Abstract
MicroRNAs (miRNAs) have been integrated into cancer development and progression, because they repress translation of target genes which can be tumor suppressors and oncogenes. A number of miRNAs have been found to be closely related to human non-small cell lung cancer (NSCLC). However, the roles of miR-136 in NSCLC are still largely unknown. Here, we show that miR-136 is significantly upregulated in human NSCLC primary tumors and cell lines compared to their nontumor counterparts. Suppression of miR-136 expression in NSCLC cell line A549 inhibited both anchorage-dependent and anchorage-independent proliferation. Further studies showed that suppression of miR-136 expression attenuated phosphorylation of extracellular-signal-regulated kinase 1/2 (Erk1/2). We found that serine/threonine protein phosphatase 2A 55 kDa regulatory subunit B α isoform (PPP2R2A, also known as B55α) was a direct target of miR-136, and suppression of miR-136 expression led to a robust increase in both mRNA and protein levels of PPP2R2A. We found that miR-136 promoted phosphorylation of Erk1/2 through inhibition of PPP2R2A expression, and forced overexpression of PPP2R2A abrogated promotion of Erk1/2 phosphorylation by miR-136. Moreover, forced overexpression of PPP2R2A abrogated the promoting effect of miR-136 on cell growth and led to a reduced growth rate of NSCLC cells. Our findings indicate that miR-136 promotes Erk1/2 phosphorylation through targeting PPP2R2A in NSCLC cells and suggest that it may serve as a therapeutic target in NSCLC therapy.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Lung cancer is a leading cause of cancer-related mortality in many countries [1], and non-small cell lung cancer (NSCLC) accounts for the majority of lung cancer cases. Surgery, chemotherapy, radiotherapy, and their combination are selected for the treatment of NSCLC depending on the histological grade, cancer stage, and patient age. Despite recent advances in clinical and experimental oncology, the prognosis of lung cancer still remains poor, with a 5-year overall survival rate of approximately 11 % [2]. Therefore, a detailed understanding of NSCLC and development of more effective treatments are the most important topics for this disease.
MicroRNAs (miRNAs) are evolutionarily conserved, endogenous noncoding RNA of about 21–24 nucleotides in length that regulate gene expression by base pairing with target mRNAs in the 3′-untranslated region (3′-UTR), leading to mRNA cleavage or translational repression [3, 4]. Through these mechanisms, miRNAs regulate up to 30 % of human genes [5] and play important parts in various biological process, including development, differentiation, apoptosis, and cell proliferation [6, 7]. According to recent findings, miRNAs are frequently dysregulated in many types of cancer [8] and involved in tumor initiation, promotion, and progression through regulation of various oncogenes or tumor suppressors [9]. Recently, accumulating evidence has shown that miRNAs are involved in NSCLC pathogenesis, providing new insights for NSCLC treatment.
miR-136 was earlier reported to be markedly upregulated in the Jurkat cell line [10], and it was also found to be associated with embryonic stem cells [11]. Latter reports showed that miR-136 was implicated in cancer biology and played roles as both tumor suppressor and oncogene in different types of cancer by targeting distinct genes. miR-136 is downregulated in human glioma and promotes apoptosis of glioma cells by targeting AEG-1 and Bcl-2 [12]. miR-136 was found to target tumor suppressor PTEN in breast cancer cells, which also showed a tumor-promoting role in cancer development [13]. Recently, miRNA microarray expression profiling showed that miR-136 was prominently overexpressed in murine and human lung cancers relative to adjacent normal lung tissues. Although miR-136 was showed to be closely related to lung cancer, its biological function in human NSCLC is largely unknown.
Mitogen-activated protein kinase (MAPK) pathways are evolutionarily conserved in eukaryotic cells and take important parts in physiological processes, such as embryonic development and immune response [14]. In mammals, four major classes of MAPKs have been identified, including the extracellular-signal-regulated kinases 1/2 (Erk1/2), the c-Jun N-terminal kinases, the p38 kinases, and ERK5. Considering the Erk1/2 pathway, the primary input activator is activated Ras, a small GTPase. It activates Raf1 (MAPK kinase kinase), which then phosphorylates and activates MEK1/2 (MAPK kinase), which finally activates Erk1/2 [15]. In its active form, Erk1/2 phosphorylates a wide range of protein substrates, including transcriptional regulators, apoptosis regulators, and steroid hormone receptors. The biological consequences of Erk1/2 substrate phosphorylation include promotion of proliferation, differentiation, survival, angiogenesis, motility, and invasion [16–18]. Hyperactivation of the Erk1/2 pathway is frequently observed in human malignancies by many reasons including aberrant activation of receptor tyrosine kinases and gain-of-function mutations in Ras, Raf, or MEK [19–22]. Therefore, components of the Erk1/2 pathway are viewed as attractive candidates for the development of targeted therapies of cancer [23], and inhibitors of the kinase function of Raf and MEK are currently evaluated in preclinical and clinical analyses [24, 25].
