Abstract
Background
Breast cancer (BC) is a common malignancy with a high mortality rate. Malignant cell transformation is associated with metabolic changes. One group of proteins that are affected is the monocarboxylate transporters (MCTs-SLC16A). The MCTs comprise 14 members, and they play an important role in the growth, proliferation, and metabolism of cancer cells by transporting monocarboxylates such as lactate, pyruvate and thyroid hormones.
Objective
We aimed to evaluate the expression of MCT3 (SLC16A8), MCT8 (SLC16A2) and MCT9 (SLC16A9) genes in breast cancer samples, comparing to normal adjacent tissues.
Methods
Forty paired breast cancer tumor samples, the adjacent non-tumor and five healthy tissues were collected. Three cancer cell lines (MCF-7, MDA-MB-231, and SKBR3) were also analyzed. The expression of SLC16A8, SLC16A2 and SLC16A9 were assessed using quantitative real-time PCR. The relationship between gene expression with the pathological features of the tumors, and the hormone receptors status of the patient’s tumors were also analyzed.
Results
There was a significantly lower expression of the MCT3 gene in tumor samples compared to adjacent normal tissue and healthy samples (p value < 0.05). There was a significant difference in the expression of all three candidate genes between the BC tissues and normal tissues, and for the, tissues with different hormone receptor status and the molecular subtypes. Altered MCT8 and MCT9 gene expression was associated with a reduced survival
Conclusion
MCT3 expression is significantly downregulated in breast cancer tissue. MCT3 may represent a novel therapeutic target in breast cancer patients, or in some hormone receptor subgroups.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Breast cancer (BC) is the second most common global cause of cancer-related mortality after lung cancer (Estiar et al. 2017; Harding et al. 2015; Moslemi et al. 2020). The mortality remains high because of the lack of effective treatments (Bogenrieder and Herlyn 2003; Harding et al. 2015; Moslemi et al. 2021; Si et al. 2015; Weigelt et al. 2005). According to the molecular classification and microarray analysis of the patient's tumors, breast cancer may be categorized into five subgroups (Liu et al. 2014; Yersal and Barutca 2014): luminal A [estrogen receptor (ER) positive, progesterone receptor (halestrap and price) positive, and human epidermal growth factor receptor 2 (HER2) negative], luminal B [ER+, PR+, and HER2low]. HER2 over-expression (HER2high, ERlow, and PRlow). Basal-like usually is Triple-negative (ER−, PR−, and HER2− basal marker) with the shortest survival. Normal breast tissue is ERlow, PRlow, and P53+ (Liu et al. 2014; Yersal and Barutca 2014).
Several factors have been reported to be risk factors for the development of some cancers, including: metabolic disorders of glucose and lactate, hormonal disorders, smoking, physical inactivity, exposure to radiation, obesity, dietary factors, alcohol, age and mutagenic substances (Stein and Colditz 2004). In cancer cells there is increased glycolysis leading to increased tissue levels of lactate, in both the absence of oxygen (anaerobic glycolysis) and in its presence (aerobic glycolysis or the Warburg effect) and this is associated with tumor progression (Draoui and Feron 2011). Increased glycolysis leads to an increased production of lactate and acidification of the tumor microenvironment (Aveseh et al. 2015), but whilst there is a reduction in extracellular pH, the intracellular pH is often normal or even in some conditions may be alkaline; this is due to expression and function of a group of membrane-proteins that called the monocarboxylate transporters (MCTs-SLC16A) and lactate dehydrogenases (LDHs) (Draoui and Feron 2011; Luz et al. 2017; Mishra and Banerjee 2019; Pinheiro et al. 2008). The MCTs family of proteins comprises 14 members, which mainly function as transporter proteins and transport monocarboxylates (lactate, pyruvate, and ketones), thyroid hormones, and carnitine (Halestrap and Meredith 2004; Halestrap and Price 1999; Van Der Deure et al. 2010).
