Abstract
CIP2A is a recently characterized oncoprotein which involves in the progression of several human malignancies. This study aimed to investigate its clinical significance and biological function in ovarian cancer. CIP2A expression was analyzed in 152 archived ovarian cancer specimens using immunohistochemistry. One hundred cases (65.79 %) showed CIP2A overexpression, including 63 of 92 serous carcinomas (68.48 %), 21 of 33 endometrioid carcinomas (63.64 %), 12 of 23 mucinous carcinomas (52.17 %), and 4 of 4 clear cell carcinomas (100 %). There is no significant difference of CIP2A expression between serous tumors and all other morphologies combined. CIP2A overexpression positively correlated with advanced FIGO stage (p = 0.0336) and tumor grade (p = 0.0213). siRNA knockdown was performed in A2780 and SKOV3 cell lines. MTT, colony formation assay, and flow cytometry were carried out to assess the role of CIP2A in proliferation, cell cycle, and apoptosis. CIP2A depletion in ovarian cancer cell lines inhibited proliferation, blocked cell cycle progression, and increased paclitaxel-induced apoptosis. Futhermore, CIP2A depletion downregulated cyclin D1, c-myc, phospho-Rb, Bcl-2, and phospho-AKT expression. These results validate the role of CIP2A as a clinically relevant oncoprotein and establish CIP2A as a promising therapeutic target of ovarian cancer.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Ovarian cancer is the leading cause of death among gynecological malignancies and represents the fifth leading cause of cancer-related deaths in women [1, 2]. Despite high incidence and mortality rates of ovarian cancer, the etiological factors involved in ovarian carcinogenesis remain poorly defined, limiting the efficacy of treatment protocols. Therefore, it is important to identify molecular mediators conferring the malignant potential to ovarian cancer cells that may be used as tumor markers in predicting risk of ovarian cancer progression [3, 4].
Cancerous inhibitor of protein phosphatase 2A (CIP2A), originally identified as a cellular PP2A inhibitor that inhibits proteolytic degradation of c-MYC, was observed to promote proliferation and was important for the maintenance of the malignant cellular phenotype [5]. In addition, CIP2A mediates the apoptotic effect of bortezomib in breast cancer [6], hepatocellular carcinoma [7], and head and neck squamous cell carcinoma [8]. Moreover, CIP2A was found to be overexpressed in various human cancers, including breast, gastric, lung, prostate, hepatocellular, colon, and renal cancers [7, 9–15]. Recently, CIP2A overexpression was observed in serous ovarian cancer and its expression was associated with poor patient survival [16]. Since ovarian epithelial tumors are morphologically heterogeneous, the expression of CIP2A in other histological types of ovarian cancer is not clear, and the biological roles of CIP2A in ovarian cancer cells remain to be examined.
In the present study, we investigated the expression pattern of CIP2A in 152 ovarian cancer specimens (including serous, endometrioid, mucinous, and clear cell carcinoma) and analyzed the correlation between its expression and clinicopathological factors. Furthermore, to clarify the role of CIP2A in ovarian cancer development, we investigated the effect of CIP2A on proliferation, cell cycle, and apoptosis in ovarian cancer cell lines.
Materials and methods
Patients and specimens
The study protocol was approved by the institutional reviewer board of China Medical University. Primary tumor specimens were obtained from 152 patients diagnosed with ovarian cancer who underwent resection in Shengjing Hospital of China Medical University between 2005 and 2008. The histological diagnosis was evaluated for sections stained with hematoxylin and eosin according to the World Health Organization (WHO) classification guidelines. Clinical and histopathological data including histopathological diagnosis, FIGO stage, and tumor grade were extracted from medical records. Six non-cancerous ovarian tissues were obtained from patients undergoing surgery as a result of suspicious ultrasound images, palpable abdominal masses, and/or a family history of ovarian cancer.
Cell culture and transfection
A2780 and SKOV3 cell lines were obtained from American Type Culture Collection (Manassas, VA, USA). A2780 and SKOV3 cells were cultured in DMEM (Invitrogen, Carlsbad, CA, USA) containing 10 % fetal calf serum (Invitrogen), 100 IU/ml penicillin (Sigma, St. Louis, MO, USA), and 100 μg/ml streptomycin (Sigma). Cells were grown on sterilized culture dishes and were passaged every 2 days with 0.25 % trypsin (Invitrogen).
