC, respectively, were used as references (kindly provided by Professor P. Busson, Villejuif, France).
All tissue samples were immediately frozen in liquid nitrogen and subsequently stored at −80 °C. Total RNA was isolated from the various tissues by TRIzol reagent (Invitrogen) following the manufacturer’s instructions and then conserved at −80 °C until use. RNA concentration and purity were measured using the NanoDrop ND-1000 UV/Vis spectrophotometer.
Stem-loop RT-qPCR
miR-10b was reverse-transcribed using a stem-looped primer (5′-CTCAACTGGTGTCGT GGAGTCGGCAATTCAGTTGAGCACAAATT-3′) which binds to six nucleotides at the 3′ side of the mature miR-10b molecule. The small nuclear RNA (snRNA) U6 was also reverse-transcribed using U6-R specific primer (5′-CGCTTCACGAATTTGCGTGTCAT-3′) and considered as an internal control. Reverse transcription (RT) reaction was performed on 50 ng of total RNA in a final volume of 10 μl containing 0.1 pmol of specific primer, 1× reaction buffer, 10 nmol of dNTP, and 100 U of M-MLV reverse transcriptase (Invitrogen). The reaction was incubated for 30 min at 16 °C followed by 50 min at 37 °C and finally stopped by incubation for 15 min at 70 °C. miR-10b level was examined by quantitative PCR (qPCR) using U6 as an endogenous control. The qPCR reaction mix includes 1 μl of RT product as template, 4 pmol of each primer, and 10 μl of 2× Evagreen super-mix (Biorad). The amplification conditions performed in Biorad-CFX96 are as follows: 95 °C for 30 s, then 45 cycles of 94 °C 10 s and 61 °C 45 s. The specificity of the PCR products was confirmed by melting curve analysis from 65 to 95 °C, with a heating rate of 0.5 °C/s. In addition, the PCR amplification efficiency of each primer pair was calculated according to the standard curve method using the equation E % = (10−1/slope − 1) × 100 by serial dilutions. Standard curves from the target miR-10b and the internal control snRNA U6 were generated using the Ct value versus log input of cDNA.
RT-PCR
Conventional RT-PCR was performed for Twist1, LMP1, HoxD4, and the GAPDH as endogenous control. RT reaction was performed on 1 μg of total RNA in a final volume of 20 μl containing 1× reaction buffer (Invitrogen), 0.1 pmol of random hexamer primers, 10 nmol of each dNTP, and 100 U of M-MLV reverse transcriptase (Invitrogen). The reactions were incubated at 37 °C for 50 min and then at 70 °C for 10 min. Following RT, 1 μl of cDNA was used as template for each PCR reaction in a final volume of 25 μl containing 1× PCR buffer, 2 mM MgCl2, 5 pmol of each appropriate primer, 200 μM of each dNTP, and 1 U of Taq DNA polymerase (Fermentas). The primer sequences, product sizes, and annealing temperatures are shown in Table 1.
Statistical analysis
All statistical analyses were performed using GraphPad Prism software 6.0. Gene expression ΔCt values of miRNA from each sample were calculated by normalizing according to internal control U6 expression, and relative quantification values were plotted. The differences between tumor and normal tissues were analyzed by Mann–Whitney test. The analysis of miR-10b levels according to clinical parameters or expression status of LMP1, Twist1, and HoxD4 was also performed using Mann–Whitney test. miR-10b expression level differences in groups expressing or not Twist1 and/or LMP1 were determined using the ANOVA test. Fisher test was applied to determine statistical correlation between miR-10b high or low level and HoxD4 expression status. The analysis of miR-10b correlation with patients’ survival was determined using Kaplan–Meier test. Statistical significance was set up to p ≤ 0.05 in each test.