An increased tissue expression of some MCTs has been demonstrated in breast cancer compared to normal breast tissue: for example, MCT1 is reported to be up-regulated in the basal (Pinheiro et al. 2010), luminal-like subtypes of breast cancer and the MCF7 cell line and also MCT4 is up-regulated in MDA-MB-231 cell line (Gallagher et al. 2007). MCT1 knockdown was found to reduce metastasis and angiogenesis of cancer cells (Hussien and Brooks 2010; Sonveaux et al. 2008; Végran et al. 2011). MCT1 and MCT4, two members of this family, have been extensively studied in various cancers and have been proposed as therapeutic targets for cancer, but the activity and expression of other MCTs in breast cancer have not been extensively studied (Luz et al. 2017; Pinheiro et al. 2012; Van Der Deure et al. 2010). These studies indicate the importance of this genes in cancer cell proliferation, metastasis, and since the expression of 3 members of this family MCT3, MCT8 and MCT9 have not been examined in breast cancer, we focused on these in this current study.
MCT3 protein is involved in the transport of ketones, pyruvate, and lactate and is expressed in most tissues. The gene for MCT3 (SCL16A8) is located on the long arm of chromosome 17 (Pérez-Escuredo et al. 2016). The MCT8 protein was discovered in 1994 and is encoded by a gene (SLC16A2) on the long arm of the X chromosome (Halestrap and Meredith 2004). It binds to thyroid hormone in mice and humans and whilst homologous to MCT10 (Friesema et al. 2003), does not play a role in the transport of lactate, aromatic amino acids and sulfonated iodothyronines (Van Der Deure et al. 2010). Decreased expression level of MCT8 in thyroid cancer has been suggested MCT8 as a marker of thyroid differentiation. It is also expressed in the brain, heart, kidney, liver, and testis (Badziong et al. 2017). Mutation in this gene results in the Allan–Herndon–Dudley syndrome (AHD) (Schwartz and Stevenson 2007). The MCT9 protein plays a role in the transport of substances such as carnitine and is expressed in most tissues (Halestrap and Meredith 2004; Pérez-Escuredo et al. 2016). It is encoded by a gene (SCL16A9) located on the long arm of chromosome 10 (Halestrap and Meredith 2004; Halestrap and Price 1999).
In this study, we evaluated the mRNA expression of these genes in breast cancer samples using the real-time PCR technique.
Materials and methods
Study population
Breast tumor tissues (n = 40), normal adjacent tissues (n = 40) and 5 healthy breast tissue samples were collected from the 2018 to 2020 from the Rasoul Akram Hospital, Tehran, Iran. All patients were identified to have breast cancer, based on the Laboratory and histological criteria, and have given informed written consent before surgery. The study was approved by the Ethics Committee of Iran University of Medical Sciences, Tehran, Iran with code number IR.IUMS.FMD.REC.1397.159. According to the patient’s history, none of the patients had received chemotherapy or radiotherapy and had no other malignancies. Healthy breast tissues obtained from normal individuals without any history of the disease. Pathological and clinical data including breast tumor ER, PR, and HER2 status by Laboratory tests (like IHC) were obtained for each patient.
Cell lines and cell culture
The following human BC cell lines were purchased from the Pasture (Iran, Tehran) MCF-7 (ER + Luminal A-like BC cell model), MDA-MB231 (Basal like BC cell model), and SKBR3 (Her2 positive BC cell model). Cells were cultured in RPMI 1640 medium (Invitrogen, Auckland, New Zealand) supplemented with 10% heat-inactivated fetal bovine serum (Invitrogen), 2 mM l-glutamine, 100 U per ml penicillin, 100 μg per ml streptomycin (Gibco BRL, Rockville, MD, USA), and 2 g/l sodium bicarbonate (Sigma, St. Louis, MO, USA). The cells were incubated at 37 °C in a humidified atmosphere with 5% CO2.
RNA extraction and cDNA synthesis
1.5 ml of cell culture medium and some (about 2–4 mg) of tissues stored at − 80 °C were used for RNA extraction. Total RNA was extracted from breast tissues and three cell line using RiboEX isolation reagent (GeneAll Biotech, Korea) according to the manufacturer’s instructions. RNA was dissolved in DEPC-treated water and its concentration was determined with a Nano Drop one C spectrophotometer (Thermo Fisher Scientific, US). 1 µg total RNA from each sample was used for cDNA synthesis with the TAKARA kit (Primer Script™ RT reagent Kit, Japan) based on the manufacturer’s protocol.