In this study, we found that miR-136 was significantly upregulated in NSCLC primary tumors and cell lines. Suppression of miR-136 led to inhibition of cell proliferation and attenuated phosphorylation of Erk1/2. Moreover, miR-136 promoted Erk1/2 phosphorylation through direct targeting of serine/threonine protein phosphatase 2A 55 kDa regulatory subunit B α isoform (PPP2R2A), while overexpression of PPP2R2A abrogated the promoting effect of miR-136 on NSCLC cell proliferation. Our research reveals miR-136 as a regulator of Erk1/2 signaling in NSCLC and provides valuable information for therapeutic development against NSCLC.
Materials and methods
Clinical specimens
NSCLC specimens, including matched nontumor tissues, were obtained from the tissue bank of The First Affiliated Hospital of Zhengzhou University. The study protocol was approved by the Institutional ethics Committee. The specimens were obtained after surgical resection and immediately frozen in liquid nitrogen until use.
Cell culture and transfection
All cell lines were obtained from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China). The normal bronchial epithelial cell line 16HBE and NSCLC cell lines A549, SPC-A1, NCI-H1650, and NCI-H1299 were maintained in DMEM media (Invitrogen, Carlsbad, USA) and supplemented with 10 % (v/v) fetal bovine serum, 100 U/ml penicillin, and 100 mg/ml streptomycin. Cell culture was conducted at 37 °C in a humidified 5 % CO2 incubator.
miR-136 mimics or anti-miR-136 oligonucleotide (Ambion, Austin, USA) was transfected using Lipofectamine 2000 regent (Invitrogen, Carlsbad, USA) following the manufacturer’s protocol.
Quantitative RT-PCR
Total RNA was isolated from NSCLC tissues and cell lines using the TRIzol regent (Invitrogen) and reverse transcribed into cDNA. miR-136 expression was detected by TaqMan® MicroRNA assay (Applied Biosystems, Foster City, USA) following the manufacturer’s instructions. U6 snRNA was used as an endogenous control. The levels of PPP2R2A were analyzed using standard quantitative RT-PCR. The following primers were used: PPP2R2A forward: 5′-TCGGATGTAAAATTCAGCCA-3′; PPP2R2A reverse: 5′-CATGCACCTGGTATGTTTCC-3′; GAPDH forward: 5′-ACGGATTTGGTCGTATTGGG-3′, and GAPDH reverse: 5′-TGATTTTGGAGG GATCTCGC-3′. The levels of miRNA and mRNA were determined using the 7500 Fast System SDS software (Applied Biosystems, Foster City, USA). Each reaction was repeated independently at least three times in triplicate. For semiquantitative RT-PCR, the number of cycles was 23 for U6 and 27 for miR-136.
Cell viability assay
Cell viability was measured using the MTT assay. MTT was diluted in PBS to a final concentration of 5 mg/ml and sterile filtered. Cells were incubated with a final concentration of 0.5 mg/ml MTT at 37 °C for 4 h. Cell culture supernatants were carefully removed, and DMSO was added. The absorbance values were determined using a microplate reader (Pharmacia Biotech, Uppsala, Sweden) at a wavelength of 490 nm. The experiments were performed three times.
Colony formation assay
The treated cells were suspended in complete medium containing 0.35 % agar and overlaid on 0.6 % agar in six-well plates (2 × 103 cells/well). Following about 4 weeks of culture, the number of colonies that include more than 50 cells was subsequently counted from three independent experiments.
Western blotting
Cells were lysed in a RAPI lysis buffer and solubilized in SDS loading buffer. Equal amounts of protein extracts were separated on a 12 % polyacrylamide gel and transferred to a nitrocellulose membrane (Amersham Biosciences). Protein expression was analyzed using standard procedures for western blotting. The following primary antibodies were used: anti-Erk1/2, Cell Signaling Technology, 1:1,000 dilution; anti-p-Erk1/2, Cell Signaling Technology, 1:1,000 dilution; anti-PPP2R2A, Abcam, 1:1,000 dilution; and anti-GAPDH, Sigma, 1:1,000 dilution. After incubation with the appropriate horseradish peroxidase-conjugated secondary antibody (Santa Cruz Biotechnology, 1:1,000 dilution), the membranes were treated with an enhanced chemiluminescence reagent (Thermo Scientific, Dreieich, Germany), exposed to X-ray film (Kodak, Rochester, USA) and quantified by densitometry (Beckman, South Pasadena, Canada).
Construction of expression vectors
The 2.2-kb 3′UTR of human PPP2R2A from a human cDNA library was amplified using PCR and cloned into a pGL3 vector (Promega, Madison, WI) to generate a Luc-PPP2R2A-wt vector. The following primers were used to clone PPP2R2A 3′UTR: forward: 5′-GGGGTACCGGTTGGCATTCCTAGCAGAAG-3′ and reverse: 5′-CCCAAGCTTGTGGCTGGGCTTAGAGTTTG-3′. Site-directed mutagenesis of the miR-133b target sites in the PPP2R2A 3′UTR was performed using a QuikChange mutagenesis kit (Stratagene, Heidelberg, Germany) to generate the Luc-PPP2R2A-mut vector.
