Abstract
Glutathione S-transferase (GST), a phase II metabolizing enzyme, plays an important role in the cellar defense system, and its activity may modulate leukemia risk. A large body of evidence has shown the possible relevance of functional polymorphisms of the genes that encode GSTs μ, π, and θ (GSTM1, GSTP1, and GST1, respectively) to the genetic susceptibility of chronic myeloid leukemia (CML). Because of the lack of available conclusive data, we performed a meta-analysis of all relevant available studies to derive a more precise estimation of the relationship. A comprehensive literature search of PubMed and Web of Knowledge electronic databases was conducted to collect relevant studies until December 20, 2013, and the extracted data were statistically analyzed using Review Manager version 5.2. Finally, 16 eligible studies were identified in the literature. The GSTT1 null genotype was associated with an increased risk of CML, as were the double null GSTT1 and GSTM1 genotypes. These findings suggest that heritable GST status influences the risk of developing CML and that more attention should be paid to carriers of these susceptibility genes.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Chronic myeloid leukemia (CML) is a malignancy of hematopoietic stem cells characterized by high levels of leukocytes, splenomegaly, myeloid hyperplasia in the bone marrow, and high levels of mature myeloid cells in the peripheral blood [1]. It was the first cancer for which a specific cytogenetic marker was found: the Philadelphia chromosome (the result of a reciprocal translocation between chromosomes 9 and 22: t(9;22)(q34,q11)) [2].
Although these clinical and biological aspects are well documented, little is known about the susceptibility of particular individuals to CML. DNA is at constant risk of being damaged by both endogenous and exogenous mechanisms. Detoxification and DNA repair enzymes protect DNA from damage. When the cellular processes of detoxification or repair are ineffective, the persisting DNA damage can cause severe failure of cellular functions, leading to either apoptosis or oncogenesis [3–5]. Thus, exposure to toxic substances could cause DNA damage, which when combined with interindividual differences in the capacity to respond to and repair that DNA damage could affect the susceptibility to CML.
It had been claimed that cytotoxic and genotoxic environmental agents—especially ionizing radiation and similar factors—increase the risk of developing CML [6]. Meanwhile, the genetic polymorphisms that have been described for multiple genes associated with DNA repair might contribute to the reported interindividual variation in the ability to detoxify [7], which could explain the mechanism of individual differences in susceptibility to CML. The glutathione S-transferases (GSTs) are a family of phase II enzymes that are involved in the detoxification of xenobiotics and had been researched widely. They catalyze the conjugation reaction between glutathione and compounds containing an electrophilic center, such as chemotherapeutic drugs, carcinogens, environmental pollutants, and a broad spectrum of other xenobiotics [8]. Hence, GSTs play significant role in cellular defense. Human cytosolic GSTs can be characterized into four distinct families according to their isoelectric points: α, μ, π, and θ [9].
Functional polymorphisms have been reported for at least three of the genes that encode GSTs: GSTM1 (μ), GSTT1 (θ), and GSTP1 (π). Both GSTM1 and GSTT1 exhibit a particularly high degree of polymorphism, one of them being the complete deletion of the gene could potentially cause a loss of enzymatic activity [10]. Approximately 20–50 % of individuals do not express the enzyme due to homozygous deletion, resulting in a diminished ability to detoxify various carcinogens; these individuals are more susceptible to DNA damage [11]. GSTP1 is located on chromosome 11q13 and is overexpressed in various tumor types [12]. The A → G polymorphism at nucleotide 313 in exon 5 of GSTP1 can lead to an amino acid substitution of isoleucine (Ile) by valine (Val) at amino acid position 105 (Ile105Val). This substitution potentially diminishes the ability to detoxify certain mutagens and carcinogens, which could result in increased DNA damage and mutation and hence a greater risk of developing cancer [13]. Biochemical studies have indicated that the conjugating activity is lower for Val homozygotes than for Ile homozygotes, with heterozygotes displaying intermediate activity [14]. Individuals with at least one Val allele might have an underlying predisposition toward cancer when they are exposed to environmentally derived or endogenously formed GSTP1 substrates [15].