On-TargetPlus SMARTpool siRNA for CIP2A (#L-014135-01) and On-TargetPlus siControl (D-001810-01-20) were purchased from Dharmacon (ThermoFisher Scientific, USA). For transfections, cells were seeded in plate 24 h before the experiment. The cells were transfected with siRNA using the DharmaFECT 1 (0.20 μL/well; ThermoFisher Scientific, USA) according to the manufacturer's protocol.
Immunohistochemistry
Surgically excised tumor specimens were fixed with 10 % neutral formalin and embedded in paraffin, and 4-μm-thick sections were prepared. Immunostaining was performed using the avidin–biotin–peroxidase complex method (Ultrasensitive™, MaiXin, Fuzhou, China). The sections were deparaffinized in xylene, rehydrated with graded alcohol, and then boiled in 0.01 M citrate buffer (pH 6.0) for 2 min in an autoclave. Hydrogen peroxide (0.3 %) was applied to block endogenous peroxide activity, and the sections were incubated with normal goat serum to reduce nonspecific binding. Tissue sections were incubated with CIP2A rabbit polyclonal antibody (1:300 dilution; Novus biologicals). Rabbit immunoglobulin (at the same concentration as for the antigen-specific antibody) was used as a negative control. Staining for both antibodies was performed at room temperature for 2 h. Biotinylated goat anti-rabbit serum IgG was used as a secondary antibody. After washing, the sections were incubated with streptavidin–biotin conjugated with horseradish peroxidase, and the peroxidase reaction was developed with 3,3′-diaminobenzidine tetrahydrochloride. Counterstaining with hematoxylin was performed, and the sections were dehydrated in ethanol before mounting.
Two independent blinded investigators examined all tumor slides randomly. Five views were examined per slide, and 100 cells were observed per view at 400× magnification. Immunostaining of CIP2A was scored on a semiquantitative scale by evaluating the intensity and percentage of tumor cells according to previous studies [12]. Cytoplasmic immunostaining in tumor cells was considered positive staining. We counted 400 tumor cells and calculated the percentage of positively stained cells. The intensity of CIP2A staining was scored as 0 (no signal), 1 (weak), 2 (moderate), and 3 (marked). Percentage scores were assigned as 1—1–25 %, 2—26–50 %, 3—51–75 %, and 4—76–100 %. The scores of each tumor sample were multiplied to give a final score of 0 to 12, and the tumors were finally determined as negative (−): score 0; lower expression (+): score ≤ 4; moderate expression (++): score 5–8; high expression (+++): score ≥ 9. Tumor sample scored (+) to (+++) were considered as overexpression.
Quantitative real-time PCR (SYBR Green method)
Quantitative real-time PCR was performed using the SYBR Green PCR master mix (Applied Biosystems) in a total volume of 20 μl on 7900HT fast real-time PCR system (Applied Biosystems) as follows: 95 °C for 30 s, 40 cycles at 95 °C for 5 s, 60 °C for 30 s. A dissociation step was performed to generate a melting curve to confirm the specificity of the amplification. β-actin was used as the reference gene. The relative levels of gene expression were represented as ΔCt = Ct gene − Ct reference, and the fold change of gene expression was calculated by the 2−ΔΔCt method. Experiments were repeated in triplicate. The primer sequences are as followers: CIP2A forward, 5′ ATACTTCAGGACCCACGTTTGAT 3′, CIP2A reverse, 5′ TCTCCAAGTACTAAAGCAGGAAAATCT 3′; β-actin forward, 5′ ATAGCACAGCCTGGATAGCAACGTAC 3′, β-actin reverse, 5′ CACCTTCTACAATGAGCTGCGTGTG 3′.
Western blot analysis
Total proteins from cells were extracted in lysis buffer (Pierce, Rockford, IL) and quantified using the Bradford method. Samples of 50 μg protein were separated by SDS-PAGE. Samples were transferred to polyvinylidene fluoride membranes (Millipore, Billerica, MA, USA) and incubated overnight at 4 °C with antibody against CIP2A (1:1000; Novus), c-myc, cyclinD1, cyclin, CDK2, CDK4, CDK6, phosphor-AKT, AKT, Bcl-2 (1:1000, Cell Signaling Technology, USA), and β-actin (1:500; Santa Cruz). After incubation with peroxidase-coupled antimouse IgG (Santa Cruz) at 37 °C for 2 h, bound proteins were visualized using ECL (Pierce) and detected using a BioImaging System (UVP Inc., Upland, CA, USA). Relative protein levels were quantified using β-actin as loading control.