PPP2R2A mRNA was amplified by PCR using the following primers: forward: 5′-CGGGATCCATGTTCCCGAAGTTTTCTC-3′ and reverse: 5′-GGAATTCCTAATTCACTTTGTCTTG-3′. The PCR products were digested with BamH I and EcoR I and inserted into a pcDNA3 vector to construct PPP2R2A overexpression vector.
Dual-luciferase reporter assay
Luciferase reporter assays were performed in A549 cells. Cells were cotransfected with pRL-TK vector, wild type (Luc-PPP2R2A-wt), or mutant (Luc-PPP2R2A-mut) reporter plasmids, along with miR-136 or control miRNA. Forty-eight hours after transfection, cells were rinsed with PBS, and dual-luciferase assay was performed (Promega). Luciferase activity was measured using a Victor luminometer (PerkinElmer, Waltham, USA). The firefly luciferase activity was normalized using cotransfected Renilla luciferase for transfection efficiency. All experiments were performed in triplicate.
Statistical analysis
Data were analyzed using the SPSS software (SPSS Inc., Chicago, IL). Quantitative data are presented as the mean ± the standard deviation. Differences between the miR-136 in tumor tissues and adjacent nontumor tissues were analyzed by the Wilcoxon matched pairs test. The Pearson test was used to identify correlation between miR-136 and PPP2R2A in NSCLC tissues. Statistical differences between groups were determined by Student’s t test. Differences were considered significant when P < 0.05.
Results
miR-136 is upregulated in NSCLC primary tumors and cell lines
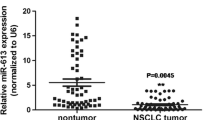
To test the expression of miR-136 in human NSCLC tissues, we examined miR-136 levels in 37 pairs of NSCLC tissues using quantitative RT-PCR analysis. Our results showed that miR-136 expression was significantly upregulated in NSCLC tissues compared to matched nontumor tissues (P < 0.01, Fig. 1a), while there was no significant difference of U6 RNA expression between NSCLC and nontumor tissues (Supplemental Fig. 1). miR-136 levels in NSCLC tissues were approximately 3.8-fold higher than those in matched nontumor tissues.
miR-136 is upregulated in NSCLC primary tumors and cell lines. a The expression level of miR-136 in 37 pairs of NSCLC tissues and matched nontumor tissues was detected by quantitative RT-PCR. U6 snRNA was used as an endogenous control. Data are presented as fold changes in tumor tissues relative to nontumor tissues. The miR-136 levels were significantly upregulated in NSCLC tissues as determined by the Wilcoxon matched pairs test, P < 0.01. b Analysis of miR-136 expression levels in NSCLC cell lines A549, SPC-A1, NCI-H1650, and NCI-H1299 compared to the normal bronchial epithelial cell line 16HBE by semiquantitative RT-PCR. U6 snRNA was used as an inner control. c Analysis of miR-136 expression levels in cell lines in b by quantitative RT-PCR. U6 snRNA was used as an inner control. d Detection of miR-136 by in situ hybridization in paired NSCLC tissue and its matched nontumor tissue. ×100 magnification
We further analyzed the correlation between the upregulated miR-136 and several clinicopathological parameters among the 37 patients. There were 26 men and 11 women included with age ranging from 35 to 74 years old (the median, 58 years). The 37 NSCLC cases can be divided into two types including 23 squamous cell carcinoma (SCC) cases and 14 adenoma cases. Our results did not reveal any significant association between miR-136 expression and the parameters including age and gender, but adenoma tended to have more increased miR-136 levels (Table 1). The tumors were also subdivided in terms of histological grades and tumor–lymph node–metastasis (TNM) stages. Among these 37 cases, 25 were well to moderately differentiated (1 grade I, 2 grade I–II, and 22 grade II), and 12 were poorly differentiated (2 grade II–III and 10 grade III). The results showed a significant correlation between miR-136 and the histological grade (P = 0.014, Table 1). According to the TNM cancer staging system, the 37 NSCLC cases were classified into TNM stage I (n = 1), II (n = 2), and II–III (n = 35). There was no significant correlation between miR-136 and the TNM stages.
We examined miR-136 expression in NSCLC cell lines A549, SPC-A1, NCI-H1650, and NCI-H1299 as well as in the normal bronchial epithelial cell line 16HBE. Compared to 16HBE cell line, miR-136 was upregulated overall but had different expression levels in all the tested NSCLC cell lines (Fig. 1b, c). No significant difference of U6 RNA expression between these cell lines was observed (Supplemental Fig. 2). Moreover, A549 cells showed relatively higher expression of miR-136 than other tested NSCLC cells. To further confirm the upregulation of miR-136 in NSCLC, we examined miR-136 expression in NSCLC tissues using in situ hybridization assay. We found that the staining signal of miR-136 was much higher in NSCLC tissue than the matched nontumor counterpart (Fig. 1d). Together, these results indicate that miR-136 is upregulated in NSCLC primary tumors and cell lines.