Numerous studies had investigated the association between these polymorphisms and the susceptibility to CML, with conflicting results [1, 6, 12, 16–28]. Clarification of this putative association was therefore necessary, and it was also the aim of the present study. To this end, data were collected from all published studies of the relationship between GSTM1, GSTT1, and GSTP1 polymorphisms and the risk of CML. After adherence to strict inclusion criteria, a meta-analysis was applied to all of the eligible studies.
Methods
Literature and search strategy
All relevant studies published before December 20, 2013 were identified through an extended computer-based search of PubMed (www.ncbi.nlm.nih.gov/pubmed) and Web of Knowledge (http://isiknowledge.com/). The search strategy was based on a combination of the following keywords: “GSTM1,” “GSTT1,” “GSTP1,” “acute myeloid leukemia” (“AML” or “acute myelocytic leukemia” or “acute myelogenous leukemia”), “chronic myeloid leukemia” (“CML” or “chronic myelocytic leukemia” or “chronic myelogenous leukemia”), “polymorphism,” “susceptibility,” and “risk.” Only journal articles were included in the analysis. All references cited in the studies were also reviewed to identify additional relevant work. Only studies involving human subjects using standard genotyping methods were considered. Cases with CML were eligible regardless of whether or not they had a first-degree relative with any cancer.
Inclusion criteria
The following inclusion criteria were applied:
-
1.
The study must have a case–control design and investigate the relationship between GSTM1, GSTT1, and GSTP1 polymorphisms and the risk of CML.
-
2.
The study must provide sufficient data on the distribution of GST gene polymorphisms in cases and in control groups of healthy subjects or sufficient information for such data to be calculated.
-
3.
The cases considered in the study must include a population of CML patients.
Data extraction
The following information was extracted for the included studies: name of the first author, year of publication, numbers of patients and controls, ethnicities of the study population, median ages of the cases and controls, sex ratios of the cases and controls, genotype distributions for the cases and controls, and main single nucleotide polymorphism (SNP) susceptibility findings of the research.
Statistical analysis
Review Manager (version 5.2) software was used for the meta-analysis. The raw data for genotype distribution were used to calculate the study-specific estimates of odds ratio (OR) and 95 % confidence interval (CI). The presence of heterogeneity was assessed using Cochran’s Q statistic and quantified using the I 2 statistic, which is proportional to the degree of heterogeneity; an I 2 value above 50 % indicates the presence of a very high degree of heterogeneity [29].
The overall pooled OR and corresponding 95 % CI were estimated using the Mantel–Haenszel method with a fixed effects model when no significant heterogeneity is present (below 50 %) [30]. When substantial heterogeneity was present, sensitivity analysis was performed by excluding individual studies. Outlying studies were identified and excluded, and the I 2 estimates for these different sets of studies were examined. When removing particular studies did not cause the heterogeneity index to fall below 50 %, the random effects model was used. This model can account for the heterogeneity in the data that undoubtedly exists due to within- and between-study variations, and thus, its estimated effect values are more conservative [29]. The significance of the pooled OR was determined by a z test. The level of statistical significance was set at P < 0.05.
Potential publication bias was estimated by constructing funnel plots. If most of the data appeared at the top of a funnel plot and was distributed roughly symmetrically, this would suggest the absence of obvious publication bias, and vice versa [31]. There was no need to construct funnel plots when there were too few analyzed studies (i.e., n < 5).
Meta-analyses
Based on the extensive data provided by the included studies, a meta-analysis was conducted to determine the influence of GSTM1, GSTT1, and GSTP1 polymorphisms on the susceptibility CML. The role of the double null GSTM1 and GSTT1 genotypes on the risk of CML was also evaluated. In addition, the effects of the various GST gene polymorphisms were analyzed relative to gender among both the controls and CML patients.