Colony formation and MTT assays
For colony formation assay, A2780 and SKOV3 cells were transfected with siRNA for 48 h and plated into three 6-cm cell culture dishes (1,000 per dish). Cells were incubated for 12 days in medium containing 10 % FBS. Plates were washed with PBS and stained with Giemsa. The number of colonies with more than 50 cells was counted. The colonies were manually counted using a microscope.
For MTT assay, cells were plated in 96-well plates in medium containing 10 % FBS at approximately 3,000 cells per well, 24 h after transfection. For quantitation of cell viability, 20 μl of 5 mg/ml MTT (thiazolyl blue) solution was added to each well and incubated for 4 h at 37 °C. The medium was removed from each well, and the resulting MTT formazan was solubilized in 150 μl of DMSO. Each solution was measured spectrophotometrically at 490 nm.
Cell cycle and apoptosis analysis
Cells (500,000) were seeded into 6-cm tissue culture dishes. Twelve hours later, cells were transfected with indicated amounts of siRNA. At the indicated time points, cells were harvested, fixed in 1 % paraformaldehyde, washed with phosphate-buffered saline (PBS), and stained in 5 mg/ml propidium iodide in PBS supplemented with RNase A (Roche, Indianapolis, IN) for 30 min at room temperature. Data were collected using BD systems.
For detection of apoptosis, adherent cells were both collected and resuspended in cold PBS for analysis. Cells were treated using 25 nM paclitaxel for 24 h. Then, cells were stained with Annexin V-FITC Apoptosis Kit (BD Pharmingen, USA) to monitor apoptosis cells and propidium iodide (PI) to detect dead cells. Data were collected using BD systems.
Statistical analysis
SPSS version 11.5 for Windows was used for all statistical analyses. A χ 2 test was used to examine possible correlations between CIP2A expression and clinicopathologic factors. Student's t test was used to compare densitometry data on focus numbers between control and CIP2A-transfected cells. All p values are based on a two-sided statistical analysis, and p < 0.05 was considered to indicate statistical significance.
Results
Expression of CIP2A in ovarian cancer tissue and ovarian cancer cells
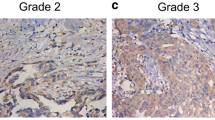
In the six normal ovarian tissues examined, negative or weak expression of CIP2A was observed (Fig. 1a). Cytoplasmic/nuclear staining was present in a subset of ovarian cancer cases (Fig. 1b), while no staining was detected in sections from the same samples subjected to immunohistochemical analysis using rabbit immunoglobulin. Of the 152 patients in the study, 100 (65.7 %) showed positive CIP2A expression. We analyzed the correlation between CIP2A protein expression and the histological staging of ovarian cancer. The positive rates of CIP2A overexpression in serous carcinoma (Fig. 1c, d), endometrioid carcinoma (Fig. 1e, f), mucinous carcinoma (Fig. 1g, h), and clear cell carcinoma (Fig. 1b) were 68.48 % (63/92), 63.64 % (21/33), 52.17 % (12/23), and 100 % (4/4), respectively (Table 1). Because of the low numbers of available tumors with less common morphologies, we compared serous tumors to all other morphologies combined, the difference of CIP2A positivity between serous tumors and all other morphologies combined was not significant (68.48 % versus 61.6 %, p = 0.2272). CIP2A expression in stage II–IV ovarian cancers (stage II, 68.00 %; stage III, 76.47 %; stage IV 80.77 %) was significantly higher than that in stage I ovarian cancers (stage I, 53.73 %, p = 0.0336). High-grade tumors (grade III, 77.61 %) were more likely to be CIP2A positive than low-grade tumors (grade II, 54.17 %; grade I, 59.46 %, p = 0.0213).