Downregulation of miR-136 reduces cell proliferation
To evaluate the biological significance of miR-136 in NSCLC, we examined whether downregulation of miR-136 could affect cell proliferation. The NSCLC cell line A549 was chosen for its highest expression of miR-136 in the tested NSCLC cell lines. Downregulation of miR-136 was examined by quantitative RT-PCR after transfection with anti-miR-136 oligonucleotide. There were approximately 75 and 64 % reductions of miR-136 expression in A549 cells after transfection with anti-miR-136 oligonucleotide on day 3 and 5, respectively, compared to control (Fig. 2a). To test the effect of miR-136 downregulation on cell proliferation, we examined cell viability using MTT assay after transfection with anti-miR-136 oligonucleotide. The results showed that downregulation of miR-136 significantly reduced viability of A549 cells (Fig. 2b). Compared to control, the viability of anti-miR-136 oligonucleotide-transfected A549 cells was decreased to 71 % (P < 0.05) on the 5th day. To further identify the effect of miR-136 downregulation on cell proliferation, the cells transfected with anti-miR-136 oligonucleotide were analyzed for colony formation ability using soft agar assay. Our results showed that miR-136 downregulation in A549 cells caused a significant decrease in the number of formed colonies (Fig. 2c). The colony numbers of anti-miR-136 oligonucleotide-transfected cells were reduced by 62 % compared to the control (P < 0.05, Fig. 2d). Taken together, our results showed that downregulation of miR-136 reduced proliferation of NSCLC cells, suggesting that miR-136 takes an important part in NSCLC cell growth.
Downregulation of miR-136 in A549 cells reduces cell proliferation. a Cells were transfected with 40 nM of anti-miR-136 or control oligonucleotide, and examined by quantitative RT-PCR. The expression levels of miR-136 after transfection on day 3 and day 5 were shown as the relative values to control. * P < 0.05, vs. control (Student’s t test). b A549 cells were transfected with anti-miR-136 or control oligonucleotide. MTT assay was performed to determine the proliferation of A549 cells. Data represent the mean ± S.D. from three independent experiments. The difference of cell growth during culture on day 5 was analyzed. *P < 0.05, vs. control (Student’s t test). (c) The proliferation of A549 cells was detected using the colony formation assay. Cells were cultured for 4 weeks after transfection with anti-miR-136 or control oligonucleotide, and counted for formed colonies. 40× magnification (d) The quantitation of relative colony numbers was shown. * P < 0.05, vs. control (Student’s t test)
Downregulation of miR-136 inhibits activation of Erk1/2
To investigate how miR-136 regulates NSCLC cell proliferation, we examined some cardinal signaling kinases which are essential for cancer cell growth in A549 cells after transfection with anti-miR-136 oligonucleotide. Our results showed that anti-miR-136 oligonucleotide caused a significant reduction of Erk1/2 phosphorylation in a dose-dependent manner (Fig. 3a and data not shown). Anti-miR-136 oligonucleotide at 40 nM concentration can inhibit 90 % of Erk1/2 phosphorylation at 72 h posttransfection (Fig. 3b). Next, we examined whether Erk1/2 phosphorylation was upregulated in NSCLC tissues in accordance with miR-136 upregulation. The phosphorylation of Erk1/2 was analyzed using immunohistochemistry assay. Our results showed that Erk1/2 phosphorylation was significantly upregulated in NSCLC tissue compared to matched adjacent nontumor tissue (Fig. 3c). These results indicate that downregulation of miR-136 inhibits phosphorylation of Erk1/2 in NSCLC cells, and there is a positive correlation between miR-136 expression and Erk1/2 phosphorylation in NSCLC.
