Abstract
Mouse mammary tumor virus (MMTV) is a well-known cause of mammary tumors in mice transmitted as endogenous proviruses or exogenously as infectious virions. The hypothesis that a retrovirus homologous to MMTV is involved in human breast cancers has resulted in renewed interest in the etiology of human breast cancer. Therefore, the detection of MMTV-like exogenous sequences in 30–40 % of invasive breast cancer has increased attention towards this hypothesis. To detect the prevalence of MMTV in Pakistani population, 666-bp-long MMTV envelop and 630-bp LTR sequences were amplified from breast cancer patient samples (tissue biopsies and peripheral blood) using mouse with mammary tumor as control. MMTV-like virus env and LTR DNA sequences were detected in 20 and 26 % of breast tumor samples, respectively, from the total of 80 breast cancer patients’ blood and tissue samples. No significant association was observed between age, grade of disease, and lymph node involvement with the prevalence of MMTV-like sequences. Our data add to the growing number of studies implicating MMTV-like virus in human breast cancer, but still clear causal association of MMTV to breast cancer remains to be reputable.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Mouse mammary tumor virus (MMTV) is a beta retrovirus first described by Bittner et al. in 1936 as an extra-chromosomal agent secreted in milk and has since been a known cause of mammary tumors in certain strains of mice [2]. MMTV-like virus has been a major suspect as a cause of some human breast cancers for over 50 years. Despite the substantial evidence that MMTV-like virus may have a role in human breast cancer, the development of conclusive evidence has been elusive [15]. MMTV itself does not have the oncogenic potential; it rather acts as an insertional mutagen. Insertion of a retroviral genome in the vicinity of a proto-oncogene has the potential to lead to an inappropriate transcriptional activation of nearby genes, causing oncogenic transformation of the infected cells [4]. Insertion of the MMTV genome within several loci designated as mt, wnt, and int has been observed, most of which encode for growth factors or other related proteins. Normally, wnt, int, and mt genes are not expressed in mammary tissue but get activated after integration of MMTV provirus into the adjacent chromosomal DNA, leading to mammary carcinogenesis [32].
MMTV-like virus has been under speculation as a possible cause of human breast cancer [26]. The hypothesis of MMTV involvement in human breast cancer was proposed based on the presence of MMTV-like retroviral particles in breast cancer biopsies and milk, supported by the detection of MMTV proteins in breast tumors by use of anti-MMTV antisera and by the presence of MMTV reactive antibodies [6, 9, 13, 20, 21, 23, 27, 33]. Molecular hybridization studies identified significant homology at the nucleic acid level between MMTV and RNA isolated from human breast tumors [27]. MMTV envelope nucleotide sequences (env) (95–99 % homologous to MMTV) were detected in 38.5 % of unselected human breast cancer samples but not in normal breast or other tissues [32]. Moreover, two complete proviral sequences were isolated from two human breast carcinomas and found to have 95 % nucleotide homology with MMTV but only 57 % homology to HERV-K [17]. MMTV gene-like sequences have been found more frequently in breast cancer patients with a family history of breast cancer, tumors discovered during pregnancy or lactation, and breast cancers from certain geographical locations [24]. The prevalence of MMTV-like gene sequences has been found to be 73.7 % in Tunisia [25], 38 % in North America [19, 31], 38 % in Italy [1, 16] and Australia [10], 31 % in Argentina [19], 16.8 % in China [18], and 4.2 % in Mexico [36]. Studies have suggested an unseen association between breast cancer development and aggressiveness and the presence of viral sequences with 80 % homology with MMTV.
Despite the considerable evidence that MMTV-like virus may be linked with breast cancer cause and tumorogenesis, the exact mechanism and relationship of MMTV and breast cancer in humans is still a subject of research [14]. Here, we extend the search for MMTV envelop and LTR-like gene sequences in a Pakistani population diagnosed with breast cancer and fibroadenoma. Our results show the presence of MMTV-like sequences in breast cancer patients and thus provide supportive evidence for the viral cause in breast cancer.