Results
Overview of the study characteristics
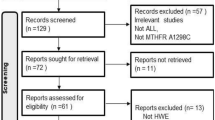
A flow chart depicting the study selection process is shown in Fig. 1. In total, 736 articles were selected based on various combinations of the keywords listed in the “Methods.” Checking for duplicates resulted in the removal of 352 articles. Of the remaining 384 articles, 309 were not on the topic of association between GST gene SNPs and the risk of myeloid leukemia, 26 were review articles, 31 focused on researching the association between GST gene SNPs and the risk of AML, and two did not provide sufficient data. After excluding these articles, only 16 studies of the relationship between GST gene SNPs and the risk of CML remained and qualified for inclusion in this meta-analysis. The basic data for every eligible study were extracted and are listed in Table 1.
Results of the meta-analysis
The literature search yielded 14 studies of the relationship between GSTM1 polymorphism and the risk of CML and provided sufficient data to be eligible for inclusion of the present study [6, 16–28]. A meta-analysis of these 14 studies was conducted, which involved data from 1,175 CML patients and 3,060 controls. The z test verified no association between GSTM1 polymorphism and the risk of CML. The I 2 value indicated the presence of a small degree of heterogeneity among the five studies, and thus, the fixed effects model was used. The funnel plot revealed good symmetry, suggesting that there was no obvious publication bias (Fig. 2a).
Thirteen studies surveyed the association between GSTT1 polymorphism and the risk of CML [6, 16–26, 28]. A meta-analysis was performed on the data of 1,164 CML patients and 2,934 controls. The I 2 value indicated a high degree of heterogeneity among the 13 studies, and the I 2 value did not fall below 50 % regardless of which study was excluded. Therefore, the association was tested using a random effects model. Ultimately, it was found that the GSTT1 null genotype significantly increased the susceptibility to CML (P = 0.004, OR = 1.57, 95 % CI = 1.15–2.14). The funnel plot suggested that no obvious publication bias was present (Fig. 2b).
Seven studies analyzed the influence of combined GSTM1 and GSTT1 null genotypes on the susceptibility to CML [6, 16, 17, 22, 25, 26, 28]. A meta-analysis was conducted with a fixed effects model using the data from 595 patients and 1,506 controls. The I 2 value indicated a high degree of heterogeneity among the seven studies. Sensitivity analysis identified the study by Ozten et al. [25] as an outlier; the removal of this study reduced the heterogeneity to 47 %. A further meta-analysis was conducted using a fixed effects model based on the data from the remaining six studies. The combined null genotype was found to increase the risk of CML (P = 0.002, OR = 1.79, 95 % CI = 1.24–2.58). No obvious publication bias was indicated by the funnel plot (Fig. 2c).
Only two studies researched the effect of the GSTP1 Ile105Val polymorphism on the risk of CML [1, 12]. While we found that controls in both of the two studies were not in accordance with the Hardy–Weinberg equilibrium, this would mean that the two studies could not be used for the meta-analysis. More researches were thus needed to clarify the association.
The distribution of GSTM1 and GSTT1 genotypes between the genders was investigated via meta-analysis using data from the three studies that provided sufficient relevant data [21–23]. The frequency of the GSTM1 null genotype was higher among the male patients than among the female cases (Fig. 3a; P = 0.009, OR = 1.91, 95 % CI = 1.18–3.09), while there was no gender difference in the frequency of the GSTT1 null genotype among CML patients (Fig. 3b). However, since only three studies were included in this gender-based analysis, more relevant research is needed to confirm this finding.
Discussion
Both individual genotypic differences and the level of expression of these carcinogen-metabolizing enzymes are crucial for determining the susceptibility of developing cancer [32]. Mutations of GSTM1, GSTT1, and GSTP1 have been linked with an increase in the number of cancers, probably due to an increased susceptibility to environmental toxins and carcinogens [33]. Many experiments have been performed with the aim of determining how these three gene polymorphisms influence the susceptibility to CML in different areas and in different ethnic groups. The findings of these studies were often contradictory, with the main sources of these differences being certain objective factors, such as race, geographic region, and age. Meta-analysis, which is recognized as one of the best methods of secondary research, can be implemented to integrate these contradictions. In the present study, meta-analysis was used to systematically summarize and analyze the relevant identified literature. The following main conclusions were drawn from the analyses:
-
1.