Results
Expression of miR-10b in primary NPC tissues and xenografts
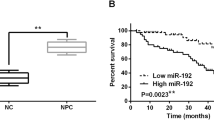
Before the analysis of miR-10b expression in NPC tissues and non-tumor nasopharyngeal tissues, the PCR amplification efficiencies of miR-10b and the internal control snRNA U6 were measured according to the standard curve method. As shown in supplementary Fig. S1, the slope values of resulting plots are approximately equal, thus implying similar amplification efficiency for miR-10b and the internal control snRNA U6 (101 and 103 %, respectively). Based on these data, the analyses of the relative fold changes in miR10b expression were then performed using the 2−∆∆Ct method, as described by Livak et al. [30]. As shown in Fig. 1a, the miR-10b level was significantly higher in primary NPC tumors compared to control tissues. The difference ranged from 0.028 to 2352 with a median of 153 fold (p = 0.004). In addition, C15 and C17 NPC xenografts show high miR-10b expression compared to non-tumor tissues (fold change = 15.67 and 21.85, respectively), contrarily to X666 (fold change = 0.028) (Fig. 1b). The relationship between the expression levels of miR-10b and the clinicopathological features of NPC patients is shown in Table 2. There were no statistically significant differences between miR-10b levels and lymph node involvement (N2–N3) and distant metastasis evolution at primary diagnosis. However, miR-10b was significantly more abundant in cases with an advanced T stage (T3–T4) than those with T1–T2 (fold change = 3.94; p = 0.038). Regarding to TNM, high expression of miR-10b was associated with advanced TNM (III and IV) (fold change = 6.77; p = 0.005). In addition, miR-10b level was correlated with patient age with a higher expression in the juvenile form (fold change = 5.35; p = 0.011). We note also that miR-10b expression was higher in tumors with undifferentiated type, although the difference did not reach statistical significance (fold change = 3.09; p = 0.082). To evaluate the association between miR-10b expression and patient’s outcome, Kaplan–Meier curves were plotted. As shown in Fig. 2, when we stratified our patients according to age of diagnosis, we showed that patients over 30 years old with high miR-10b expression had a significantly shorter overall survival compared to those with low miR-10b expression (p = 0.048). This correlation was not observed when all NPC patients were considered (both age groups) (p = 0.88).
a The relative levels of miR-10b (2−∆∆Ct) in each tumor and non-tumor samples normalized with the median of non-tumor samples. miR-10b is statistically over-expressed in tumor samples with a fold change = 153 (p = 0.004). b miR-10b relative expression in C15, C17, and X666 NPC xenografts normalized with the median of non-tumor samples. Data are presented in a logarithmic scale to better visualize over- and under-expression. The p-value was calculated using the non-parametric Mann–Whitney U test
Correlation of miR-10b level and Twist1 and LMP1 mRNA
The positive expression of Twist1 and LMP1 was, respectively, detected in 23 and 24 cases out of the 43 primary NPC biopsies as well as in C15 and C17 NPC xenografts. In the non-tumor samples, LMP1 expression was negative in all six cases, while Twist1 was detected in only two cases (Fig. 3). Using Mann–Whitney test, we show that miR-10b over-expression was significantly correlated with Twist1 and LMP1 positive cases (fold change = 6.32; p = 0.014 and 6.58; p = 0.010, respectively). According to the functional link reported in NPC cell lines showing that LMP1 induces the over-expression of Twist1 which in turn activates the transcription of miR-10b, we subdivided our cases in three groups. The first group (namely, group 1) encloses the 27 cases exhibiting a negative expression of Twist1 independently of the expression profile of LMP1. The second group (namely, group 2) includes the 11 cases showing a positive expression of Twist1 and a negative expression of LMP1. Finally, the third group (namely, group 3) concerns the 14 cases showing simultaneous expression of Twist1 and LMP1. As shown in Fig. 4, miR-10b expression was significantly different according to the expression pattern of LMP1 and Twist1 (p = 0.033). Indeed the higher level of miR-10b was found in cases with simultaneous expression of LMP1 and Twist1 (group 3), compared to those with only Twist1 positive expression (group 2; fold change 2.49) and to those with Twist1 negative expression (group 1; fold change 8.81).