Downregulation of miR-136 attenuates Erk1/2 activation. a A549 cells were transfected with anti-miR-136 (20, 40, or 80 nM) or control oligonucleotide (80 nM). The levels of Erk1/2 and p-Erk1/2 were analyzed by western blotting at 72 h posttransfection. Representative results from three independent experiments are shown. b Quantitative analysis of p-Erk1/2 protein normalized to Erk1/2 levels. Values represent the mean ± S.D. from three independent experiments. c Detection of Erk1/2 phosphorylation by immunohistochemistry in the same NSCLC tissue and its matched nontumor tissue. ×100 magnification
miR-136 directly targets PPP2R2A in A549 cells
We used two programs, PicTar and TargetScan, to predict target genes of miR-136. A gene termed PPP2R2A (also known as B55α) was identified as a potential target of miR-136. The 3′UTR of PPP2R2A mRNA contains a putative binding site of miR-136 (Fig. 4a). To confirm that this site was responsible for the regulation by miR-136, we integrated wide-type PPP2R2A 3′UTR or PPP2R2A 3′UTR with mutation of miR-136 binding site into a luciferase reporter vector, and tested the effect of miR-136 on luciferase activity using dual-luciferase assay. The results showed that miR-136 significantly repressed the luciferase activity of WT-3′UTR reporter vector in A549 cells compared to control miRNA (P < 0.05), but barely affected the luciferase activity of Mut-3′UTR reporter vector (Fig. 4b). To determine whether down-regulation of miR-136 affects the expression of endogenous PPP2R2A, we transfected anti-miR-136 oligonucleotide into A549 cells and examined PPP2R2A expression using western blotting analysis. Our results showed that transfection with anti-miR-136 oligonucleotide caused a significant increase of endogenous expression of PPP2R2A (Fig. 4c). Because miRNAs inhibit expression of target gene through translation repression or cleavage of mRNA, we examined whether downregulation of miR-136 affects mRNA levels of PPP2R2A. Our results showed that PPP2R2A mRNA was also significantly increased after anti-miR-136 oligonucleotide transfection (Fig. 4d). It indicates that miR-136 can directly target PPP2R2A and cause cleavage of PPP2R2A mRNA. To confirm this hypothesis, we examined PPP2R2A mRNA levels in the same 37 pairs of NSCLC tissues which miR-136 was examined in. We found that PPP2R2A mRNA expression was significantly downregulated in NSCLC tissues compared to nontumor tissues (P < 0.01, Fig. 4e). Moreover, there is a reverse correlation between miR-136 and PPP2R2A expression in NSCLC tissues (Pearson test, r = −0.377, P < 0.01). Taken together, our results demonstrated that miR-136 directly targeted PPP2R2A in NSCLC.
miR-136 directly targets PPP2R2A in A549 cells. a Schematic representation of PPP2R2A 3′UTR showing a predicted target site for miR-136. A PPP2R2A 3′UTR containing the mutant miR-136 binding sequence was cloned for luciferase reporter assay. b The dual-luciferase reporter assay was performed in A549 cells. The luciferase reporter plasmid containing wild-type or mutant PPP2R2A 3′UTR was cotransfected into A549 cells with miR-136 or control miRNA. Luciferase activity was determined at 48 h posttransfection using the dual-luciferase assay and shown as the relative firefly activity normalized to renilla activity. Bars indicate the relative luciferase activities of three experiments. *P < 0.05, vs. control (Student’s t test). c A549 cells were transfected with miR-136 or control miRNA. The PPP2R2A protein levels were examined by western blotting at 72 h posttransfection. GAPDH was used as a loading control. d A549 cells were treated as c. PPP2R2A mRNA levels were detected at 72 h posttransfection by quantitative RT-PCR. GAPDH was used as an endogenous control. *P < 0.05, vs. control (Student’s t test). e The expression level of PPP2R2A in 37 pairs of NSCLC tissues and matched nontumor tissues was detected by quantitative RT-PCR. U6 snRNA was used as an endogenous control. Data are presented as fold changes in tumor tissues relative to nontumor tissues. The PPP2R2A levels were significantly downregulated in NSCLC tissues as determined by the Wilcoxon matched pairs test, P < 0.01
We further analyzed the correlation between the downregulated PPP2R2A and several clinicopathological parameters among our 37 NSCLC tissues. The results showed no significant association between miR-136 expression and the parameters including age, gender, and TNM stages (Table 2). However, we found that there is a significant correlation between PPP2R2A and the histological grade (P = 0.013, Table 2).
PPP2R2A is responsible for miR-136-induced Erk1/2 phosphorylation and cell proliferation
Because PPP2R2A can cause dephosphorylation of some kinases including Erk1/2, we investigated whether PPP2R2A plays essential roles in miR-136-regulated Erk1/2 activation. We transfected miR-136 alone or together with PPP2R2A into A549 cells and then examined Erk1/2 phosphorylation using western blotting. Our results showed that miR-136 inhibited PPP2R2A expression and promoted Erk1/2 phosphorylation without affecting total levels of Erk1/2 protein (Fig. 5a). However, when miR-136 was transfected together with PPP2R2A into A549 cells, miR-136-promoted Erk1/2 phosphorylation was significantly inhibited; meanwhile, the miR-136 levels were identical in miR-136 transfected cells and cells transfected with miR-136 along with PPP2R2A (Fig. 5b). It indicated that PPP2R2A can block miR-136-promoted phosphorylation of Erk1/2, suggesting that PPP2R2A is responsible for miR-136-induced Erk1/2 phosphorylation. We further examined whether forced overexpression of PPP2R2A can neutralize the promoting effect of miR-136 on cell proliferation. A549 cells were transfected with miR-136 alone or together with PPP2R2A, and cell viability was examined using MTT assay. The results showed that miR-136 promoted cell proliferation, while PPP2R2A in reverse significantly inhibited cell proliferation and abrogated the promoting effect of miR-136 on NSCLC cell proliferation (Fig. 5c). miR-136 induced a 32 % increase of cell growth compared to control on the 5th day (P < 0.05), while the growth of cells cotransfected with miR-136 and PPP2R2A showed a 44 % reduction compared to that transfected with miR-136 alone on the 5th day (P < 0.05), which is also much lower than that of control cells. Meanwhile, on the 5th day, there were identical levels of miR-136 in miR-136-transfected cells and cells transfected with miR-136 along with PPP2R2A, supporting that PPP2R2A can neutralize the promoting effect of miR-136 on cell proliferation (Fig. 5d). Taken together, our results demonstrate that PPP2R2A is important for miR-136-induced Erk1/2 phosphorylation and cell proliferation.