Quantitative real-time polymerase chain reaction
Quantitative real-time PCR (qtrs.-PCR) was performed on ABI Step One Sequence Detection System (Applied Biosystems, USA) using a SYBR Green-based PCR kit (Ampliqon, Denmark). The reaction was performed using a single cycle in 95 °C for 15 min followed by continual 45 cycles (95 °C for 10 s, 60 °C for 20 s and 72 °C for 30 s). The primer sequences for each gene are listed in Table 1. Each reaction was followed by melting curve analysis Involving heating of the PCR product from 60 to 95 °C. The melt curve in the samples was compared with melt curve of positive and negative control to recognize specific amplicon from primer dimer formation.
Hypoxanthine phosphoribosyltransferase1 (HPRT1) was amplified as internal control that showed the least variation in gene expression between our normal and tumor samples.
MRNA microarray profiling data analysis
The UALCAN (http://ualcan.path.uab.edu/index.html), an interactive web resource (Chandrashekar et al. 2017) was used to explore the mRNA expression levels of the candidate genes in tumor and normal breast tissues data.
Breast Cancer Gene-Expression Miner v4.4 (bc-GenExMiner v4.4) (http://bcgenex.centregauducheau.fr), is a statistical mining tool of published annotated breast cancer transcriptomic data (DNA microarrays [n = 10001]) (Jézéquel et al. 2012, 2013). It offers the possibility to explore gene-expression of genes of interest in breast cancer. Targeted expression analysis performed with DNA microarrays data and different populations such as receptor statuses (ER, PR and HER2 by IHC), Age and molecular subtype (Table 2).
Survival analysis
To further validate, we used the Kaplan–Meier plotter database, which includes gene chip and RNA-seq data—sources for the databases include GEO, EGA and TCGA. The primary purpose of these tools are a meta-analysis based discovery and validation of survival biomarkers (Györffy et al. 2010). Kaplan–Meier plotter database is a pan-cancer tool to analyze the relationship between gene expression of candidate genes and overall survival of 3955 BC patients. Hazard ratio (HR), 95% confidence intervals (CI) and log-rank p values were calculated.
Statistical analysis
All statistical analyses were performed using SPSS v.25 software (SPSS Inc., Chicago, IL, USA), Rest 2009 software (version 2.0.13) and Graph Pad Prism v.7.0 (Graph Pad Prism version 5.0, San Diego, USA). Rest is software for determining the relative expression results of real-time PCR that uses pair-wise fixed reallocation randomization tests to analyze data (Pfaffl et al. 2002). Descriptive statistical analysis was used to evaluate the expression of genes in the samples. p values < 0.05 were considered to be statistically significant.
Results
Clinic-pathological assessment
The mean age of patients at the time of diagnosis was 51.93 ± 2.23 (range between 30 and 73 years). According to TNM classifications, the tumors were staged by a pathologist, who had no prior knowledge of the test results. Clinicopathological factors including: age, tumor size, histological type, lymph node metastasis and TNM staging, were analyzed for evaluation of a correlation with considered genes expression (2). The subtypes were classified as follows according to ER, PR, and HER2: Triplepositive (ER positive, PR positive, HER2 positive); HER2positive (ER negative, PR negative, and HER2 positive); HER2negative (ER positive, PR positive, and HER2 negative); and Triplenegative (ER negative, PR negative, and HER2 negative) subtypes. Most samples were of the Invasive ductal carcinoma type (93.4%). 80% of patients were involved in pathologic stage I and II (32 of 40) and 70% of samples were invasive ductal carcinoma. Immunohistochemically subtypes of the 40 samples were categorized in four groups' including: triplepositive (17%); HER2positive (14%); HER2negative (37%) and triplenegative (32%).
Expression of MCT3, MCT8 and MCT9 genes in tissue samples and their relationship to tumor subtypes
According to the obtained data, there was a significant down regulation of the MCT3 gene in the tumor tissues compared to healthy (p value = 0.0001) and adjacent (p value = 0.004) tissues (Fig. 1). Expression level of MCT3 gene in four common subgroups of breast cancer showed that triple negative compared with triple positive was not significant decreased in expression level, but in HER2 negative comparison with HER2 positive subtype and HER2 positive comparison with triple negative subtype, the expression level of MCT3 gene over expressed non-significantly (Fig. 2).