Materials and methods
Patients and controls
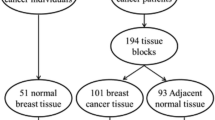
Peripheral blood samples and formalin-fixed paraffin-embedded (FFPE) tissue biopsies of a total of 80 clinically diagnosed breast cancer patients were taken from different hospitals of Rawalpindi, Islamabad, and Lahore after taking informed consent. Blood samples from one healthy male and five healthy female volunteers (without having a history of any type of cancers reported in their families) were taken as control. Blood samples were collected in sterile vacutainers containing EDTA as anti-coagulant and were stored at 4 °C. To conduct this study, approval from Institutional Review Board (IRB) of Atta-ur-Rahman School of Applied Biosciences (ASAB) was taken prior to the start of the project.
Animals
Balb/C mice strain generously provided by AMSON Vaccine & Pharma, Pakistan, was maintained and bred in ASAB animal house at NUST H-12 campus. Few of the mice developed spontaneous mammary tumor, and those mice were then kept in separate chambers to avoid spread of the tumor. Mice were monitored for mammary tumors by palpation. Tumor-bearing mice were held 3 to 4 weeks before sacrifice. The tumor mass was isolated through biopsy while blood samples were taken through heart puncturing.
DNA isolation from FFPE tissue and blood samples
The samples were processed for DNA extraction from formalin-fixed paraffin embedded (FFPE) tissue samples as described by Chan et al. [3]. The major steps involved de-parafinization, xylene removal, tissue lysis, phenol-chloroform extraction, and resuspension of isolated DNA.
The DNA from human and mice blood samples were collected using phenol-chloroform extraction method. Quality of the DNA was checked on 0.8 % agarose gel, followed by ethidium bromide staining. Genomic DNA concentration was estimated using a spectrophotometer by measuring the optical density of DNA at 260 and 280 nm and ratio of absorbance at 260/280 nm was used to assess the purity of DNA. A ratio of ~1.8 is generally accepted as “pure” for DNA; so the samples having the ratio below 1.7 were treated with phenol-chloroform to remove the contaminating proteins.
Checking the DNA integrity using polymerase chain reaction (PCR) for β-globin gene was performed as described by Daniel et al. [5] using forward primer 5′-ACACAACTGTGTTCACTAGC-3′ and reverse primer 5′-CAACTTCATCCACGTTCACC-3′. PCR was performed using standard PCR procedures to avoid contamination, and DNA quality was assessed by amplifying a 119-bp fragment of the β-globin gene under the PCR profile; denaturing at 95 °C for 40 s, annealing at 58 °C for 40 s, and extension at 72 °C for 50 s for 35 cycles, and a final extension at 72 °C for 7 min, keeping the number of cycles 35.
A 119-bp DNA fragment on agarose gel depicted the amplification of β-globin gene (Figs. 2 and 3). Only those samples which gave amplification of β-globin gene were selected, and the rest of the samples were discarded.
Screening of MMTV-like env and LTR sequence
Blood as well as tissue biopsies were taken from the mice with tumor, and histopathology of the mice tissue samples was done to confirm the mammary tumor. DNA from both tissue and blood sample were extracted, and PCR was performed to detect env and LTR-like MMTV DNA sequences [32]. MMTV-env gene of 660 bp was amplified using forward primer 5′-CCTCACTGCCAGATC-3′ and reverse primer 5′-CTATCTGTGGCATACCT-3′. MMTV-LTR gene of 630 bp was amplified using forward primer 5′-GGTGGCAACCAGGGACTTAT-3′ and reverse primer 5′-CGTGTGTTTGTGTCTGTTCG-3′. PCR was performed in 50-μL reaction mixture which consisted of 1× polymerase buffer, 2 mM MgCl2, 2 mM dNTPs (dATP, dGTP, dTTP, dCTP), 20 pmol of each forward and reverse primers, 1 unit of thermostable Taq polymerase, and 20 ng of DNA as template. Nuclease-free water was added to make the volume up to 50 μL. Thermal profile for PCR reaction consisted of 35 cycles of denaturation at 95 °C for 40 s, annealing at 50 °C for 40 s, and extension at 72 °C for 50 s for 35 cycles, and a final extension at 72 °C for 7 min. To avoid false positive PCR amplification results, PCR was performed using standard PCR procedures. Amplified PCR products were resolved on 1.5 % agarose gel followed by ethidium bromide staining and analysis through UV transilluminator. DNA fragments referring to ~660-bp-long MMTV-env and ~630-bp-long MMTV-LTR were eluted from gel and cloned in pCR II vector (Invitrogen Life Technologies, Inc., Carlbad, CA, USA) containing LacZ as reporter gene, allowing blue-white selection, following manufacturer’s protocol. A total of eight transformed white colonies of Escherichia coli were screened through restriction digestion with EcoR1. Clones were subjected to commercial DNA sequencing (Operon, Canada) and sequences were analyzed through BLAST homology search and CLC Bio software. The resulting sequences (accession numbers KC179710–KC179713) were compared to known published sequences and to sequences in the GenBank, with accession ID as AY659980–AY659985 for env sequences and AY652964–AY652978 for LTR sequences. The confirmed clones were then used as positive controls in all PCR reactions.