PPP2R2A is responsible for miR-136-promoted Erk1/2 phosphorylation and cell proliferation. a A549 cells were transfected with miR-136 as well as PPP2R2A or control vector. At 72 h posttransfection, the levels of PPP2R2A, Erk1/2, and p-Erk1/2 were assessed by western blotting. b The expression levels of miR-136 in the cells treated as a were examined by quantitative RT-PCR. The relative expression of miR-136 at 72 h posttransfection compared to control was shown. *P < 0.05, vs. control (Student’s t test). c A549 cells were treated with a. MTT assay was performed to determine cell proliferation. Data represent the mean ± S.D. from three independent experiments. The difference of cell growth during culture on day 5 was analyzed. *P < 0.05, vs. miR-136 (Student’s t test). d A549 cells were treated as c. The relative expression of miR-136 in A549 cells during culture on day 5 was shown. *P < 0.05, vs. control (Student’s t test)
Discussion
With the advent of miRNA expression profiling, significant effort has been made to correlate miRNA expression with tumor progression and prognosis. In addition, the Dicer enzymes, necessary for the maturation of the miRNAs, have been reported to be downregulated in the tumor tissue of patients with lung cancer in whom it had a prognostic impact on survival [26], suggesting that miRNAs take important part in lung cancer. To date, a number of dysregulated miRNAs found in lung cancer correlate with patient survival [27, 28] and therapeutic response [29]. Some of them has been showed to be implicated in the therapeutic resistance mechanisms in lung cancer [30]. Therefore, understanding the roles of miRNA in lung cancer might provide valuable information for therapeutic strategies in lung cancer treatment. Here, we focused on the roles of miR-136 in NSCLC.
We examined miR-136 expression in NSCLC primary tumors and cell lines through quantitative RT-PCR assay. Our results showed that miR-136 was upregulated in NSCLC tissues and cell lines compared to their nontumor counterparts. The dysregulation of miR-136 has been reported in some types of cancer. Those reports have indicated that miR-136 is downregulated in human glioma, but upregulated in leukemia cell line Jurkat and breast cancer cell lines. Moreover, miRNA microarray expression profiling in murine and human lung cancers recently has showed some upregulated miRNAs in lung tumor tissues including miR-136. Their results and ours all support the upregulation of miR-136 in lung cancer and suggest that miR-136 might play an important role in lung cancer oncogenesis.
There is little information available for the function of miR-136, particularly in cancer. The existing studies have shown the complexity of miR-136 in different types of cancer. It acts as a tumor suppressor by targeting Bcl-2 in glioma, but acts as an oncogene by targeting PTEN in breast cancer cells. However, its role in cancer is likely correlated with its expression levels. Considering its upregulation in lung cancer, we posited that it might promote lung cancer development. Our results showed that suppression of miR-136 resulted in inhibition of NSCLC cell viability as indicated by MTT assay. We also found that the anchorage-independent proliferation was also inhibited by miR-136 suppression. These results indicate that miR-136 takes an important role in NSCLC cell growth.
To determine how suppression of miR-136 causes growth inhibition of NSCLC cells, we examined several kinases which are essential for cancer cell growth after anti-miR-136 transfection, and we found that Erk1/2 was significantly inhibited by miR-36 suppression in a dose-dependent manner. miRNAs typically function through inhibiting expression of target genes. We hypothesized that miR-136 probably inhibited expression of certain genes which could inhibit Erk1/2 phosphorylation. We searched for its potential targets using bioinformatics analysis and identified a phosphatase subunit PPP2R2A. There is one miR-136 binding site in the 3′UTR of PPP2R2A mRNA. In our luciferase report assay, we showed that Luc-PPP2R2A-wt was specifically responsive to miR-136 overexpression. In contrast, the Luc-PPP2R2A-mut, which contained the mutation of miR-136 binding site in the PPP2R2A 3′UTR, successfully abolished the effect of miR-136 on luciferase activity. We also demonstrated that miR-136 likely inhibited PPP2R2A expression through cleavage of PPP2R2A mRNA, because the mRNA and protein levels of PPP2R2A were both significantly increased when miR-136 expression was suppressed.
