Abstract
Myopodin is an actin-binding protein believed to play a tumor suppressor role in several solid neoplasias. We evaluated the potential differential myopodin methylation and expression and their clinical relevance in colon cancer. The epigenetic silencing of myopodin by hypermethylation was tested in colon cancer cells (n = 5) before and after azacitidine treatment. Myopodin methylation status was evaluated by methylation-specific PCR in colon cancer cells and colorectal tissues (n = 210) grouped in a training set (n = 62) and two independent validation series (n = 100 and n = 48) collected at independent clinical settings. Myopodin expression patterns were analyzed by immunohistochemistry on tissue arrays. Myopodin hypermethylation correlated with gene and protein expression loss, being increased in vitro by azacitidine. Myopodin was frequently methylated in colon cancer cells (four out of five). Methylation rates were 90.3%, 70.0%, and 47.8% in the training and validation sets, respectively. Myopodin methylation rendered a diagnostic accuracy of 83.9% (p < 0.0005). Cytoplasmic myopodin expression was significantly higher in non-neoplastic biopsies compared to colon tumors (p < 0.0005). Loss of myopodin expression correlated with increasing tumor stage (p = 0.011), methylation (p = 0.005), and poor overall survival (p = 0.003). In the first validation set (n = 100), myopodin methylation predicted disease-free (p = 0.046) and overall survival (p = 0.031). In the second validation cohort, myopodin methylation and protein expression patterns predicted disease-specific (p = 0.012 and p = 0.001, respectively) and overall survival (p = 0.009 and p = 0.043, respectively). Thus, myopodin was revealed to be epigenetically modified in colon cancer. The diagnostic and prognostic clinical utility of myopodin methylation and expression patterns suggest considering their assessment for the clinical management of colon cancer patients.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Colorectal cancer (CRC) is one of the most frequent and aggressive solid tumors [1, 2]. Colon tumors arise as a consequence of the accumulation of genetic (e.g., gene amplification or mutations) and epigenetic alterations (e.g., aberrant DNA methylation, chromatin modifications, or microRNAs), among others, in colonic epithelial cells during neoplastic transformation [3–5]. The advent of high-throughput approaches is accelerating the discovery of genetic and epigenetic cancer targets and biomarkers in human tumors [3–8], including colon cancer. Epigenetic modifications, particularly DNA methylation in selected gene promoters, are recognized as the most common epigenetic alterations in colon tumors [9, 10]. Transcriptional inactivation by promoter hypermethylation of critical cancer-related genes, including tumor suppressors [10], represents a frequent event in colon cancer [9–12]. Substantial efforts have been made to determine the role of aberrant DNA methylation in colon carcinogenesis and the biomarker utility of methylation patterns for the clinical management of colon cancer patients.
Myopodin is an actin-bundling protein redistributing between the nucleus and the cytoplasm in a differentiation-dependent and stress-induced manner [13–18]. Initial studies in uroepithelial neoplasias showed that loss of myopodin expression was useful for staging prostate and bladder tumors and predicting clinical outcome [19–21]. In vitro and in vivo studies suggested a tumor suppressor role of myopodin in uroepithelial cancer [19–24]. Myopodin was recently shown epigenetically silenced by hypermethylation in bladder cancer [25]. The methylation status of myopodin was clinically useful as a tumor stratification biomarker, clinical outcome prognosticator, and predictor of BCG response for bladder cancer patients [25, 26]. In this report, we evaluated whether myopodin expression and methylation patterns differed along colon cancer initiation and progression and their potential clinical relevance. The novelty of our findings is high because (a) myopodin was not previously described in colorectal malignancies and (b) both the differential expression and methylation myopodin patterns provided a diagnostic and prognostic utility for the clinical management of colon cancer patients.