Expression of CIP2A protein in ovarian cancer tissues. a Negative CIP2A expression in normal ovary specimen. b Strong cytoplasmic staining of CIP2A in clear cell carcinoma. c Weak CIP2A expression in serous carcinoma. d Strong CIP2A expression in serous carcinoma. e Negative CIP2A expression in endometrioid carcinoma. f Strong CIP2A expression in endometrioid carcinoma. g Negative CIP2A immunostaining in mucinous carcinoma. h Strong CIP2A immunostaining in mucinous carcinoma (magnification, 200×)
CIP2A depletion in ovarian cancer cell lines inhibits cell proliferation
To determine whether CIP2A enhances the proliferation of ovarian cancer cells, we checked CIP2A expression level in several ovarian cancer cell lines and found that A2780 and SKOV3 cells had relative high CIP2A expression (Fig. 2a). siRNA knockdown was performed in these two cell lines. As shown in Fig. 2b, c, CIP2A siRNA decreased CIP2A protein and mRNA in A2780 and SKOV3 cells (knockdown efficiency in A2780, 85 %; SKOV3, 76 %, Fig. 2b). The proliferation rate was determined by MTT assay. A significant decrease in proliferation was observed in A2780 and SKOV3 cells treated with CIP2A siRNA compared with cells treated with negative siRNA (Fig. 3a). Consistent with the MTT results, colony formation assay showed that CIP2A knockdown in A2780 and SKOV3 cells led to a significant decrease in focus numbers (A2780, control 349 ± 19 vs. CIP2A siRNA 207 ± 12, p < 0.05; SKOV3, control 288 ± 16 vs. CIP2A siRNA 106 ± 8, p < 0.05, Fig. 3b).
CIP2A expression and knockdown efficiency in ovarian cancer cells. a Endogenous expression of CIP2A in four ovarian cancer cell lines. b Real-time PCR analysis and western blot showed that siRNA treatment of CIP2A markedly decreased CIP2A levels in A2780 and SKOV3 cells in comparison with cells transfected with negative siRNA
CIP2A knockdown inhibited cell proliferation in ovarian cancer cell lines. a MTT assay showed that CIP2A knockdown inhibited cell proliferation. b Colony formation assay was performed in cells transfected with CIP2A siRNA. A decrease was seen in the groups with siRNA treatment. Asterisk indicates p < 0.05
CIP2A regulates cell cycle progression and paclitaxel-induced apoptosis in ovarian cancer cells
The cell cycle analyses were performed in cancer cells with or without CIP2A knockdown, the percentage of G1 phase was increased in cells with CIP2A knockdown (A2780, control vs. CIP2A siRNA, 45.5 ± 2.8 vs. 55.8 ± 3.1, p < 0.05; SKOV3, control vs. CIP2A siRNA, 55.2 ± 2.3 vs. 65.6 ± 3.2, p < 0.05), whereas the percentage of S phase was decreased in these cells compared with control cells (A2780, control vs. CIP2A siRNA, 38.6 ± 1.9 vs. 31.3 ± 2.1; SKOV3, control vs. CIP2A siRNA, 29.5 ± 1.7 vs. 22.3 ± 1.5, p < 0.05) (Fig. 4a). These results indicated that CIP2A regulated cell cycle progression at the G1/S boundary. Also, the level of G2 phase cells was slightly changed. In addition, 25 nM paclitaxel was used to treat A2780 and SKOV3 cells with CIP2A knockdown for 24 h. Annexin V kit was employed to characterize the rate of apoptosis. As shown in Fig. 4b, a significant population of early and late apoptosis (A2780, 23.63 %; SKOV3, 20.69 %) was observed in cells with CIP2A knockdown compared with scramble controls (A2780, 14.25 %; SKOV3, 12.75 %), demonstrating that CIP2A knockdown results in apoptosis of the ovarian cancer cells.