Statistical analysis
The prevalence of MMTV-like env and LTR DNA sequences in Pakistani breast cancer patients was correlated with age, grade of the disease, and lymph node presence or absence by using Statistical Product and Service Solutions (SPSS) correlation test.
Results
Amplification and cloning of MMTV envelop and LTR genes from mice
Amplification of MMTV 666-bp envelope and 630-bp LTR gene sequences from the blood and tissue of infected mice with mammary tumors was performed, as shown in Fig. 1. Amplified sequences were ligated in pCRII vector for TA cloning, separately, and then used to transform cells of DH5 alpha strain of E. coli. Transformed colonies were screened for the presence of positive clones by EcoRI digestion. After restriction digestion, positive clones obtained were subjected to automated sequencing. Sequencing results showed a >98 % homology of amplified MMTV envelope and LTR gene fragment from mice with MMTV proviral genome.
Amplification of MMTV envelop and LTR genes from Pakistani normal women and breast cancer patients
Normal women and breast cancer patients’ tissue biopsies were tested for the presence of MMTV sequences. MMTV 666-bp envelope and 630-bp LTR-like sequences were detected using a positive control of envelope and LTR genes cloned from mice source and a reagent control. Figures 2 and 3 represent MMTV envelope and LTR sequences, respectively, amplified from Pakistani normal and breast cancer patients along with negative and positive controls. No amplification was observed in negative controls and normal women breast tissue samples. We have found 16 out of 80 samples (20 %) positive for the presence of Envelope gene, whereas 21 samples (26 %) were positive for the presence of LTR gene in breast cancer patients. None of the control samples were positive for any of env or LTR gene.
Statistical analysis using SPSS software
SPSS (Statistical Product and Service Solutions) was used to analyze the prevalence of MMTV envelope and LTR genes in Pakistani breast cancer patients. The results show that 20 and 26.3 % of the samples were positive for the presence of MMTV envelope and LTR sequences, respectively, from the total samples, as shown in Tables 1 and 2. No significant correlation was observed in any case of these comparisons. No significant correlation was found while comparing different pathological parameters: age of the patient, type of breast cancer, grade of the disease, lymph node involvement, etc., with the prevalence of MMTV-like DNA sequences with p value >0.05 (Table 3).
Sequencing and homology results
Homology results show 100 % match between MMTV envelope sequence from Pakistani human and murine sources, with 99 % similarity to MMTV proviral genome. However, in the case of MMTV LTR-like sequence, >95 % similarity was observed.
Mus musculus envelope protein-like (Env) gene isolate (partial sequence KC179713) was observed to have 100 % homology with Homo sapiens isolate B-env-3 envelope protein-like (Env) gene (partial sequence KC179710) isolated from a Pakistani breast cancer female patient, as shown in Fig. 4.
Homology results indicated a similarity of 99 % between Pakistani M. musculus isolate (KC179712) and M. musculus DNA, clone lambda5, endogenous mouse mammary tumor virus LTR region (AB049194) (Fig. 5). H. sapiens isolate B-LTR-3 retrotransposon (partial sequence KC179711) showed 100 % homology with mouse mammary tumor virus complete proviral genome AF033807 (Fig. 6).