Materials and methods
Tumor samples
Primary colon tissues were collected following the protection guidelines of human subjects at participating hospitals. Inclusion criteria of newly diagnosed colon cancer patients were based on the histopathologic information, covering from early to advanced stages. It was also required to have tissue material available for obtaining high-quality DNA for methylation analyses. An initial training set included paraffin blocks of benign intestine disease (n = 31) and colon tumors (n = 31) for which matching normal intestine was available. This set served to (a) screen methylation rates in matching pairs of normal and colon tumors and in benign intestinal disease, (b) evaluate the association of myopodin expression with its methylation status, and (c) evaluate the association of methylation and expression with clinicopathologic variables. Demographic information of colon cancer patients indicated the presence of 23 males and 8 females, with a median age of 68.2 years (range, 38.0–92.0 years). In control individuals without colon cancer, there were 24 males and 7 females, with a median age of 61.6 years (range, 40.0–82.0 years). Two independent validation sets were analyzed. The first validation cohort included a series of 100 Dukes B frozen colon tumors. This set served to (a) validate methylation rates and (b) validate the association of methylation with clinicopathologic variables and prognostic analyses. Demographic information of colon cancer patients indicated the presence of 54 males and 46 females, with a median age of 72.3 years (range, 35.0–92.0 years).The second validation cohort included a series of 48 paraffin blocks. This set served to (a) validate methylation rates, (b) validate the association of myopodin expression with its methylation status, and (c) validate the association of methylation and expression with clinicopathologic variables and prognostic analyses. Histopathologic staging of colon tumors was defined under standard criteria [27, 28] after subsequent surgical interventions. Demographic information of colon cancer patients indicated the presence of 27 males and 21 females, with a median age of 69.0 years (range, 36.0–84.0 years). Distribution of clinical variables of all series is provided in Table 1.
Methylation analyses of the promoter of myopodin
The methylation status of myopodin (Fig. 1a) was analyzed by two PCR analysis strategies of bisulfite-modified genomic DNA, which induces chemical conversion of unmethylated, but not methylated, cytosine to uracil. First, genomic sequencing of both DNA strands of myopodin was analyzed after bisulfite treatment of at least three clones each of the cancer cell lines analyzed [25]. Confirmation on at least two independent clones was required to assign methylation sites. A second strategy used PCR with primers specific for either the methylated or the modified unmethylated DNA [methylation-specific PCR (MS-PCR)] [25]. Primer sequences for bisulfite sequencing and unmethylated and methylated reactions were designed encompassing its transcription start site distant at 322 bp before its ATG starting codon, as previously reported [25]. OCT and paraffin-embedded tissue material were macrodissected based on hematoxylin–eosin evaluations to ensure a minimum of 75% of tumor cells before DNA extraction [25, 26]. Genomic DNA was extracted using standard methods. DNA from normal lymphocytes (NL) treated in vitro with SssI methyltransferase was used as a positive control for methylated alleles. DNA from NL was used as a positive control for unmethylated alleles. PCR products were loaded onto nondenaturing 2% agarose gels, stained with Gel Stain (Lonza, Rockland, ME, USA), and visualized under a UV transilluminator.
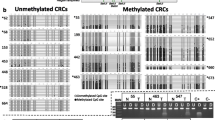
Identification of myopodin methylation in colon cancer cell lines. a Schematic depiction of myopodin CpG islands around its transcription start site. Nucleotide sequences of the CpG island region analyzed by bisulfite sequencing, highlighting in yellow and red the sequencing primers and underlining the primers utilized for MS-PCR. b Analysis of CpG islands methylation status of the promoter of myopodin by bisulfite genomic sequencing in colon human cancer cell lines. CpG dinucleotides were represented in squares. The presence of “Cs” in the dinucleotide CpG reflects methylated cytosines (black squares), while the presence of “Ts” in the dinucleotide CpG reflects unmethylated cytosines (white squares). The presence of methylation was confirmed in at least three of the clones that were sequenced for the cell lines under analyses
RNA and protein analysis of myopodin in colon cancer cell lines
Human colon cancer cell lines (n = 5) were treated with 1 and 5 μmol/L azacitidine (AZA; Sigma, St Louis, MO, USA) for 72 h to achieve demethylation [25]. RNA was isolated using standard methods. RNA (1 μg) was reverse transcribed using AMV reverse transcriptase (Promega, Fitchburg, MI, USA) and amplified using specific primers and conditions for myopodin, as previously reported [25]. PCR was done using a final volume of 15 μL containing 1× PCR Ecostart buffer (Ecogen, Truro, England), 1.5 mmol/L MgCl2, 0.2 mmol/L deoxynucleotide triphosphate, 0.25 μmol/L of each primer, 1.5 U Ecostart Taq polymerase (Ecogen, Truro, England), and 0.4 μg cDNA as template. Glyceraldehyde 3-phosphate dehydrogenase was used as internal control to ensure cDNA quality and loading accuracy. The amplification product was resolved by 2% agarose gel electrophoresis and visualized by ethidium bromide staining. Cell lysates for protein analysis were analyzed by Western blotting using an antimyopodin antibody (rabbit polyclonal; Sigma, St Louis, MO, USA; 1:500 dilutions). Equal loading was tested by reprobing with an α-tubulin antibody (mouse monoclonal; Sigma, St Louis, MO, USA; 1:4,000 dilutions).