CIP2A regulates cell cycle progression and paclitaxel-induced apoptosis in ovarian cancer cells. a CIP2A knockdown increased G1 phase cells (A2780, control vs. CIP2A siRNA, 45.5 ± 2.8 vs. 55.8 ± 3.1, p < 0.05; SKOV3, control vs. CIP2A siRNA, 55.2 ± 2.3 vs. 65.6 ± 3.2, p < 0.05) and decreased S phase cells (A2780, control vs. CIP2A siRNA, 38.6 ± 1.9 vs. 31.3 ± 2.1; SKOV3, control vs. CIP2A siRNA, 29.5 ± 1.7 vs. 22.3 ± 1.5, p < 0.05). b Apoptotic cell death was determined by flow-cytometric analysis with Annexin V and PI staining. A2780 and SKOV3 cells were transfected with CIP2A siRNA and control siRNA and then subjected to apoptosis assay. The percentage of apoptosis, including early and late stage of apoptotic cell death in each group, is shown
CIP2A depletion downregulates c-myc, cyclin D1, Bcl-2 expression and inhibits AKT phosphorylation in ovarian cancer cells
To investigate the mechanism underlying cell cycle arrest, we tested the effect of CIP2A knockdown on cyclin D1, c-myc, CDK2, CDK4, and CDK6 levels. As shown in Fig. 5, western blotting analysis revealed that knockdown of CIP2A decreased the protein levels of cyclin D1 and c-myc expression in both cell lines, while the levels of CDK2, CDK4, and CDK6 were not changed. The level of Rb phosphorylation was also decreased in both cell lines. We also examined the level of apoptosis-related Bcl-2 protein and found that CIP2A depletion led to decreased Bcl-2 expression. In addition, the level of AKT phosphorylation was significantly inhibited in cells with siRNA treatment. Together, these results suggested that CIP2A induced cell proliferation by cyclin D1 and c-myc regulation; meanwhile, CIP2A regulated ovarian cancer cell apoptosis through Bcl-2 and AKT pathway.
CIP2A depletion downregulated c-MYC, cyclin D1, Bcl-2 expression and Rb, AKT phosphorylation in ovarian cancer cells. Western blot analysis revealed that knockdown of CIP2A decreased the protein levels of c-MYC, cyclin D1, and Bcl-2 in both cell lines and resulted in a reduction in the phosphorylation of Rb and AKT
Discussion
The present study demonstrated that CIP2A was overexpressed in several types of ovarian cancer, including serous, endometrioid, mucinous, and clear cell carcinoma. This is consistent with previous study in serous ovarian cancer [16] and suggests that CIP2A may be involved in the pathogenesis of these types of ovarian carcinomas. Ovarian epithelial tumors are morphologically heterogeneous and are classified by pathologists into serous, clear cell, endometrioid, and mucinous subtypes on the basis of histopathological examination. Each subtype is characterized by a specific mRNA profile, genetic risk factors, and molecular features [17, 18], suggesting that ovarian carcinoma is a heterogeneous disease. Despite this heterogeneity, we found no significant difference of CIP2A levels among serous, endometrioid, and mucinous carcinoma in our study. However, we found that CIP2A overexpression rate was high in clear cell carcinoma, which is known to show relatively strong resistance to carboplatin and paclitaxel regimen and thus poor prognosis compared to serous adenocarcinoma [19]. The relatively higher overexpression rate in clear cell carcinoma suggested CIP2A may reflect morphological heterogeneity. In accordance with previous reports [16], CIP2A overexpression was found to be positively associated with FIGO stage and tumor grade, suggesting the role of CIP2A in ovarian cancer progression. Different tumorigenic pathways have been proposed in the development of low-grade carcinomas (type I) versus high-grade carcinomas (type II), which evolve rapidly, disseminate early, and are highly aggressive [20]. The rate of CIP2A overexpression was slightly higher in high-grade carcinomas than low-grade carcinomas, indicating its potential association with aggressiveness of tumor cells.
To gain insight into the role of CIP2A in ovarian tumorigenesis, we knocked down CIP2A expression in A2780 and SKOV3 cell lines. Consistent with former reports [5, 14], we found that CIP2A depletion caused significant decrease in the proliferation rate and colony formation ability of A2780 and SKOV3 cell lines. To examine the potential mechanism, we examined the effect of CIP2A knockdown on cell-cycle progression and found that CIP2A facilitated G1-S transition in these cell lines. Furthermore, we checked the expression of a series of cell-cycle-related molecules including cyclin D1, cyclin E, and CDK4/6. We found that the levels of cyclinD1 were decreased after CIP2A knockdown. The level of Rb phosphorylation was also decreased. Cyclin E expression was downregulated only in A2780 cell line with siRNA treatment, which may be a cell-type-specific phenomenon. Cyclin D1 interacts with Cdk4/6 to form a complex, which regulates cell proliferation by controlling progression through the restriction point within the G1 phase of the cell cycle [21]. Cyclin D1 was overexpressed in a variety of cancers and associated with proliferation [22–24], including ovarian cancers [25]. Cyclin D1 degradation is sufficient to induce G1 cell cycle arrest despite constitutive expression of cyclin E2 in ovarian cancer cells [26]. In addition, we demonstrated that CIP2A depletion significantly downregulated c-Myc expression. c-Myc is a cellular proto-oncogene associated with a variety of human cancers and is strongly implicated in the control of cellular proliferation. Loss of c-Myc is associated with reduction of cyclin D1–Cdk4 and –Cdk6 complexes during the cell cycle transition [27]. Thus, our results showed that decreased level of cyclin D1 correlated with the fact of decreased level of S phase cells and increased G1 phase cells after CIP2A depletion, suggesting that CIP2A plays an important role in cell cycle control of ovarian cancer cells.