The relative gene expressions of MCT3, MCT8 and MCT9 in different tissues. MCT-3, MCT-8 and MCT-9 genes are down regulated in tumor and adjacent samples compared to healthy tissues. Data shown are mean [± SD] OD values. *Value shown p value < 0.05, **value shown p value < 0.01 and ***value shown p value < 0.001
Expression of genes in subtype of breast cancer. Comparing breast cancer subtypes with healthy tissue, the MCT3 gene in three subtypes of negative triple (p = 0.03), positive triple (p = 0.031) and Her2 negative (p = 0.0001) had a significant down regulation. Data shown are mean [± SD] OD values. *Value shown p value < 0.05, **value shown p value < 0.01 and ***value shown p value < 0.001
MCT8 gene showed a non-significant down regulation in tumor tissues compared to adjacent and healthy tissues (Fig. 1). Expression level of this gene in subgroups of breast cancer showed that in HER2 negative comparison with HER2 positive subtype, the expression level of MCT8 was significantly lower (p value = 0.001). We also showed that MCT8 was significantly overexpression in comparison between HER2 positive with the triple negative subtype (p value = 0.027). The expression level of MCT8 gene in the triple negative compared with triple positive tumor was not significantly lower in expression level (Fig. 2).
The MCT9 gene showed a non-significant down regulation in tumor tissues compared to adjacent and healthy tissues (Fig. 1), in subgroups of breast cancer the triple positive compared with triple negative tissue was not significant reduced in expression level, but in the HER2 negative comparison with the HER2 positive subtype and HER2 positive comparison with triple negative subtype, the expression level of MCT9 gene was over expressed non-significantly (Fig. 2).
Expression level of MCT3, MCT8 and MCT9 genes in tumors in relation to the clinical stage and grade.
In order to evaluate the expression of MCTs genes in different stages and grades of breast cancer, in comparison to healthy tissue and each other, the samples were divided into six groups (stage I–III and grade I–III). The MCTs genes have down regulation in most stages compared to healthy tissue and down regulation of MCT3 genes in stage I (p value = 0.009), stage II (p value = 0.0001) and stage III (p value = 0.013) has been significantly (Fig. 3). Also, the MCT8 gene showed a non-significant over expression in stage I compared to healthy tissue. Compared in the different stages, MCT3 and MCT8 in stage II compared to stage I had a significant over expression (Table 3).
Also, The MCTs genes have down regulation in all grades compared to healthy tissue and down regulation of MCT3 genes in grade I (p = 0.027), grade II (p = 0.0001) and grade III (p = 0.005) has been significantly (Fig. 4). Compared in the different grades, none of the genes showed significant expression (Table 4).
Expression level of MCT3, MCT8 and MCT9 genes in three cell lines
MCF7 is representative of primary breast cancer cells [invasive ductal carcinoma (IDC) OR luminal A] that express E-cadherin, ER and PR receptors (Hiscox et al. 2012; Zheng et al. 2007). MDA-MB-231 is representative of metastatic, aggressive and invasive adenocarcinoma cells that are Vimentin- positive and does not express E-cadherin, ER, PR and HER2 receptors (TNBC) (Lacroix et al. 2004). SKBR3 is representative of adenocarcinoma that over-expresses the HER2 gene product (HER2 positive) (Dai et al. 2017). All the genes in the three cell lines had non-significant decrease in expression compared to healthy tissues, Also our results showed that MCT-9 in MDA-MB-231 (p value = 0.0001) and SKBR3 (p value = 0.0001) cell lines down regulated compared to MCF-7 cell line, but MCT-3 in SKBR3 cell line (p value = 0.0001) and MCT-8 in both SKBR3 (p value = 0.0001) and MDA-MB-231 cell lines (p value = 0.0001), up regulated compared to MCF-7 cell line (Fig. 5).