Tissue microarrays
Two tissue microarrays were constructed at the Spanish National Cancer Center and used in this study, including paraffin tissues belonging to patients and controls recruited under institutional review board-approved protocols at collaborating institutions. Five-micrometer sections of paraffin-embedded tissues were stained with hematoxylin and eosin to identify viable, morphologically representative areas of the specimen from which needle core samples were taken, using a precision instrument (Beecher Instruments, Silver Spring, MD, USA). From each specimen, triplicate cores with diameters of 1.0 mm were punched and arrayed on the recipient paraffin block. Five-micrometer sections of these tissue array blocks were cut and placed on charged polylysine-coated slides and used for immunohistochemistry analysis. The two tissue microarrays included the training set (n = 62) and the second validation set (n = 48) specimens. Clinicopathologic and annotated follow-up information allowed the evaluation of associations of myopodin methylation and protein expression patterns among them and with clinicopathologic variables.
Immunohistochemistry
Protein expression patterns of myopodin were assessed at the microanatomic level by immunohistochemistry on these tissue microarrays following standard avidin–biotin immunoperoxidase procedures [25]. Antigen retrieval methods (0.01% citric acid for 15 min under microwave treatment) were utilized prior to incubation overnight at 4°C with the primary antibody mentioned above at 1:1,500 dilutions. The biotinylated secondary antibody (Vector Laboratories, Burlingame, CA, USA) was used at 1:1,000 dilutions. The absence of primary antibody was used as negative control, while kidney and muscle were used as positive controls. Diaminobenzidine was utilized as the final chromogen and hematoxylin as the nuclear counterstain [25].
Serum tumor markers
Serum was extracted preoperatively to evaluate the diagnostic and prognostic value of CEA by electrochemiluminescence immunoassay using the Cobas® 6000 autoanalyzer (Elecsys Cobas e601; Roche Diagnostics, Indianapolis, IN, USA).
Statistical analysis
The consensus value of the scores of two independent observers of the three representative cores from each tumor sample arrayed was used for statistical analyses. Associations among myopodin methylation and protein expression patterns of myopodin with clinicopathological variables were evaluated using nonparametric Wilcoxon–Mann–Whitney and Kruskal–Wallis tests [29]. Myopodin expression was evaluated as a continuous variable based on the number of cells expressing the protein in the cytoplasm. The intensity of the staining was categorized from negative (−) to low (+), intermediate (++), and high (+++). The diagnostic performance of myopodin methylation measured by MS-PCR in tissue specimens was evaluated through the estimations of its sensitivity, specificity, and diagnostic accuracy. Sensitivity was defined by the number of tumors methylated for myopodin among cases with confirmed colon cancer by histopathology. Specificity was defined by the number of tissue specimens unmethylated for myopodin among tissues belonging to controls without colon cancer histopathologically confirmed. Diagnostic accuracy was defined by the number of correctly classified cases and controls based on the area observed under the curve using receiver operative curve (ROC) analyses, evaluating the 95% confidence interval (95% CI) and statistical significance [29]. Associations of methylation and protein expression patterns with clinical outcome were evaluated in those cases for which follow-up information were available using the log-rank test [29]. Expression cutoffs for prognostic evaluation were selected based on the median values of expression among the groups under analyses. Disease-specific survival time was defined as the months elapsed between surgery and death as a result of disease (or the last follow-up date). Overall survival time was defined as the months elapsed between surgery and death (or the last follow-up date). Patients who were alive at the last follow-up or lost to follow-up were censored. Survival curves were plotted using the standard Kaplan–Meier methodology [29]. Statistical analyses were performed using the SPSS statistical package (version 17.0).
Results
Myopodin is frequently methylated in colon cancer cells and epigenetically silenced in vitro
Myopodin was initially tested to be a candidate gene for hypermethylation in colon cancer cells by means of bisulfite genomic sequencing and MS-PCR targeted to the areas surrounding its transcription start site (Fig. 1a). Bisulfite sequencing revealed the myopodin methylation for four (80.0%) of the five colon cancer cell lines analyzed (Fig. 1b). Among the normal tissues analyzed, normal intestine and NL were found unmethylated at the myopodin CpG island promoter. Myopodin methylation patterns observed by sequencing highly confirmed the methylation found by MS-PCR (Fig. 2a).