It is reported that overexpression of CIP2A up-regulated Akt and protected cells from bortezomib-induced apoptosis in hepatocellular carcinoma cells and in head and neck squamous cell carcinoma cells [7, 8]. We found that CIP2A knockdown facilitated paclitaxel-induced apoptosis of ovarian cancer cells. Bcl-2 prevents cells from undergoing apoptosis in response to a variety of stimuli and is thought to be involved in resistance to conventional cancer treatment. It is reported that AKT pathway activation modulates Bcl-2 expression [28]. In the present study, we found that CIP2A depletion decreased Bcl-2 protein and AKT phosphorylation. These results suggest that CIP2A regulates cell apoptosis via Bcl-2 and AKT activation.
In conclusion, CIP2A is overexpressed in ovarian cancer and correlated with tumor-grade and advanced FIGO stage. CIP2A depletion inhibits cell proliferation and facilitates paclitaxel-induced apoptosis. In addition, CIP2A depletion inhibits cyclin D1, c-Myc, phospho-Rb, Bcl-2, and phospho-AKT expression. These results provide an important point for elucidation of the functional importance of CIP2A in the progression of ovarian cancer. CIP2A might serve as a molecular target closely associated with cell proliferation and apoptosis for future development of ovarian cancer therapeutics.
References
Jemal A, Siegel R, Xu J, Ward E. Cancer statistics. CA Cancer J Clin. 2010;60(5):277–300.
Permuth-Wey J, Sellers TA. Epidemiology of ovarian cancer. Methods Mol Biol. 2009;472:413–37.
Darcy KM and Birrer MJ. 2010.Translational research in the Gynecologic Oncology Group: evaluation of ovarian cancer markers, profiles, and novel therapies. Gynecol Oncol. 117(3): p. 429–39.
Despierre E, Lambrechts D, Neven P, Amant F, Lambrechts S and Vergote I. 2010. The molecular genetic basis of ovarian cancer and its roadmap towards a better treatment. Gynecol Oncol. 117(2): p. 358-65
Junttila MR, Puustinen P, Niemela M, Ahola R, Arnold H, Bottzauw T, et al. CIP2A inhibits PP2A in human malignancies. Cell. 2007;130(1):51–62.
Tseng LM, Liu CY, Chang KC, Chu PY, Shiau CW and Chen KF. 2012. CIP2A is a target of bortezomib in human triple negative breast cancer cells. Breast Cancer Res. 14(2): p. R68.
Chen KF, Liu CY, Lin YC, Yu HC, Liu TH, Hou DR, et al. 2010. CIP2A mediates effects of bortezomib on phospho-Akt and apoptosis in hepatocellular carcinoma cells. Oncogene. 29(47): p. 6257-66.
Lin YC, Chen KC, Chen CC, Cheng AL and Chen KF. 2012. CIP2A-mediated Akt activation plays a role in bortezomib-induced apoptosis in head and neck squamous cell carcinoma cells. Oral Oncol 48(7):585-93
Come C, Laine A, Chanrion M, Edgren H, Mattila E, Liu X, et al. CIP2A is associated with human breast cancer aggressivity. Clin Cancer Res. 2009;15(16):5092–100.
Niemela M, Kauko O, Sihto H, Mpindi JP, Nicorici D, Pernila P, et al. 2012. CIP2A signature reveals the MYC dependency of CIP2A-regulated phenotypes and its clinical association with breast cancer subtypes. Oncogene (in press)
Li W, Ge Z, Liu C, Liu Z, Bjorkholm M, Jia J, et al. CIP2A is overexpressed in gastric cancer and its depletion leads to impaired clonogenicity, senescence, or differentiation of tumor cells. Clin Cancer Res. 2008;14(12):3722–8.