Breast cancer microarray profiling data analysis
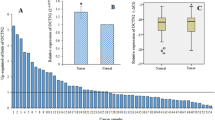
The UALCAN analysis showed that while the expression level candidate genes decreased in BC primary tumors compared with normal cases (p value < 0.001) (Fig. 6). The downregulation of candidate genes, have often been shown in a variety of cancers compared with normal cases (Fig. 7).
Expression validation in the UALCAN database for downregulated MCT8 (A), MCT3 (B) and MCT9 (C) genes in breast cancer samples. Blue box plot represent Normal samples (n = 114) and red box plot represent primary tumor samples (n = 1097). (Data from The Cancer Genome Atlas (TCGA) database) (***p value < 0.001)
bc-GenExMiner analysis showed that MCT8 (SLC16A2) has significant difference in ER negative versus ER positive (p value = 0.012) and HER2 negative versus HER2 positive (p value < 0.0001) status. MCT3 (SLC16A8) has significant difference in ER negative versus ER positive (p value < 0.0001), PR negative versus PR positive (p value = 0.002) and HER2 negative versus HER2 positive (p value = 0.004) status. MCT9 (SLC16A9) has significant difference in ER negative versus ER positive (p value < 0.0001), PR negative versus PR positive (p value < 0.0001) and HER2 negative versus HER2 positive (p value = 0.002) status.
Also All three candidate genes have significant differences between age before and after 51, and in each intrinsic molecular subtype with significantly p value (Figs. 8, 9, 10).
bc-GenExMiner data analysis of the mRNA level of MCT8 (SLC16A2) gene in different Receptor, age and subtype statuses in DNA microarray data. Box and whisker plot of SLC16A2 (MCT8) expression according to ER (A), PR (B) and HER2 (C) by IHC. Beeswarm plot of MCT8 expression in different age (D). Violin plot of MCT8 expression in different intrinsic molecular subtype according to PAM50 subtypes in DNA microarray data (E)
bc-GenExMiner data analysis of the mRNA level of MCT3 (SLC16A8) gene in different Receptor, age and subtype statuses in DNA microarray data. Box and whisker plot of SLC16A8 (MCT3) expression according to ER (A), PR (B) and HER2 (C) by IHC. Beeswarm plot of MCT3 gene in different age (D). Violin plot of MCT3 gene in different intrinsic molecular subtype according to PAM50 subtypes in DNA microarray data (E)
bc-GenExMiner data analysis of the mRNA level of MCT9 (SLC16A9) gene in different Receptor, age and subtype statuses in DNA microarray data. Box and whisker plot of MCT9 (SLC16A9) expression according to ER (A), PR (B) and HER2 (C) by IHC. Beeswarm plot of MCT9 gene in different age (D). Violin plot of MCT9 gene in different intrinsic molecular subtype according to PAM50 subtypes in DNA microarray data (E)
The expression level of MCT8 and MCT9 were associated with poor overall survival in Breast cancer samples
Using the Kaplan–Meier plotter database, we determined that the overall survival of 3955 BC patients (from the Kaplan–Meier plotter database) is low in cases with decreased expression of candidate genes. The follow-up time was determined in 250 months. The plots showed that low expression of SLC16A9 (MCT9) and SLC16A2 (MCT8) was associated with poor overall survival (Fig. 11).
A Kaplan–Meier (https://kmplot.com/analysis/) survival curve showing the prognostic value of SLC16A9 (MCT9) in predicting BC using the Pan-cancer analysis. Low expression of mRNA indicated poor overall survival. B Low expression of SLC 16A8 (MCT3) showed no significant effect on survival (HRs; 95% confidence interval in parentheses). C Low expression of SLC16A2 (MCT8) showed poor overall survival
Discussion
Nearly one in nine women develops breast cancer, and breast cancer accounts for about one-third of all cancers in women, the second most common cancer after lung cancer and the most common cause of cancer deaths among women (Wolman 2014). As a result, it is important to better understand and evaluate the genes and molecular markers involved in this cancer.
The important features of transporters in MCT super family are their ability to transport across membranes and their wide substrates, making them essential for many cellular processes and cancers (Halestrap 2013). MCT3, MCT8 and MCT9 genes and their substrates not yet well understood and there is very limited information on the expression and their potential roles. That's why for the first time, we examined the expression of the MCT3, MCT8 and MCT9 genes in the patients with breast cancer and MCF7, SKBR3 and MDA-MB-231 cell lines.