CpG island methylation was associated with myopodin silencing. a MS-PCRs for myopodin in human colon cancer cell lines. The presence of a PCR band under the lane M indicates a methylated myopodin, while the presence of a PCR band under the lane U indicates an unmethylated gene. Normal lymphocytes (NL) and in vitro methylated DNA (IVD) were used as negative and positive controls for unmethylated and methylated myopodin, respectively. b The treatment with the demethylating agent AZA reactivated the transcript expression of myopodin. Reverse transcription polymerase chain reaction analysis of myopodin expression. GAPDH expression is shown as transcript loading control. Myopodin transcript expression increased in the methylated cell lines SW620, SW480, and HCT15 after AZA exposure. The unmethylated cell line (RKO) did not show changes in myopodin expression. c The treatment with the demethylating agent AZA reactivated the protein expression of myopodin. Immunoblotting of myopodin protein expression in colon cancer cells after AZA exposure. Tubulin expression is shown as loading control. Myopodin protein expression increased in the methylated cell lines SW620, SW480, and HCT15 after AZA exposure. The unmethylated cell line (RKO) did not show changes in myopodin expression
A further link between hypermethylation and gene silencing was established by the treatment of methylated and unmethylated colon cancer cell lines with a DNA demethylating agent. Exposure of the methylated cell lines to the demethylating drug, 5-AZA-2′-deoxycytidine (AZA), increased the expression of myopodin at the transcript level (Fig. 2b). Immunoblotting analyses were performed to further confirm that myopodin protein expression was also increased after AZA exposure (Fig. 2c). RKO was used as a control cell line to assess the specificity of AZA exposure not to modify myopodin expression in unmethylated colon cancer cells. Overall, AZA reactivation analyses indicated a high correlation of increasing transcript and protein expression estimates and myopodin methylation status.
Myopodin is frequently hypermethylated in primary colon tumors and hypermethylation segregated colon cancer patients from controls
Once the functional consequences of myopodin hypermethylation were determined in vitro, we evaluated its impact in human tissue material by MS-PCR (Fig. 3a). First of all, we tested whether myopodin hypermethylation was a cancer-specific event in colon cancer. Comparison of methylation of 10 colon tumors and their respective pairs of normal intestine revealed that myopodin methylation was found in 80.0% of the colon tumors and in 10% of the matching normal intestine analyzed by MS-PCR. Initial methylation screening was performed in DNA from paraffin-embedded material belonging to the training set of benign intestine diseases (n = 31) and primary colon tumors (n = 31). Myopodin sensitivity was 90.3% in colon cancer patients with a specificity of 74.2% in benign intestinal diseases. ROC analyses provided a global diagnostic accuracy of 83.9%, as shown by the area under the curve (AUC = 0.839, 95% CI = 0.720–0.958, p ≤ 0.0005). Importantly, myopodin methylation showed a higher diagnostic accuracy than preoperative serum CEA levels measured in these patients (Fig. 3b; AUC = 0.563, 95% CI = 0.416–0.711, p = 0.404). These findings supported a diagnostic role for the methylation of myopodin by means of MS-PCR to discriminate between colon cancer patients and controls with higher diagnostic accuracy than current serum biomarkers.