PPP2R2A is inferred to be a candidate tumor suppressor gene because it belongs to the protein phosphatase 2 (PP2A) regulatory subunit B family. PP2A refers to a large family of heterotrimeric Ser/Thr phosphatases and is implicated in the negative control of cell growth and division through regulating a variety of cellular processes, including signal transduction, DNA replication, apoptosis, and cell cycle progression. The PP2A core enzyme consists of a catalytic C subunit and a structural A subunit. The AC dimer recruits a third regulatory B subunit, which is responsible for the substrate specificity and function of the PP2A heterotrimeric complex. Four unrelated families of B subunits have been identified to date: B/B55/PPP2R2A, B′/B56/PR61/PPP2R5, B″/PR72/PPP2R3, and Striatin/STRN. PPP2R2A gene is located at the 8p21 chromosome region, which is frequently deleted in a wide range of epithelial cancers, and PPP2R2A was found to be frequently inactivated in multiple cancer types by various mechanisms including loss of heterozygosity and gene truncation [31]. Many reports showed that reduced PPP2R2A expression was found in prostate adenocarcinoma samples, acute myelogenous leukemia, as well as in lung cancer. Accumulating evidence indicates that PPP2R2A targets PP2A to numerous kinases and signaling pathways that are in involved in cell proliferation, DNA replication, apoptosis, and cell migration. PPP2R2A acts to antagonize activation of FoxM1 and p107, thus playing roles in cell cycle progression [32, 33]. PPP2R2A is also the major regulatory subunit mediating dephosphorylation of FOXO1 and is associated with oxidative and apoptotic stress [34]. PPP2R2A directly interacts with beta-catenin, which is important for cell migration, and specifically regulates PP2A-mediated dephosphorylation of beta-catenin [35]. Moreover, PPP2R2A had been found to contribute to tumor suppressor activity by negatively regulating pathways such as Akt and Erk1/2 signaling pathways to ensure normal growth of cells [36, 37]. Silencing of PPP2R2A leads to hyperactivation of Erk stimulated by constitutively active MEK1.
In this study, we showed that miR-136 was upregulated in NSCLC and had an important part in NSCLC cell proliferation. We found that miR-136 directly targeted PPP2R2A, a protein phosphatase regulatory subunit which is important for dephosphorylation of Erk1/2, and contributed to elevated phosphorylation of Erk1/2 in NSCLC cells. Our study showed the important roles of miR-136 in NSCLC and provided valuable information for therapeutic development against NSCLC.
References
Jemal A, Siegel R, Xu J, Ward E. Cancer statistics, 2010. CA Cancer J Clin. 2010;60:277–300.
Verdecchia A, Francisci S, Brenner H, et al. Recent cancer survival in Europe: a 2000–02 period analysis of EUROCARE-4 data. Lancet Oncol. 2007;8:784–96.
Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T. Identification of novel genes coding for small expressed RNAs. Science. 2001;294:853–8.
Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–97.
Petrocca F, Visone R, Onelli MR, et al. E2F1-regulated microRNAs impair TGFbeta-dependent cell-cycle arrest and apoptosis in gastric cancer. Cancer Cell. 2008;13:272–86.
Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–5.
Farh KK, Grimson A, Jan C, et al. The widespread impact of mammalian microRNAs on mRNA repression and evolution. Science. 2005;310:1817–21.
Croce CM. Causes and consequences of microRNA dysregulation in cancer. Nat Rev Genet. 2009;10:704–14.
Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–66.
Yu J, Wang F, Yang GH, et al. Human microRNA clusters: genomic organization and expression profile in leukemia cell lines. Biochem Biophys Res Commun. 2006;349:59–68.
Liu SP, Fu RH, Yu HH, et al. MicroRNAs regulation modulated self-renewal and lineage differentiation of stem cells. Cell Transplant. 2009;18:1039–45.
Yang Y, Wu J, Guan H, et al. MiR-136 promotes apoptosis of glioma cells by targeting AEG-1 and Bcl-2. FEBS Lett. 2012;586:3608–12.
Lee DY, Jeyapalan Z, Fang L, et al. Expression of versican 3′-untranslated region modulates endogenous microRNA functions. PLoS One. 2010;5:e13599.
Chang L, Karin M. Mammalian MAP kinase signalling cascades. Nature. 2001;410:37–40.
Krishna M, Narang H. The complexity of mitogen-activated protein kinases (MAPKs) made simple. Cell Mol Life Sci. 2008;65:3525–44.
Pearson G, Robinson F, Beers Gibson T, et al. Mitogen-activated protein (MAP) kinase pathways: regulation and physiological functions. Endocr Rev. 2001;22:153–83.
Pages G, Milanini J, Richard DE, et al. Signaling angiogenesis via p42/p44 MAP kinase cascade. Ann N Y Acad Sci. 2000;902:187–200.