Some members of the MCT family can be grouped together, for example MCT1 to MCT4 are involved in the transfer of monocarboxylates (like lactate and pyruvate) and also MCT8 and MCT10 are involved in the transfer of aromatic amino acids and thyroid hormones. But other components of this family are not included in a particular group due to the lack of complete substrate recognition (Halestrap 2013). MCT1 and MCT4, are two members of this family that have been extensively studied in various cancers such as breast cancer and have been introduced as therapeutic targets for cancer, but the activity and expression of other members of this family in breast cancer have not been studied (Halestrap 2013). MCT1 up-regulated in the basal (Pinheiro et al. 2010), luminal-like subtypes of breast cancer and MCF7 cell line and targeted MCT1 can cause decrease in metastasis and angiogenesis of the cancer cells (Hussien and Brooks 2010; Sonveaux et al. 2008; Végran et al. 2011). Also, the MCT4 and MCT2 up-regulated in MDA-MB-231 and MCF-7 cell lines compared to HMEC 184 cells (Gallagher et al. 2007). Zhu and et al. (2005) demonstrated decreased expression of MCT3, and this was associated with increased severity of atherosclerosis. The MCT3 downregulation has also been observed in the non-small cell lung cancer (NSCLC) (Eilertsen et al. 2014). Our results are also consistent with these results, showing a significant down regulation of this gene in tumor tissues compared to normal tissues, but MCT-3 in SKBR3 cell line over expressed compared to MCF-7 cell line. MCT3 gene expression in stage II disease was significantly different from stage I and stage III disease. These data suggest that in addition to being detectable in various breast cancer subtypes, these markers like HER2 (Wong and Hurvitz 2014), and EphA2 (Rezaie et al. 2020, 2019) may be used to predict the response to the treatment or disease progression.
MCT8 and MCT10 are expressed in heart, kidney, liver, skeletal muscle, placenta, testis and appears to perform a particularly useful function in the brain where it is necessary for the thyroid hormone uptake that is important for normal brain development. Badziong et al. (2017) showed that the MCT8 expression was downregulated in both benign and the malignant thyroid tumors as compared to the normal thyroid tissue, In contrast, significant upregulation of MCT8, LAT2 and LAT4 genes were found in Graves' disease, So in that research they suggested MCT8 is a appropriate object for thyroid differentiation and MCT8 upregulation occurs in hyper functional thyroid tissue (Badziong et al. 2017). Significant down regulation of MCT8 gene was also confirmed by Nwosu et al. (2017), with analysis of eight microarray datasets in human hepatocellular carcinoma. In agreement with these studies, we found a significant reduction in the expression of this gene in the tumor tissues compared to the normal tissues. MCT8 is significantly overexpression in the comparison to HER2 positive with triple negative subtype; this means that HER2 expression is positively associated with increased expression of this gene. Also, in this study we reported upregulation of the MCT8 gene in the tumor tissue compared with the tumor adjacent tissues. This may indicate that the MCT8 gene could help differentiate tumor cells from adjacent cells and could be the possible mechanism for these associations.
So far, the MCT9 gene expression in various human cancers has not been studied by molecular methods such as real time PCR, but significantly down regulation in breast cancer has been identified using microarray analysis (Kim et al. 2017). The previous studies reported down regulation of the MCT9 gene in invasive breast cancer and also CNS diffuse large B-cell lymphoma compared to non-CNS diffuse large B-cell lymphoma by analyzing microarray data (do Hyoung, 2015). MCT9 may be involved in aerobic glycolysis of malignant tumors and may be increased in certain types of lymphomas. MCT9 expression is also significantly down-regulated in MDA-MB-231 and SKBR3 cell lines compared to MCF-7 cell line.
Conclusion
Since the expression MCT3, MCT8 and MCT9 have not been examined in breast cancer previously this study may help to understand the importance of these genes in the breast cancer progression. The expression of MCT3 may be used as a prognostic marker; however, further research is required to confirm this.