Dong QZ, Wang Y, Dong XJ, Li ZX, Tang ZP, Cui QZ, et al. 2011. CIP2A is overexpressed in non-small cell lung cancer and correlates with poor prognosis. Ann Surg Oncol. 18(3): p. 857-65.
Vaarala MH, Vaisanen MR and Ristimaki A. 2010. CIP2A expression is increased in prostate cancer. J Exp Clin Cancer Res. 29: p. 136.
Bockelman C, Koskensalo S, Hagstrom J, Lundin M, Ristimaki A and Haglund C. 2012. CIP2A overexpression is associated with c-Myc expression in colorectal cancer. Cancer Biol Ther. 13(5):289–95.
Ren J, Li W, Yan L, Jiao W, Tian S, Li D, et al. 2011. Expression of CIP2A in renal cell carcinomas correlates with tumour invasion, metastasis and patients' survival. Br J Cancer. 105(12): p. 1905-11.
Bockelman C, Lassus H, Hemmes A, Leminen A, Westermarck J, Haglund C, et al. 2011. Prognostic role of CIP2A expression in serous ovarian cancer. Br J Cancer. 105(7): p. 989-95.
Schwartz DR, Kardia SL, Shedden KA, Kuick R, Michailidis G, Taylor JM, et al. Gene expression in ovarian cancer reflects both morphology and biological behavior, distinguishing clear cell from other poor-prognosis ovarian carcinomas. Cancer Res. 2002;62(16):4722–9.
Zorn KK, Bonome T, Gangi L, Chandramouli GV, Awtrey CS, Gardner GJ, et al. Gene expression profiles of serous, endometrioid, and clear cell subtypes of ovarian and endometrial cancer. Clin Cancer Res. 2005;11(18):6422–30.
Kim A, Ueda Y, Naka T and Enomoto T. 2012. Therapeutic strategies in epithelial ovarian cancer. J Exp Clin Cancer Res. 31: p. 14.
Levanon K, Crum C, Drapkin R. New insights into the pathogenesis of serous ovarian cancer and its clinical impact. J Clin Oncol. 2008;26(32):5284–93.
Knudsen KE, Diehl JA, Haiman CA, Knudsen ES. Cyclin D1: polymorphism, aberrant splicing and cancer risk. Oncogene. 2006;25(11):1620–8.
Roy PG, Thompson AM. Cyclin D1 and breast cancer. Breast. 2006;15(6):718–27.
Ratschiller D, Heighway J, Gugger M, Kappeler A, Pirnia F, Schmid RA, et al. Cyclin D1 overexpression in bronchial epithelia of patients with lung cancer is associated with smoking and predicts survival. J Clin Oncol. 2003;21(11):2085–93.
Keum JS, Kong G, Yang SC, Shin DH, Park SS, Lee JH, et al. Cyclin D1 overexpression is an indicator of poor prognosis in resectable non-small cell lung cancer. Br J Cancer. 1999;81(1):127–32.
Bali A, O'Brien PM, Edwards LS, Sutherland RL, Hacker NF, Henshall SM. Cyclin D1, p53, and p21Waf1/Cip1 expression is predictive of poor clinical outcome in serous epithelial ovarian cancer. Clin Cancer Res. 2004;10(15):5168–77.
Masamha CP, Benbrook DM. Cyclin D1 degradation is sufficient to induce G1 cell cycle arrest despite constitutive expression of cyclin E2 in ovarian cancer cells. Cancer Res. 2009;69(16):6565–72.
Mateyak MK, Obaya AJ, Sedivy JM. c-Myc regulates cyclin D-Cdk4 and -Cdk6 activity but affects cell cycle progression at multiple independent points. Mol Cell Biol. 1999;19(7):4672–83.
Pugazhenthi S, Nesterova A, Sable C, Heidenreich KA, Boxer LM, Heasley LE, et al. Akt/protein kinase B up-regulates Bcl-2 expression through cAMP-response element-binding protein. J Biol Chem. 2000;275(15):10761–6.
Acknowledgment
This work was supported by grants from the National Natural Science Foundation of China (No. 30973191).
Conflicts of interest
None.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Fang, Y., Li, Z., Wang, X. et al. CIP2A is overexpressed in human ovarian cancer and regulates cell proliferation and apoptosis. Tumor Biol. 33, 2299–2306 (2012). https://doi.org/10.1007/s13277-012-0492-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13277-012-0492-2