Discussion
Breast cancer is the most frequently diagnosed malignancy of women in developed and developing countries especially in Pakistan. It has been estimated that one in every nine Pakistani women is likely to suffer from breast cancer, one of the highest incidence rates in Asia. [29]. In Pakistan, the reasons behind these relatively high rates of breast cancer are unknown.
To date, at least seven viruses have been confirmed to contribute significantly to human cancers [22]. MMTV, one of the oncoviruses, has also been strongly linked to breast cancer because of two main reasons: being established as a causative agent of mammary cancer in the most well-known animal model, i.e., mouse, and most of the studies on the association of MMTV-like sequences and human breast cancers have found a strong correlation with very few studies for null association. The detection of MMTV-like env sequences has been reported in variable proportions that did not exceed 40 % of BC cases in several countries, with an exception of higher proportion (74 %) of MMTV-like sequences in Tunisian women diagnosed with BC during the 1970s [11]. MMTV-like env gene sequences indicate the presence of a replication-competent MMTV-like virus [14].
Ford et al. have also reported premalignant breast lesions (20–28 %) and male gynecomastia (19 %) samples with significant rates of MMTV positivity, but we did not find any prevalence of MMTV LTR and Envelop sequences in any of breast lesions and male gynecomastia samples [10]. A MMTV-like long-terminal repeat super antigen in human breast cancer has been sequenced by Wang et al. in 2004 from human breast cancer and has been found highly homologous to those of MMTV [30].
We have conducted this study independent of earlier studies reporting a high prevalence of MMTV-like sequences in human breast cancer, and it is in direct contrast to two recent negative reports questioning the association of MMTV-like sequences with breast cancer [35, 34]. The ethnic background of healthy controls and breast tumor patients are quite similar. However, in our case, the median age of patients with normal breast tissue and patients with env and LTR-positive and -negative tumors differs (35 and 45 years, respectively), with over half of our normal controls being under the age of 30 years, whereas in the study of Etkind et al. [7], the median age of patients with normal breast tissue and patients with Envelop-positive and -negative tumors differs (15 and 32 years, respectively), with over half of their normal controls being under the age of 40 years. However, for our entire breast tumor samples, we have found 20 % env-positive samples and 26 % LTR-positive samples as reported by Wang et al. [31, 32] and Etkind et al. [7].
Although such data are not enough to establish the causal role of this virus, at least three different studies have tried to establish MMTV as an exogenous source. Firstly, Melana et al. [19] used paired samples and found MMTV in 30 % of the cancerous cases but only in one of the 106 normal tissue samples from the same patients [19]. Secondly, Etkind et al. found MMTV-homologous sequences in father, mother, and daughter living together for years and who all developed breast cancers [8]. Thirdly, Stewart et al. associated MMTV-positive breast cancer prevalence to the population density of M. musculus [28]. The abovementioned three researches showed the variation in prevalence in different regions, the variation of results in different populations living in the same region [10], and the establishment of the fact that MMTV can infect human cells in culture [12].
We have correlated THE presence of MMTV env and LTR gene-like sequences with special clinical disease status, grading, and lymph node presence, but we did not find any significant correlation. With a greater number of samples in the future, we may determine MMTV-like sequences as another potential molecular marker and may distinguish a subset of human breast cancer for which viral etiology is tenable; the further findings may have implications for epidemiology and potentially for therapy and prevention.
Conclusion
MMTV-like Envelop and LTR sequences have been amplified and sequenced from infected mice with mammary tumor. DNA samples of Pakistani breast cancer patients were screened for the prevalence of MMTV Envelop and LTR gene sequences in the Pakistani population, and it was found that 26 % of the samples were positive for LTR gene, whereas 20 % of the samples were positive for the presence of envelop gene. However, no correlation was observed for prevalence of MMTV gene sequences with age, gender, disease type, lymph node involvement, and grade of the disease. By this study, MMTV cannot be said to be an etiological agent to cause breast cancer, but further studies can provide more insights into the association of MMTV with breast cancer in the Pakistani population.