Joslin EJ, Opresko LK, Wells A, Wiley HS, Lauffenburger DA. EGF-receptor-mediated mammary epithelial cell migration is driven by sustained ERK signaling from autocrine stimulation. J Cell Sci. 2007;120:3688–99.
Hynes NE, Lane HA. ERBB receptors and cancer: the complexity of targeted inhibitors. Nat Rev Cancer. 2005;5:341–54.
Schubbert S, Shannon K, Bollag G. Hyperactive Ras in developmental disorders and cancer. Nat Rev Cancer. 2007;7:295–308.
Davies H, Bignell GR, Cox C, et al. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949–54.
Marks JL, Gong Y, Chitale D, et al. Novel MEK1 mutation identified by mutational analysis of epidermal growth factor receptor signaling pathway genes in lung adenocarcinoma. Cancer Res. 2008;68:5524–8.
Sebolt-Leopold JS, Herrera R. Targeting the mitogen-activated protein kinase cascade to treat cancer. Nat Rev Cancer. 2004;4:937–47.
Sebolt-Leopold JS, Dudley DT, Herrera R, et al. Blockade of the MAP kinase pathway suppresses growth of colon tumors in vivo. Nat Med. 1999;5:810–6.
Solit DB, Garraway LA, Pratilas CA, et al. BRAF mutation predicts sensitivity to MEK inhibition. Nature. 2006;439:358–62.
Karube Y, Tanaka H, Osada H, et al. Reduced expression of Dicer associated with poor prognosis in lung cancer patients. Cancer Sci. 2005;96:111–5.
Yanaihara N, Caplen N, Bowman E, et al. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell. 2006;9:189–98.
Navarro A, Diaz T, Gallardo E, et al. Prognostic implications of miR-16 expression levels in resected non-small-cell lung cancer. J Surg Oncol. 2011;103:411–5.
Garofalo M, Quintavalle C, Di Leva G, et al. MicroRNA signatures of TRAIL resistance in human non-small cell lung cancer. Oncogene. 2008;27:3845–55.
Skrzypski M, Dziadziuszko R, Jassem J. MicroRNA in lung cancer diagnostics and treatment. Mutat Res. 2011;717:25–31.
Mao X, Boyd LK, Yanez-Munoz RJ, et al. Chromosome rearrangement associated inactivation of tumour suppressor genes in prostate cancer. Am J Cancer Res. 2011;1:604–17.
Jayadeva G, Kurimchak A, Garriga J, et al. B55alpha PP2A holoenzymes modulate the phosphorylation status of the retinoblastoma-related protein p107 and its activation. J Biol Chem. 2010;285:29863–73.
Alvarez-Fernandez M, Halim VA, Aprelia M, Laoukili J, Mohammed S, Medema RH. Protein phosphatase 2A (B55alpha) prevents premature activation of forkhead transcription factor FoxM1 by antagonizing cyclin A/cyclin-dependent kinase-mediated phosphorylation. J Biol Chem. 2011;286:33029–36.
Yan L, Guo S, Brault M, et al. The B55alpha-containing PP2A holoenzyme dephosphorylates FOXO1 in islet beta-cells under oxidative stress. Biochem J. 2012;444:239–47.
Zhang W, Yang J, Liu Y, et al. PR55 alpha, a regulatory subunit of PP2A, specifically regulates PP2A-mediated beta-catenin dephosphorylation. J Biol Chem. 2009;284:22649–56.
Kuo YC, Huang KY, Yang CH, Yang YS, Lee WY, Chiang CW. Regulation of phosphorylation of Thr-308 of Akt, cell proliferation, and survival by the B55alpha regulatory subunit targeting of the protein phosphatase 2A holoenzyme to Akt. J Biol Chem. 2008;283:1882–92.
Strack S. Overexpression of the protein phosphatase 2A regulatory subunit Bgamma promotes neuronal differentiation by activating the MAP kinase (MAPK) cascade. J Biol Chem. 2002;277:41525–32.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (no. 91071909).
Conflicts of interest
None
Author information
Authors and Affiliations
Corresponding author
Additional information
Sining Shen and Han Yue contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplemental Fig. 1
Typical amplification plot of the U6 RNA in 37 pairs of NSCLC tissues and matched nontumor tissues obtained using quantitative RT-PCR assay. Equal amounts of RNA were used for all the samples (JPEG 24 kb)
Supplemental Fig. 2
Typical amplification plot of the U6 RNA in NSCLC cell lines A549, SPC-A1, NCI-H1650, NCI-H1299 and the normal bronchial epithelial cell line 16HBE obtained using quantitative RT-PCR assay. Equal amounts of RNA were used for all the samples (JPEG 21 kb)
Rights and permissions
About this article
Cite this article
Shen, S., Yue, H., Li, Y. et al. Upregulation of miR-136 in human non-small cell lung cancer cells promotes Erk1/2 activation by targeting PPP2R2A. Tumor Biol. 35, 631–640 (2014). https://doi.org/10.1007/s13277-013-1087-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13277-013-1087-2