The GSTT1 null genotype is a risk factor for CML. Furthermore, the double null GSTM1 and GSTT1 genotypes can also further increase the risk of CML.
-
2.
The impact of the GSTM1 null genotype on the risk of CML did not reach statistical difference. This is consistent with the conclusions of most of the included studies.
A meta-analysis similar to that presented herein was performed by Zintzaras et al. in 2009, who investigated the influence of GSTM1 and GSTT1 polymorphisms on the susceptibility to CML, with similar conclusions [34]. The contrast models for GSTM1 and GSTT1 polymorphisms were the same between the two studies of Zintzaras et al. and ours. The main differences between the two studies came from three sides. Firstly, the work of Zintzaras et al. assessed the association between the combined GSTM1 null/GSTT1 null genotype and the risk of developing CML relative to the GSTM1 normal/GSTT1 normal genotype and they got nonsignificant results, while our analysis used different contrast models of combined GSTM1 null/GSTT1 null genotype versus other genotype, and significant result was achieved that the double null genotype increased the risk of CML compared with other genotypes. Secondly, our study also considered the role of GSTP1 Ile105Val SNP on the risk of CML, which was absent in the other study, although the meta-analysis was not available for the limitation of included studies. Thirdly, the literature search in the meta-analysis of Zintzaras et al. was conducted before 1 January 2009. Several more studies were published regarding the GST gene polymorphisms and susceptibility to CML. Therefore, the sample was larger (i.e., number of studies included), and thus, the statistical power was greater in our meta-analysis.
The results of this study have important practical significance. Individual genotypic differences and also the level of expression of carcinogen-metabolizing enzymes are crucial in determining the susceptibility of developing the cancer [32]. Although there is a considerable amount of research on this topic, the results have not received sufficient attention in clinical settings. The present meta-analysis revealed that the GSTT1 and double null genotypes can significantly increase the risk of CML, while the GSTM1 polymorphism had no effect on the susceptibility to CML. Thus, genotype testing is very important, since it can identify people who are carrying the risk genotypes that would make them more susceptible to CML induced by environmental carcinogens. If identified, these people will be able to take the necessary protective measures, which is particularly important for those with long-term exposure to environmental pollutants.
In this study, we just analyzed the role of phase II enzymes, based on the theory that individuals may be more or less susceptible to developing CML as a result of DNA variants in the genes encoding xenobiotic-metabolizing enzymes; we supposed phase I enzymes, mainly the cytochrome P450 (CYP) superfamily, might exert some effect on the risk of suffering CML. Several studies had been performed from that point, focusing on two genes: CYP1A1 and CYP2D6. The study of Taspinar et al. revealed that persons carrying CYP1A1 Val allele had an increased risk of CML [6], while no significant difference was observed between the healthy individuals and CML patients in the frequency of polymorphic variants of CYP1A1 genes in the work of Ovsepian [24]. Another study researched both CYP1A1 and CYP2D6, and the combined genotypes study of CYP1A1 and CYP2D6 was also performed. No significant results were found [18]. These controversial results did not support a clear association between the phase I enzyme gene polymorphisms and susceptibility to CML, and we hence suggested that further studies should be done in this respect.
Some problems arose during the process of data integration. First, the number of studies included in some meta-analyses was low, as was the number of cases. It is recognized that sample size plays an important role in predicting the association between genotypes and risk of cancer in case–control studies. Therefore, the inclusion of studies with very small samples may lead to an overestimation of the true association [35]. The results of meta-analyses that are based on relatively small numbers of studies should be interpreted with caution. In addition, there was some heterogeneity between several of the studies as a result of uncontrolled confounding factors and internal selection bias. Heterogeneity cannot be avoided. We solve this problem by adopting sensitivity analysis and the random effects model. The former reduced the heterogeneity among studies, and the latter fully allowed for the diversity between studies. Meanwhile, two limitations of this meta-analysis should be considered when interpreting its findings. First, the results were based on unadjusted estimates; a more precise analysis should be conducted using data from individuals, which would allow researchers to adjust for covariates including age, ethnicity, family history, environmental factors, and lifestyle. Second, only published studies were included in this meta-analysis. There is always a certain degree of publication bias, and nonsignificant or negative findings may be unpublished.