Overall results of miR-10b levels according to the expression pattern of LMP1 and Twist1 genes. Group 1 encloses the 27 cases exhibiting a negative expression of Twist1 independently of the expression profile of LMP1. Group 2 includes the 11 cases showing a positive expression of Twist1 and a negative expression of LMP1. Group 3 concerns the 14 cases showing simultaneous expression of Twist1 and LMP1
Correlation between miR-10b and HoxD4 expression
To assess whether hsa-miR-10b gene regulation is associated with the adjacent gene HoxD4, we examined the corresponding mRNA expression in tumor and non-tumor tissues. HoxD4 expression was detected in the three NPC xenografts as well as in ten out of 43 primary NPC biopsies. In addition, HoxD4 expression was detected in three out of the six non-tumor nasopharyngeal tissues. No significant correlation between miR-10b and HoxD4 expression was found (fold change 1.04; p = 0.676). To apply Fisher test, all cases were classified into high and low miR-10b levels based on the mean fold relative level of miR-10b (=201.03). In fact, a high level of miR-10b was found in only four out of the 16 HoxD4 positive cases (three NPC xenografts, ten NPC biopsies, and three non-tumor tissues), showing no significant correlation between these two genes (p = 0.205).
Discussion
Our data showed significant over-expression of miR-10b in primary NPC biopsies compared to non-tumor tissues (fold change 153; p = 0.004), which is in line with previous studies on patients with esophageal cancer [21], breast cancer [31], and gliomas [32]. The down-regulation of miR-10b was reported by Gee et al., using microarray method, in patients with head and neck cancer excluding NPC (fold change = 0.1) [33]. Besides the technique used for miR-10b quantification, our finding on NPC patients is in line with the general assumption which classifies this malignancy as a distinct entity from the other head and neck cancers, that being related to latent EBV infection.
The functional investigation of miR-10b in NPC cells has been firstly reported by Li et al., showing in C666.1 that the up-regulation of miR-10b increases the mobility and promotes the migration and invasiveness in vitro as well as the death of tumor-bearing nude mice in vivo [19]. Later, Sun et al. reported the same in vitro observation on the CN-2Z cell line [22]. Based on these data, we expected to find that the high levels of miR-10b would be associated to lymph node and/or extra-nodal metastases. However, our findings show no statistical correlation with these clinical parameters (p = 0.682 and p = 0.499, respectively). Meanwhile, miR-10b over-expression was correlated with the advanced T3–T4 stage (fold change 3.94; p = 0.038), suggesting that the miR-10b over-expression could be involved in cell proliferation rather than in migration and invasion processes of NPC tumors. Based on the miROB online database (http://mirob.interactome.ru/microRNA_databases), miR-10b appears to target different genes and thus controls various cellular pathways (supplementary Fig. S2). Among them, we note several key cell cycle inhibitors and pro-apoptotic genes such as BCL2L11, TFAP2C, CDKN1A, CDKN2A, TP53, CYLD, and FOXO3. The validation of these miR-10b targets in glioblastoma cells [24, 34] could support our above suggestion that remains to be verified by further functional studies. According to the patients’ age, our study shows that the miR-10b expression levels tend to increase in the juvenile form of North African NPCs. We and others have previously shown that this NPC form has specific clinical and biological characteristics compared to adult form. In the juvenile form, immunohistochemistry assay has shown that the two key proteins controlling the apoptotic survival balance p53 and Bcl2 are less abundantly expressed, whereas the C-kit receptor and the EBV oncoprotein LMP1 are more frequently detected [35, 36]. There are also reports showing that IgA anti-EA and anti-VCA antibodies are less abundant in juvenile form [37, 38]. Regarding the clinical features, young patients have a high rate of lymph node metastasis and are subjected to earlier recurrences; however, their outcome is significantly better than older patients [39, 40]. In our cohort, the seven younger patients were classified as TNM III and IV and had prolonged overall survival rate reaching 124 months, with no cases of death caused by NPC. In addition, our findings on survival parameters show that the high level of miR-10b is significantly related to a patient’s shortened survival, especially in adult NPC patients. Our data showed that young NPC patients had a high amount of miR-10b and a prolonged overall survival rate, which support the previous data suggesting a distinct oncogenic pathway of these two age groups. Similar to our results on the NPC adult form, the prognosis value of miR-10b has been also reported in other cancer types such as hepatocellular, gastric, and colorectal cancers [41–43].