Myopodin hypermethylation plays a role as a diagnostic and prognostic biomarker in colon cancer. a Representative MS-PCR analyses for myopodin using primary colon tumors. A subset of these tumors was shown to be unmethylated and the majority of them were methylated. Normal lymphocytes (NL) and in vitro methylated DNA (IVD) were used as negative and positive control for unmethylated and methylated myopodin, respectively. b Training set: diagnostic role. ROC of myopodin methylation as a detection biomarker for colon cancer on a series of 61 tissue specimens collected during colonoscopy or intestinal open surgery. Among these, 31 had colon tumors. The AUC obtained was 0.839 (95% CI, 0.720–0.958; p < 0.0005). c First validation set: disease-specific survival prognosis. Kaplan–Meier curve survival analysis indicating that tumors with a methylated myopodin had poorer disease-specific survival than those with unmethylated gene (log-rank, p = 0.046). d First validation set: overall survival prognosis. Kaplan–Meier curve survival analysis indicating that tumors with a methylated myopodin had shorter overall survival than those with unmethylated gene (log-rank, p = 0.031). e Second validation set: disease-specific survival prognosis. Kaplan–Meier curve survival analysis indicating that tumors with a methylated myopodin had poorer disease-specific survival than those with unmethylated gene (log-rank, p = 0.012). f Second validation set: overall survival prognosis. Kaplan–Meier curve survival analysis indicating that tumors with a methylated myopodin had shorter overall survival than those with unmethylated gene (log-rank, p = 0.009)
Myopodin methylation is correlated with poor clinical outcome
The methylation status of myopodin was then linked to clinicopathologic variables (Table 1). The limited number of cases analyzed did not allow reaching statistically significant associations between methylation and histopathological variables in the training set. However, it was observed that those cases with adenomas who developed adenocarcinomas showed higher methylation rates than those free of disease after 3 years follow-up (Mann–Whitney, p = 0.014). In this initial small subset of tumor cases (n = 31), patients with methylation for myopodin showed poorer disease-specific survival, but this trend did not reach statistically significant associations (log-rank, p = 0.132). These findings prompted us to perform further MS-PCR analyses on two independent validation cohorts of colon tumors with available follow-up. In the first verification cohort of Dukes B tumors, from which genomic DNA was extracted from frozen material (n = 100), myopodin methylation was also shown to be a frequent event (73.0%). Importantly, myopodin methylation significantly correlated with a poorer disease-specific survival (log-rank, p = 0.046; Fig. 3c) and poorer overall survival (log-rank, p = 0.031; Fig. 3d). In a second independent validation set from another clinical institution consisting of 48 paraffin-embedded tumors, the myopodin methylation rate was 47.8%, which correlated with poor disease-specific survival (log-rank, p = 0.012; Fig. 3e) and overall survival (log-rank, p = 0.009; Fig. 3f). Overall, these results indicated that myopodin hypermethylation was a frequent event in colon cancer with prognostic clinical utility for patients affected with colon cancer.
Loss of cytoplasmic myopodin protein expression patterns correlated with methylation, tumor progression, and clinical outcome in colon cancer patients
The next set of analyses dealt with the evaluation of the protein expression patterns of myopodin in colon tissues by immunohistochemistry in the tissue arrays constructed for this study. The goals were (a) to evaluate the differential expression of myopodin in colon cancer, (b) to link myopodin methylation with the silencing of the gene in human material, and (c) to evaluate the association of the expression patterns with clinicopathological variables. Protein expression of myopodin by immunohistochemistry was mainly observed in the cytoplasm. Certain tumors also showed staining in the nucleus and membrane reinforcement. In the array containing the training set with those patients with benign intestine diseases and colon tumors (n = 62), high cytoplasmic myopodin protein expression was found in normal intestine, patients with benign intestine disease, and in early stage adenocarcinomas compared with advanced colon tumors (Fig. 4a, b) (Mann–Whitney, p ≤ 0.0005). Tumors methylated for myopodin had lower cytoplasmic protein expression than unmethylated cases (Mann–Whitney, p = 0.005). The loss of cytoplasmic myopodin protein expression significantly correlated with increasing tumor stage (Kruskal–Wallis, p = 0.011). In colon cancer patients with available follow-up, the presence of a cytoplasmic protein expression of myopodin lower than 60% significantly correlated with shorter overall survival (log-rank, p = 0.003; Fig. 4c). In the array containing the second validation set, low cytoplasmic protein expression also correlated with myopodin methylation (p = 0.038) and tumor stage (p = 0.019). Tumors with low cytoplasmic intensity showed shorter disease-specific survival (log-rank, p = 0.001; Fig. 4d), and those with a cytoplasmic protein expression of myopodin lower than 60% showed shorter overall survival (log-rank, p = 0.043; Fig. 4e). Overall, these analyses revealed that the loss of myopodin cytoplasmic protein correlated with methylation, tumor stage, and poor clinical outcome for colon cancer patients in two independent cohorts. Thus, myopodin methylation and its cytoplasmic expression could be considered likely prognosticators of tumor progression and poor outcome in patients with colon cancer.