References
Bindra A, Muradrasoli S, Kisekka R, Nordgren H, Warnberg F, Blomberg J. Search for DNA of exogenous mouse mammary tumor virus-related virus in human breast cancer samples. J Gen Virol. 2007;88(Pt 6):1806–9.
Callahan R. MMTV-induced mutations in mouse mammary tumors: their potential relevance to human breast cancer. Breast Cancer Res Treat. 1996;39(1):33–44.
Chan PKS, Chan DPC, To KF, Yu MY, Cheung JLK, Cheng AF. Evaluation of extraction methods from paraffin wax embedded tissues for PCR amplification of human and viral DNA. J Clin Pathol. 2001;54(5):401–3.
Coffin JM, Hughes SH, Varmus HE. Retroviruses. Cold Spring Harbor: Cold Spring Harbor Laboratory; 1997.
Daniel RL, Daniel Z, Bin W, Jeffrey AC, Robert HS. Real-time observation of transcription initiation and elongation on an endogenous yeast gene. Science. 2011;332:475–8. doi:10.1126/science.1202142.
Day NK, Witkin SS, Sarkar NH, Kinne D, Jussawalla DJ, Levin A, et al. Antibodies reactive with murine mammary tumor virus in sera of patients with breast cancer: geographic and family studies. Proc Natl Acad Sci U S A. 1981;78:2483–7.
Etkind P, Du J, Khan A, Pillitteri J, Wiernik PH. Mouse mammary tumor virus-like ENV gene sequences in human breast tumors and in a lymphoma of a breast cancer patient. Clin Cancer Res. 2000;6(4):1273–8.
Etkind P, Stewart A, Wiernik PH. Mouse mammary tumor virus (MMTV)-like DNA sequences in the breast tumors of father, mother, and daughter. Infectious Agents and Cancer 2008. 2008;3:2. doi:10.1186/1750-9378-3-2.
Feldman SP, Schlom J, Spiegelman S. Further evidence for oncornaviruses in human milk: the production of cores. Proc Natl Acad Sci U S A. 1973;70:1976–80.
Ford CE, Faedo M, Crouch R, Lawson JS, Rawlinson WD. Progression from normal breast pathology to breast cancer is associated with increasing prevalence of mouse mammary tumor virus-like sequences in men and women. Cancer Res. 2004;64(14):4755–9.
Hachana M, Trimeche M, Ziadi S, Amara K, Gaddas N, Mokni M, et al. Prevalence and characteristics of the MMTV-like associated breast carcinomas in Tunisia. Cancer Lett. 2008;271(2):222–30.
Hohenadl C, Gunzburg WH, Salmons B, Indik S. The 5′ leader sequence of mouse mammary tumor virus enhances expression of the envelope and reporter genes. J Gen Virol. 2012;93(Pt 2):308–18. doi:10.1099/vir.0.035196-0.
Holder Jr WD, Wells Jr SA. Antibody reacting with the murine mammary tumor virus in the serum of patients with breast carcinoma: a possible serological detection method for breast carcinoma. Cancer Res. 1983;43:239–44.
Lawson JS, Gunzburg WH, Whitaker NJ. Viruses and human breast cancer. Future Microbiol. 2006;1(1):33–51.
Lawson JS, Glenn WK, Salmons B, Ye Y, Heng B, Moody P, et al. Mouse mammary tumor virus-like sequences in human breast cancer. Cancer Res. 2010. doi:10.1158/0008-5472.
Levine PH, Pogo BG, Klouj A, Coronel S, Woodson K, Melana SM, et al. Increasing evidence for a human breast carcinoma virus with geographic differences. Cancer. 2004;101(4):721–6.
Liu B, Wang Y, Melana SM, Pelisson I, Najfeld V, Holland JF, et al. Identification of a proviral structure in human breast cancer. Cancer Res. 2001;61(4):1754–9.
Luo T, Wu XT, Zhang MM, Qian K. Study of mouse mammary tumor virus-like gene sequences expressing in breast tumors of Chinese women. Sichuan Da Xue Xue Bao Yi Xue Ban. 2006;37(6):844–6. 851.
Melana SM, Holland JF, Pogo BG. Search for mouse mammary tumor virus-like env sequences in cancer and normal breast from the same individuals. Clin Cancer Res. 2001;7(2):283–4.