In the second part of this study, we focused on the correlation between miR-10b level and its upstream regulators LMP1 and Twist1. In fact, Ma et al. have defined Twist1 as the transcription factor of miR-10b which binds to E-box sequence in its putative promoter region (−460, −111 bp) [20]. In addition, Horikawa et al. have shown, in NPC cells, that the viral oncoprotein LMP1 induces the over-expression of Twist1 via the NF-κB pathway [44]. More recently, in C666.1 NPC cell line, the functional link between these three molecular actors has been confirmed by Li et al., concluding that LMP1 induces Twist1, which in turn up-regulates miR-10b [19]. Our data showed that the over-expression of miR-10b correlates with the expression of Twist1 (fold change 6.32; p = 0.014). In addition, the higher level of miR-10b was observed in samples expressing simultaneously LMP1 and Twist1 compared to those expressing only Twist1. This association suggests that LMP1 improves the over-expression of miR-10b, which is in agreement with the previous findings on NPC cell line [19].
The hsa-miRNA-10b gene is embedded in HOXD cluster on chromosome 2q31, at 1 kb upstream of HOXD4 gene [20]. Since the co-localization was conserved during evolution, Foley et al. have proposed that these two genes would share a common regulation region located at 1.5 kb upstream of hsa-miR-10b, which encompasses the RARE elements. In fact, the authors have demonstrated the effect of a trans-retinoic acid treatment on improving the simultaneous expression of miR-10b and HoxD4 in neuroblastoma cells [26]. Furthermore, Kim et al. have concluded that the hypermethylation of CpG islands, located at the previous DNA control region, would exhibit a transcriptional repression of these two neighbor genes in gastric cancer cells [45]. These data led us to examine whether miR-10b and HoxD4 expressions are correlated in NPC biopsies, suggesting another regulation pathway of miR-10b expression. Our results showed that the expression of these two genes was not correlated, suggesting that probably both genes are subjected to distinct control pathways. However, further studies are warranted on the transcriptional regulation of hsa-miR-10b in NPC cell lines by analyzing, for example, the effect of DNA demethylating agent or trans-retinoic acids on miR-10b expression.
In conclusion, we present here the first evidence of miR-10b over-expression in NPC patients which correlates with the advanced clinical stages and the juvenile form of North African NPC. In addition, our study shows that the elevated miR-10b level was associated with the simultaneous expression of LMP1 and Twist1, supporting the previous in vitro data showing the impact of LMP1 on miR-10b up-regulation through the transcription factor Twist1.
References
Yu MC, Yuan JM. Epidemiology of nasopharyngeal carcinoma. Semin Cancer Biol. 2002;12(6):421–9.
Boussen H, Bouaouina N, Mokni-Baizig N, Gamoudi A, Chouchane L, Benna F, et al. Nasopharyngeal carcinoma. Recent data. Pathol Biol. 2005;53(1):45–51.
Wang HY, Sun BY, Zhu ZH, Chang ET, To KF, Hwang JS, et al. Eight-signature classifier for prediction of nasopharyngeal carcinoma survival. J Clin Oncol. 2011;29:4516–25.
Chen J, Hu CF, Hou JH, Shao Q, Yan LX, Zhu XF, et al. Epstein–Barr virus encoded latent membrane protein 1 regulates mTOR signaling pathway genes which predict poor prognosis of nasopharyngeal carcinoma. J Transl Med. 2010;8:30.