Protein expression patterns of myopodin correlated with tumor progression and clinical outcome in colon cancer. a and b Representative immunostainings of the differential protein expression patterns of myopodin at the tissue level by immunohistochemistry on tissue arrays in a normal intestinal epithelium and proliferative adenomatous polyps and b colon tumors. As previously reported [13, 21], myopodin showed positive staining in muscle stromal cells, which served as positive control for immunohistochemistry. c Training set: overall survival. Kaplan–Meier curve survival analysis indicating that cytoplasmic protein expression of myopodin in <60% of cancer cells measured by immunohistochemistry on tissue arrays was associated with poor overall survival (log-rank, p = 0.003). d Validation set: disease-specific survival. Kaplan–Meier curve survival analysis indicating that patients with low intensity in the cytoplasmic protein expression of myopodin in cancer cells measured by immunohistochemistry on tissue arrays was associated with poor disease-specific survival (log-rank, p = 0.001). The intensity of the staining was categorized from negative (−) to low (+), intermediate (++), and high (+++). e Validation set: overall survival. Kaplan–Meier curve survival analysis indicating that cytoplasmic protein expression of myopodin in <60% of cancer cells measured by immunohistochemistry on tissue arrays was associated with poor overall survival (log-rank, p = 0.043)
Discussion
This study identified the differential expression and the epigenetic silencing of myopodin in colon cancer. Importantly, myopodin methylation and its expression were clinically relevant for the diagnostic and prognostic assessment for patients affected with colon tumors. The consequences of the CpG island hypermethylation of myopodin along colon cancer initiation and progression require to be assessed from the standpoint of mechanistic and translational implications. Mechanistically, it is important to link the promoter methylation status to expression estimates of myopodin. AZA exposure experiments confirmed the effect of methylation in myopodin expression by restoring myopodin transcript and protein expression, specifically in methylated colon cancer cells. In concordance, myopodin cytoplasmic expression was shown to be significantly lower in methylated than in unmethylated colon tumors in two independent human tissue cohorts. Thus, methylation correlated with loss of myopodin expression also in human clinical material. Overall, in vitro and in vivo human analyses revealed that myopodin was aberrantly silenced by CpG island promoter hypermethylation.
Colon cancer represents the third human neoplasia displaying loss of myopodin expression. In prostate cancer, losses of myopodin protein expression correlated with aggressive prostate cancer using in vitro, in vivo, and clinical material and were explained by chromosomal deletions at 4q26 [19]. In bladder tumors, the loss of myopodin expression correlated with increased tumor staging and poor clinical outcome and was attributed to epigenetic silencing by hypermethylation [20]. The epigenetic silencing identified in colon cancer is consistent with the mechanism by which myopodin is lost in bladder progression [25]. In this report, the impact and clinical relevance of myopodin methylation along colon cancer development was evaluated using in vitro strategies and on tissue specimens. Our results revealed that myopodin was hypermethylated and potentially useful as a diagnostic and tumor stratification biomarker and clinical outcome prognosticator for patients with colon cancer.
The translational implications of the discovery of myopodin methylation in colon cancer have strongly been addressed in this work. The association of myopodin with colon cancer progression can be justified as follows. The cancer specificity evaluation by MS-PCR in pairs of colon tumors and matching normal intestinal epithelium related myopodin methylation to colon cancer progression at initial tumorigenic steps. Second, it was shown that myopodin methylation was a frequent event in three independent series of tumors, regardless of the source of genomic DNA from frozen or paraffin-embedded colon tissue material. Third, myopodin methylation significantly discriminated colon cancer from benign intestinal disease. Fourth, it correlated with tumor staging. Furthermore, it added prognostic information of clinical outcome. In addition to the clinicopathologic stratification of colon cancer patients, a relevant translational point relates to treatment. In this new scenario, myopodin represents a potential therapeutic target whose expression could be potentially reactivated by demethylating drugs.
A critical step in the clinical evaluation of myopodin along colon cancer progression deals with the analyses of myopodin protein expression patterns by immunohistochemistry on colon tumors of known myopodin methylation status. The loss of myopodin cytoplasmic protein expression was correlated with increasing methylation rates and tumor stage. Interestingly, low cytoplasmic myopodin protein levels were correlated with poor clinical outcome under different clinical endpoints, suggesting its potential role as a prognostic marker for the clinical management of colon cancer patients. Thus, the combination of epigenetic and protein analyses revealed that myopodin is differentially expressed along colon cancer progression in association with clinicopathologic variables and clinical outcome. The epigenetic silencing of myopodin might aid in understanding as to how it may contribute to tumorigenesis and colon cancer progression. Future studies are warranted to dissect such specific mechanisms in the context of colon cancer and other solid human neoplasias.
The clinical diagnostic utility of myopodin methylation patterns in tissue specimens represents a relevant finding. Myopodin methylation discriminated colon cancer patients from benign intestinal disease with higher diagnostic accuracy than preoperative CEA serum levels, which is recognized as the serum reference diagnostic biomarker for colon cancer [30]. Assessing myopodin methylation represents a potential alternative adjunct for the early detection of patients under suspicion of colon cancer and potentially for the follow-up of colon cancer patients.