In summary, although studies investigating the association between GST gene polymorphisms and the risk of CML arrive at different conclusions, this meta-analysis suggests that the GSTT1 null genotype is a risk factor for CML. Other studies will need to be conducted to investigate the relationship between the SNPs within the phase I enzyme gene and GSTP1 and CML susceptibility.
References
Karkucak M et al. Investigation of GSTP1 (Ile105Val) gene polymorphism in chronic myeloid leukaemia patients. Int J Hum Genet. 2012;12(3):145–9.
Nowicki MO et al. BCR/ABL oncogenic kinase promotes unfaithful repair of the reactive oxygen species-dependent DNA double-strand breaks. Blood. 2004;104(12):3746–53.
Ishikawa K, Ishii H, Saito T. DNA damage-dependent cell cycle checkpoints and genomic stability. DNA Cell Biol. 2006;25(7):406–11.
Kennedy RD, D’Andrea AD. DNA repair pathways in clinical practice: lessons from pediatric cancer susceptibility syndromes. J Clin Oncol. 2006;24(23):3799–808.
Voso MT et al. Increased risk of acute myeloid leukaemia due to polymorphisms in detoxification and DNA repair enzymes. Ann Oncol. 2007;18(9):1523–8.
Taspinar M et al. CYP1A1, GST gene polymorphisms and risk of chronic myeloid leukemia. Swiss Med Wkly. 2008;138(1–2):12–7.
Bhatla D et al. DNA repair polymorphisms and outcome of chemotherapy for acute myelogenous leukemia: a report from the Children’s Oncology Group. Leukemia. 2008;22(2):265–72.
Hayes JD, Pulford DJ. The glutathione S-transferase supergene family: regulation of GST and the contribution of the isoenzymes to cancer chemoprotection and drug resistance. Crit Rev Biochem Mol Biol. 1995;30(6):445–600.
Mannervik B et al. Nomenclature for mammalian soluble glutathione transferases. Methods Enzymol. 2005;401:1–8.
Alves S et al. The GSTM1 and GSTT1 genetic polymorphisms and susceptibility to acute lymphoblastic leukemia in children from north Portugal. Leukemia. 2002;16(8):1565–7.
Strange RC et al. Glutathione-S-transferase family of enzymes. Mutat Res. 2001;482(1–2):21–6.
Sailaja K et al. Association of the GSTP1 gene (Ile105Val) polymorphism with chronic myeloid leukemia. Asian Pac J Cancer Prev. 2010;11(2):461–4.
Dunna NR et al. Association of GSTP1 gene (I105V) polymorphism with acute leukaemia. J Genet. 2012;91(1):e60–3.
Watson MA et al. Human glutathione S-transferase P1 polymorphisms: relationship to lung tissue enzyme activity and population frequency distribution. Carcinogenesis. 1998;19(2):275–80.
Harries LW et al. Identification of genetic polymorphisms at the glutathione S-transferase Pi locus and association with susceptibility to bladder, testicular and prostate cancer. Carcinogenesis. 1997;18(4):641–4.
Bajpai P, Tripathi AK, Agrawal D. Increased frequencies of glutathione-S-transferase (GSTM1 and GSTT1) null genotypes in Indian patients with chronic myeloid leukemia. Leuk Res. 2007;31(10):1359–63.
Bhat G et al. Polymorphic variation in glutathione-S-transferase genes and risk of chronic myeloid leukaemia in the Kashmiri population. Asian Pac J Cancer Prev. 2012;13(1):69–73.
Chen HC et al. Genetic polymorphisms of metabolic enzymes CYP1A1, CYP2D6, GSTM1 and GSTT1 and leukemia susceptibility. Eur J Cancer Prev. 2008;17(3):251–8.