Dawson CW, Port RJ, Young LS. The role of the EBV-encoded latent membrane proteins LMP1 and LMP2 in the pathogenesis of nasopharyngeal carcinoma (NPC). Semin Cancer Biol. 2012;22:144–53.
Mosialos G, Birkenbach M, Yalamanchili R, VanArsdale T, Ware C, Kieff E. The Epstein–Barr virus transforming protein LMP1 engages signaling proteins for the tumor necrosis factor receptor family. Cell. 1995;80:389–99.
Zhao Y, Wang Y, Zeng S, Hu X. LMP1 expression is positively associated with metastasis of nasopharyngeal carcinoma: evidence from a meta-analysis. J Clin Pathol. 2012;65:41–5.
Kondo S, Wakisaka N, Muramatsu M, Zen Y, Endo K, Murono S, et al. Epstein–Barr virus latent membrane protein 1 induces cancer stem/progenitor-like cells in nasopharyngeal epithelial cell lines. J Virol. 2011;85:11255–64.
Bartel DP, Chen CZ. Micromanagers of gene expression: the potentially widespread influence of metazoan microRNAs. Nat Rev Genet. 2004;5(5):396–400.
Brennecke J, Hipfner DR, Stark A, Russell RB, Cohen SM. Bantam encodes a developmentally regulated microRNA that controls cell proliferation and regulates the proapoptotic gene hid in Drosophila. Cell. 2003;113:25–36.
Chen CZ, Li L, Lodish HF, Bartel DP. MicroRNAs modulate hematopoietic lineage differentiation. Science. 2004;303:83–6.
Galasso M, Sandhu SK, Volinia S. MicroRNA expression signatures in solid malignancies. Cancer J. 2012;18(3):238–43.
Tran N, McLean T, Zhang X, Zhoao CJ, Thomson JM, O'Brien C, et al. MicroRNA expression profiles in head and neck cancer cell lines. Biochem Biophys Res Commun. 2007;358:12–7.
Chen HC, Chen GH, Chen YH, Liao WL, Liu CY, Chang KP, et al. MicroRNA deregulation and pathway alterations in nasopharyngeal carcinoma. Br J Cancer. 2009;24(6):1002–11.
Yu H, Lu J, Zuo L, Yan Q, Yu Z, Li X, et al. Epstein–Barr virus downregulates microRNA 203 through the oncoprotein latent membrane protein 1: a contribution to increased tumor incidence in epithelial cells. J Virol. 2012;86:3088–99.
Anastasiadou E, Boccellato F, Vincenti S, Rosato P, Bozzoni I, Frati L, et al. Epstein–Barr virus encoded LMP1 downregulates TCL1 oncogene through miR-29b. Oncogene. 2010;29:1316–28.
Du ZM, Hu LF, Wang HY, Yan LX, Zeng YX, Shao JY, et al. Upregulation of miR-155 in nasopharyngeal carcinoma is partly driven by LMP1 and LMP2A and downregulates a negative prognostic marker JMJD1A. PLoS One. 2011;6(4):e19137.
Yang GD, Huang TJ, Peng LX, Yang CF, Liu RY, Huang HB, et al. Epstein–Barr Virus_Encoded LMP1 upregulates microRNA-21 to promote the resistance of nasopharyngeal carcinoma cells to cisplatin-induced apoptosis by suppressing PDCD4 and Fas-L. Plos One. 2013;8(10):e78355.
Li G, Wu Z, Peng Y, Liu X, Lu J, Wang L, et al. MicroRNA-10b induced by Epstein–Barr virus-encoded latent membrane protein-1 promotes the metastasis of human nasopharyngeal carcinoma cells. Cancer Lett. 2010;299(1):29–36.