Mesa-Tejada R, Keydar I, Ramanarayanan M, Ohno T, Fenoglio C, Spiegelman S. Detection in human breast carcinomas of an antigen immunologically related to a group-specific antigen of mouse mammary tumor virus. Proc Natl Acad Sci U S A. 1978;75:1529–33.
Michalides R, Spiegelman S, Schlom J. Biochemical characterization of putative subviral particulates from human malignant breast tumors. Cancer Res. 1975;35:1003–8.
Moore PS, Chang Y. Why do viruses cause cancer? Highlights of the first century of human tumour virology. Nat Rev Cancer. 2010;10(12):878–89. doi:10.1038/nrc2961.
Ohno T, Mesa-Tejada R, Keydar I, Ramanarayanan M, Bausch J, Spiegelman S. Human breast carcinoma antigen is immunologically related to the polypeptide of the group-specific glycoprotein of mouse mammary tumor virus. Proc Natl Acad Sci U S A. 1979;76:2460–4.
Pogo BG, Melana SM, Holland JF, Mandeli JF, Pilotti S, Casalini P, et al. Sequences homologous to the mouse mammary tumor virus env gene in human breast carcinoma correlate with overexpression of laminin receptor. Clin Cancer Res. 1999;5(8):2108–11.
Pogo BG, Holland JF, Levine PH. Human mammary tumor virus in inflammatory breast cancer. Cancer. 2010;116(11):2741–4.
Salmons B, Gunzburg WH. Current perspectives in the biology of mouse mammary tumour virus. Virus Res. 1987;8(2):81–102.
Spiegelman S, Axel R, Schlom J. Virus-related RNA in human and mouse mammary tumors. J Natl Cancer Inst. 1972;48:1205–11.
Stewart TH, Sage RD, Stewart AF, Cameron DW. Breast cancer incidence highest in the range of one species of house mouse, Mus domesticus. Br J Cancer. 2000;82(2):446–51.
Sohail S, Alam SN. Breast cancer in Pakistan: awareness and early detection. J Coll Phys Surg Pak. 2007;17:711–2.
Wang Y, Jiang JD, Xu D, Li Y, Qu C, Holland JF, et al. A mouse mammary tumor virus-like long terminal repeat superantigen in human breast cancer. Cancer Res. 2004;64(12):4105–11.
Wang Y, Pelisson I, Melana SM, Go V, Holland JF, Pogo BG. MMTV-like env gene sequences in human breast cancer. Arch Virol. 2001;146(1):171–80.
Wang Y, Holland JF, Bleiweiss IJ, Melana S, Liu X, Pelisson I, et al. Detection of mammary tumor virus env gene-like sequences in human breast cancer. Cancer Res. 1995;55:5173–9.
Witkin SS, Sarkar NH, Kinne DW, Breed CN, Good RA, Day NK. Antigens and antibodies cross-reactive to the murine mammary tumor virus in human breast cyst fluids. J Clin Invest. 1981;67:216–22.
Witt A, Hartmann B, Marton E, Zeillinger R, Schreiber M, Kubista E. The mouse mammary tumor virus-like env gene sequence is not detectable in breast cancer tissue of Austrian patients. Oncol Rep. 2003;10(4):1025–9.
Zangen R, Harden S, Cohen D, Parrella P, Sidransky D. Mouse mammary tumor-like env gene as a molecular marker for breast cancer? Int J Cancer. 2002;102(3):304–7.
Zapata-Benavides P, Saavedra-Alonso S, Zamora-Avila D, Vargas-Rodarte C, Barrera-Rodriguez R, Salinas-Silva J, et al. Mouse mammary tumor virus-like gene sequences in breast cancer samples of Mexican women. Intervirology. 2007;50(6):402–7.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Naushad, W., bin Rahat, T., Gomez, M.K. et al. Detection and identification of mouse mammary tumor virus-like DNA sequences in blood and breast tissues of breast cancer patients. Tumor Biol. 35, 8077–8086 (2014). https://doi.org/10.1007/s13277-014-1972-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13277-014-1972-3