Hishida A et al. GSTT1 and GSTM1 deletions, NQO1 C609T polymorphism and risk of chronic myelogenous leukemia in Japanese. Asian Pac J Cancer Prev. 2005;6(3):251–5.
Loffler H et al. Reduced risk for chronic myelogenous leukemia in individuals with the cytochrome P-450 gene polymorphism CYP1A1*2A. Blood. 2001;98(13):3874–5.
Lordelo GS et al. Association between methylene tetrahydrofolate reductase and glutathione S-transferase M1 gene polymorphisms and chronic myeloid leukemia in a Brazilian population. Genet Mol Res. 2012;11(2):1013–26.
Lourenco GJ et al. Polymorphisms of glutathione S-transferase mu1 (GSTM1) and theta 1 (GSTT1) genes in chronic myeloid leukaemia. Eur J Haematol. 2005;75(6):530–1.
Mondal BC et al. Glutathione S-transferase M1 and T1 null genotype frequency in chronic myeloid leukaemia. Eur J Cancer Prev. 2005;14(3):281–4.
Ovsepian VA, Vinogradova E, Sherstneva ES. Cytochrome P4501A1, glutathione S-transferase M1 and T1 gene polymorphisms in chronic myeloid leukemia. Genetika. 2010;46(10):1360–2.
Ozten N, Sunguroglu A, Bosland MC. Variations in glutathione-S-transferase genes influence risk of chronic myeloid leukemia. Hematol Oncol. 2012;30(3):150–5.
Souza CL et al. Polymorphisms in the glutathione S-transferase theta and mu genes and susceptibility to myeloid leukemia in Brazilian patients. Genet Mol Biol. 2008;31(1):39–41.
Lemos MC et al. Genetic polymorphism of CYP2D6, GSTM1 and NAT2 and susceptibility to haematological neoplasias. Carcinogenesis. 1999;20(7):1225–9.
Ouerhani S et al. Influence of genetic polymorphisms of xenobiotic metabolizing enzymes on the risk of developing leukemia in a Tunisian population. Bull Cancer. 2011;98(12):95–106.
Lopez-Lopez, E., et al., A systematic review and meta-analysis of MTHFR polymorphisms in methotrexate toxicity prediction in pediatric acute lymphoblastic leukemia. Pharmacogenomics J, 2012.
Gurion R et al. Has the time for first-line treatment with second generation tyrosine kinase inhibitors in patients with chronic myelogenous leukemia already come? Systematic review and meta-analysis. Haematologica. 2013;98(1):95–102.
Egger M et al. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315(7109):629–34.
Kawajiri K et al. The CYP1A1 gene and cancer susceptibility. Crit Rev Oncol Hematol. 1993;14(1):77–87.
Tan W et al. Impact of genetic polymorphisms in cytochrome P450 2E1 and glutathione S-transferases M1, T1, and P1 on susceptibility to esophageal cancer among high-risk individuals in China. Cancer Epidemiol Biomarkers Prev. 2000;9(6):551–6.
Zintzaras E. Glutathione S-transferase M1 and T1 genes and susceptibility to chronic myeloid leukemia: a meta-analysis. Genet Test Mol Biomarkers. 2009;13(6):791–7.
Das P, Shaik AP, Bammidi VK. Meta-analysis study of glutathione-S-transferases (GSTM1, GSTP1, and GSTT1) gene polymorphisms and risk of acute myeloid leukemia. Leuk Lymphoma. 2009;50(8):1345–51.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (No. 81071765; 81372379) and the Fundamental Research Funds for the Central Universities (No. 08143047) of China.
Conflicts of interest
None
Author information
Authors and Affiliations
Corresponding authors
Additional information
H.-r. He and X.-x. Zhang contributed equally to this manuscript
Rights and permissions
About this article
Cite this article
He, Hr., Zhang, Xx., Sun, Jy. et al. Glutathione S-transferase gene polymorphisms and susceptibility to chronic myeloid leukemia. Tumor Biol. 35, 6119–6125 (2014). https://doi.org/10.1007/s13277-014-1810-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13277-014-1810-7