Ma L, Teruya-Feldstein J, Weinberg RA. Tumour invasion and metastasis initiated by microRNA-10b in breast cancer. Nature. 2007;449(7163):682–8.
Tian Y, Luo A, Cai Y, Su Q, Ding F, Chen H, et al. MicroRNA-10b promotes migration and invasion through KLF4 in human esophageal cancer cell lines. J Biol Chem. 2010;285(11):7986–94.
Sun XJ, Liu H, Zhang P, Zhang XD, Jiang ZW, Jiang CC. miR-10b promotes migration and invasion in nasopharyngeal carcinoma cells. Asian Pac J Cancer Prev. 2013;14(9):5533–7.
Chai G, Liu N, Ma J, Li H, Oblinger JL, Prahalad AK, et al. MicroRNA-10b regulates tumorigenesis in neurofibromatosis type 1. Cancer Sci. 2010;101:1997–2004.
Gabriely G, Yi M, Narayan RS, Niers JM, Wurdinger T, Imitola J, et al. Human gliomas growth is controlled by microRNA-10b. Cancer Res. 2011;71(10):3563–72.
Mussnich P, D’Angelo D, Leone V, Croce CM, Fusco A. The high mobility group A proteins contribute to thyroid cell transformation by regulating miR-603 and miR-10b expression. Mol Oncol. 2013;7(3):531–42.
Foley NH, Bray I, Watters KM, Das S, Bryan K, Bernas T, et al. microRNAs 10a and 10b are potent inducers of neuroblastoma cell differentiation through targeting of nuclear receptor corepressor 2. Cell Death Differ. 2011;18(7):1089–98.
Fleming ID, Cooper JS, Henson DE, Hutter RV, Kennedy BJ, et al. American Joint Committee on Cancer “cancer staging manual”. 5th ed. Philadelphia: Lippincott-Raven; 1997. p. pp. 31–9.
Sobin LH and Wittekind CH, Eds. International Union Against Cancer. TNM classification of malignant tumours. Wiley-Liss, New York, 5th edition, 1997. pp. 25–32.
Chan JKC, Pilch BZ, Kuo TT, Wenig BM, Lee AWM. Tumors of the nasopharynx: introduction. In: Barnes L, Eveson JW, Reichart P, Sidransky D, editors. Pathology and genetics of head and neck tumours (World Health Organization classification of tumours). Lyon: IARC; 2005. p. 82–4.
Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔ C T method. Methods. 2001;25(4):402–8.
Hafez MM, Hassan ZK, Zekri AR, Gaber AA, Al Rejaie SS, Sayed-Ahmed MM, et al. MicroRNAs and metastasis-related gene expression in Egyptian breast cancer patients. Asian Pac J Cancer Prev. 2012;13(2):591–8.
Sasayama T, Nishihara M, Kondoh T, Hosoda K, Kohmura E. MicroRNA-10b is overexpressed in malignant gliomas and associated with tumor invasive factors, uPAR and RhoC. Int J Cancer. 2009;125(6):1407–13.
Gee HE, Camps C, Buffa FM, Patiar S, Winter SC, Betts G, et al. hsa-mir-210 is a marker of tumor hypoxia and a prognostic factor in head and neck cancer. Cancer. 2010;116(9):2148–58.
Lin J, Teo S, Lam DH, Jeyaseelan K, Wang S. MicroRNA-10b pleiotropically regulates invasion, angiogenecity and apoptosis of tumor cells resembling mesenchymal subtype of glioblastoma multiforme. Cell Death Dis. 2012;3:e398.
Khabir A, Sellami A, Sakka M, Ghorbel AM, Daoud J, Frikha M, et al. Contrasted frequencies of p53 accumulation in the two age groups of North African nasopharyngeal carcinoma. Clin Cancer Res. 2000;6(10):3932–6.
Khabir A, Ghorbel A, Daoud J, Frikha M, Drira MM, Laplanche A, et al. Similar BCL-X but different BCL-2 levels in the two age groups of North African nasopharyngeal carcinomas. Cancer Detect Prev. 2003;27(4):250–5.