It is important to be aware that three isoforms (Myo1–3), translated into full-length proteins of 1,093, 1,109, and 1,261 amino acids, have been described for myopodin [31]. We have observed that the antibody we utilized for immunoblotting and immunohistochemistry recognizes the longest isoform 3 of myopodin when overexpressed in bladder cancer cells (unpublished data). Further investigation is warranted to address whether methylation of myopodin promoter impacts in a different manner to the expression of each isoform, with specific antibodies against each of them. Moreover, it is important to mention that, in addition to the CpG islands encompassing its transcription start site distant at 322 bp before its ATG starting codon analyzed in this report [25, 26], other CpG islands either in the promoter region or other gene shores could also be involved in the epigenetic regulation of myopodin expression.
In summary, our study revealed that myopodin is differentially expressed in colon cancer. It provided a mechanistic explanation for the observed loss of myopodin cytoplasmic expression in colorectal malignancies by epigenetic silencing. Hypermethylation emerged as a strong indicator of tumor aggressiveness for colon cancer patients. The loss of cytoplasmic myopodin protein expression also stratified colon tumors histopathologically and predicted clinical outcome. Interestingly, myopodin methylation in tissue specimens played a higher diagnostic accuracy than CEA for colon cancer. These observations support introducing myopodin assessment for the stratification and clinical management of colon cancer patients.
References
Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90.
Cunningham D, Atkin W, Lenz HJ, Lynch HT, Minsky B, Nordlinger B, Starling N. Colorectal cancer. Lancet. 2010;375:1030–47.
Grady WM, Carethers JM. Genomic and epigenetic instability in colorectal cancer pathogenesis. Gastroenterology. 2008;135:1079–99.
Carmona FJ, Esteller M. Epigenetics of human colon cancer. Mutat Res. 2010;693:53–60.
Rashid A, Issa JP. CpG island methylation in gastroenterologic neoplasia: a maturing field. Gastroenterology. 2004;127:1578–88.
Sánchez-Carbayo M, Cordon-Cardo C. Applications of array technology: identification of molecular targets in bladder cancer. Br J Cancer. 2003;89:2172–7.
Jordà M, Peinado MA. Methods for DNA methylation analysis and applications in colon cancer. Mutat Res. 2010;693:84–93.
Sánchez-Carbayo M. Use of high-throughput DNA microarrays to identify biomarkers for bladder cancer. Clin Chem. 2003;49:23–31.
Herman JG, Baylin SB. Gene silencing in cancer in association with promoter hypermethylation. N Engl J Med. 2003;349:2042–54.
Esteller M. CpG island hypermethylation and tumor suppressor genes: a booming present, a brighter future. Oncogene. 2002;21:5427–40.
Kim MS, Lee J, Sidransky D. DNA methylation markers in colorectal cancer. Cancer Metastasis Rev. 2010;29:181–206.
Cheng YW, Pincas H, Bacolod MD, Schemmann G, Giardina SF, Huang J, Barral S, Idrees K, Khan SA, Zeng Z, Rosenberg S, Notterman DA, Ott J, Paty P, Barany F. CpG island methylator phenotype associates with low-degree chromosomal abnormalities in colorectal cancer. Clin Cancer Res. 2008;14:6005–13.
Weins A, Schwarz K, Faul C, Barisoni L, Linke WA, Mundel P. Differentiation- and stress-dependent nuclear cytoplasmic redistribution of myopodin, a novel actin-bundling protein. J Cell Biol. 2001;155:393–404.
Van Impe K, De Corte V, Eichinger L, Bruyneel E, Mareel M, Vandekerckhove J, Gettemans J. The Nucleo-cytoplasmic actin-binding protein CapG lacks a nuclear export sequence present in structurally related proteins. J Biol Chem. 2003;278:17945–52.
Faul C, Hüttelmaier S, Oh J, Hachet V, Singer RH, Mundel P. Promotion of import alpha-mediated nuclear import by the phosphorylation-dependent binding of cargo protein to 14-3-3. J Cell Biol. 2005;169:415–24.
De Ganck A, Hubert T, Van Impe K, Geelen D, Vandekerckhove J, De Corte V, Gettemans J. A monopartite nuclear localization sequence regulates nuclear targeting of the actin binding protein myopodin. FEBS Lett. 2005;579:6673–80.