Karray H, Ayadi W, Fki L, Hammami A, Daoud J, Drira MM, et al. Comparison of three different serological techniques for primary diagnosis and monitoring of nasopharyngeal carcinoma in two age groups from Tunisia. J Med Virol. 2005;75(4):593–602.
Ayadi W, Karray-Hakim H, Feki L, Khabir A, Boudawara T, Ghorbel A, et al. IgA antibodies against the Epstein–Barr nuclear antigen1 as a valuable biomarker for the diagnosis of nasopharyngeal carcinoma in Tunisian patients. J Med Virol. 2009;81(8):1412–21.
Downing NL, Wolden S, Wong P, Petrik DW, Hara W, Le QT. Comparison of treatment results between adult and juvenile nasopharyngeal carcinoma. Int J Radiat Oncol, Biol, Phys. 2009;75(4):1064–70.
Sultan I, Casanova M, Ferrari A, Rihani R, Rodriguez-Galindo C. Differential features of nasopharyngeal carcinoma in children and adults: a SEER study. Pediatr Blood Cancer. 2010;55(2):279–84.
Nishida N, Yamashita S, Mimori K, Sudo T, Tanaka F, Shibata K, et al. miR-10b is a prognostic indicator in colorectal cancer and confers resistance to the chemotherapeutic agent 5-fluorouracil in colorectal cancer cells. Ann Surg Oncol. 2012;19(9):3065–71.
Li QJ, Zhou L, Yang F, Wang GX, Zheng H, Wang DS, et al. microRNA-10b promotes migration and invasion through CADM1 in human hepatocellular carcinoma cells. Tumor boil. 2012;33(5):1455–65.
Wang YY, Ye ZY, Zhao ZS, Li L, Wang YX, Tao HQ, et al. Clinicopathologic significance of miR-10b expression in gastric carcinoma. Hum Pathol. 2013;44(7):1278–85.
Horikawa T, Yang J, Kondo S, Yoshizaki T, Joab I, Furukawa M, et al. Twist and epithelial–mesenchymal transition are induced by the EBV oncoprotein latent membrane protein 1 and are associated with metastatic nasopharyngeal carcinoma. Cancer Res. 2007;67(5):1970–8.
Kim K, Lee HC, Park JL, Kim M, Kim SY, Noh SM, et al. Epigenetic regulation of microRNA-10b and targeting of oncogenic MAPRE1 in gastric cancer. Epigenetics. 2011;6(6):740–51.
Acknowledgments
This work was supported by a grant of the Tunisian Ministry of High Education and Scientific Research. We wish to thank Pr Adnane Hammami for allowing us to perform the RT-q-PCR experiments in the microbiology laboratory at the School of Medicine of University of Sfax.
Conflicts of interest
None.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary Fig. S1
Generating a standard curve to assess reaction efficiency. A Amplification curves and their corresponding melt curves of qPCR reactions using a series of tenfold dilutions of specific cDNA (1, 1/10, 1/100, 1/1000, and 1/10000) of miR-10b (A1) and U6 (A2). B Standard curve with the Ct plotted against the log of the starting quantity of template for each dilution. The equation of the generated curve is shown above the graph. The calculated efficiency was 103 and 101 % for miR10b and U6, respectively (TIFF 2886 kb)
Supplementary Fig. S2
Regulatory network of miR-10b. This schematic representation is created within the open informatic’s site MirOB (http://mirob.interactome.ru/microRNA_databases) (TIFF 4773 kb)
Rights and permissions
About this article
Cite this article
Allaya, N., Khabir, A., Sallemi-Boudawara, T. et al. Over-expression of miR-10b in NPC patients: correlation with LMP1 and Twist1. Tumor Biol. 36, 3807–3814 (2015). https://doi.org/10.1007/s13277-014-3022-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13277-014-3022-6