Faul C, Dhume A, Schecter AD, Mundel P. Protein kinase A, Ca2+/calmodulin-dependent kinase II, and calcineurin regulate the intracellular trafficking of myopodin between the Z-disc and the nucleus of cardiac myocytes. Mol Cell Biol. 2007;27:8215–27.
Liang J, Ke G, You W, Peng Z, Lan J, Kalesse M, Tartakoff AM, Kaplan F, Tao T. Interaction between importin 13 and myopodin suggests a nuclear import pathway for myopodin. Mol Cell Biochem. 2008;307:93–100.
Lin F, Yu YP, Woods J, Cieply K, Gooding B, Finkelstein P, Dhir R, Krill D, Becich MJ, Michalopoulos G, Finkelstein S, Luo JH. Myopodin, a synaptopodin homologue, is frequently deleted in invasive prostate cancers. Am J Pathol. 2001;159:1603–12.
Yu YP, Tseng GC, Luo JH. Inactivation of myopodin expression associated with prostate cancer relapse. Urology. 2006;68:578–82.
Sanchez-Carbayo M, Schwarz K, Charytonowicz E, Cordon-Cardo C, Mundel P. Tumor suppressor role for myopodin in bladder cancer: loss of nuclear expression of myopodin is cell-cycle dependent and predicts clinical outcome. Oncogene. 2003;22:5298–305.
Jing L, Liu L, Yu YP, Dhir R, Acquafondada M, Landsittel D, Cieply K, Wells A, Luo JH. Expression of myopodin induces suppression of tumor growth and metastasis. Am J Pathol. 2004;164:1799–806.
Yu YP, Luo JH. Myopodin-mediated suppression of prostate cancer cell migration involves interaction with zyxin. Cancer Res. 2006;66:7414–9.
Yu YP, Luo JH. Phosphorylation and interaction of myopodin by integrin-link kinase lead to suppression of cell growth and motility in prostate cancer cells. Oncogene. 2011;30:4855–63.
Cebrian V, Alvarez M, Aleman A, Palou J, Bellmunt J, Gonzalez-Peramato P, Cordón-Cardo C, García J, Piulats JM, Sánchez-Carbayo M. Discovery of myopodin methylation in bladder cancer. J Pathol. 2008;216:111–9.
Alvarez-Múgica M, Cebrian V, Fernández-Gómez JM, Fresno F, Escaf S, Sánchez-Carbayo M. Myopodin methylation is associated with clinical outcome in patients with T1G3 bladder cancer. J Urol. 2010;184:1507–13.
Ogino S, Chan AT, Fuchs CS, Giovannucci E. Molecular pathological epidemiology of colorectal neoplasia: an emerging transdisciplinary and interdisciplinary field. Gut. 2011;60:397–411.
Puppa G, Sonzogni A, Colombari R, Pelosi G. TNM staging system of colorectal carcinoma: a critical appraisal of challenging issues. Arch Patol Lab Med. 2010;134:837–52.
Dawson-Saunders B, Trapp RG. Basic & clinical biostatistics. 2nd ed. Norwalk: Appleton & Lange; 1994.
Duffy MJ, van Dalen A, Haglund C, Hansson L, Holinski-Feder E, Klapdor R, Lamerz R, Peltomaki P, Sturgeon C, Topolcan O. Tumour markers in colorectal cancer: European Group on Tumour Markers (EGTM) guidelines for clinical use. Eur J Cancer. 2007;43:1348–60.
De Ganck A, De Corte V, Staes A, Gevaert K, Vandekerckhove J, Gettemans J. Multiple isoforms of the tumor suppressor myopodin are simultaneously transcribed in cancer cells. Biochem Biophys Res Commun. 2008;370:269–73.
Acknowledgments
We thank all members of the laboratory of M. Sánchez-Carbayo for the technical support and constructive suggestions in the preparation of this manuscript. We also would like to thank the Tumor Bank belonging to the Molecular Pathology Program at the Spanish National Cancer Center and all the members of our clinical collaborators at the different institutions involved in this study for the support in facilitating the tumor specimens as well as the clinical follow-up of the colon cancer cases analyzed in this study.
Grant support
Spanish Ministry of Science and Innovation grant SAF2009-13035 and Mutua Madrileña 2010 (to M. Sánchez-Carbayo).
Conflicts of interest
None.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Esteban, S., Moya, P., Fernandez-Suarez, A. et al. Diagnostic and prognostic utility of methylation and protein expression patterns of myopodin in colon cancer. Tumor Biol. 33, 337–346 (2012). https://doi.org/10.1007/s13277-012-0320-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13277-012-0320-8