Limitations
This study had some limitations, like that sample size. In other studies with larger sample size, it might be feasible to investigate a number of issues in detail, which needs a more homogenous clinical and molecular population. Also for validating these results, the complementary techniques such as western blotting could be explored.
In conclusion, MCT3 gene showed low expressions in tumor samples compared to adjacent normal and healthy mammary tissues. Besides, MCT3 can represent a novel therapeutic target in breast cancer patients or some of the following subgroups such as HER2-negative or triple-negative.
Availability of data and material
The data that support the findings of this study are available.
Abbreviations
- BC:
-
Breast cancer
- ER:
-
Estrogen receptor
- HER2:
-
Human epidermal growth factor receptor 2
- MCTs:
-
Monocarboxylate transporters
- LDHs:
-
Lactate dehydrogenases
- AHD:
-
Allan–Herndon–Dudley syndrome
- HPRT1:
-
Hypoxanthine phosphoribosyltransferase1
References
Aveseh M, Nikooie R, Aminaie M (2015) Exercise-induced changes in tumour LDH-B and MCT1 expression are modulated by oestrogen-related receptor alpha in breast cancer-bearing BALB/c mice. J Physiol 593:2635–2648
Badziong J, Ting S, Synoracki S, Tiedje V, Brix K, Brabant G, Moeller LC, Schmid KW, Fuhrer D, Zwanziger D (2017) Differential regulation of monocarboxylate transporter 8 expression in thyroid cancer and hyperthyroidism. Eur J Endocrinol 177:243–250
Bogenrieder T, Herlyn M (2003) Axis of evil: molecular mechanisms of cancer metastasis. Oncogene 22:6524–6536
Chandrashekar DS, Bashel B, Balasubramanya SAH, Creighton CJ, Ponce-Rodriguez I, Chakravarthi BV, Varambally S (2017) UALCAN: a portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia 19:649–658
Dai X, Cheng H, Bai Z, Li J (2017) Breast cancer cell line classification and its relevance with breast tumor subtyping. J Cancer 8:3131
Do Hyoung L, Kim WS, Kim SJ, Yoo HY, Ko YH (2015) Microarray gene-expression profiling analysis comparing PCNSL and non-CNS diffuse large B-cell lymphoma. Anticancer Res 35:3333–3340
Draoui N, Feron O (2011) Lactate shuttles at a glance: from physiological paradigms to anti-cancer treatments. DMM 4:727–732
Eilertsen M, Andersen S, Al-Saad S, Kiselev Y, Donnem T, Stenvold H, Pettersen I, Al-Shibli K, Richardsen E, Busund L-T (2014) Monocarboxylate transporters 1–4 in NSCLC: MCT1 is an independent prognostic marker for survival. PLoS ONE 9:e105038
Estiar MA, Esmaeili R, Zare A-A, Farahmand L, Fazilaty H, Zekri A, Jafarbeik-Iravani N, Majidzadeh-a K (2017) High expression of CEACAM19, a new member of carcinoembryonic antigen gene family, in patients with breast cancer. Clin Exp Med 17:547–553
Friesema EC, Ganguly S, Abdalla A, Fox JEM, Halestrap AP, Visser TJ (2003) Identification of monocarboxylate transporter 8 as a specific thyroid hormone transporter. J Biol Chem 278:40128–40135
Gallagher SM, Castorino JJ, Wang D, Philp NJ (2007) Monocarboxylate transporter 4 regulates maturation and trafficking of CD147 to the plasma membrane in the metastatic breast cancer cell line MDA-MB-231. Cancer Res 67:4182–4189
Györffy B, Lanczky A, Eklund AC, Denkert C, Budczies J, Li Q, Szallasi Z (2010) An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients. Breast Cancer Res Treat 123:725–731
Halestrap AP (2013) The SLC16 gene family–structure, role and regulation in health and disease. Mol Aspects Med 34:337–349
Halestrap AP, Meredith D (2004) The SLC16 gene family—from monocarboxylate transporters (MCTs) to aromatic amino acid transporters and beyond. Pflugers Arch 447:619–628
Halestrap AP, Price NT (1999) The proton-linked monocarboxylate transporter (MCT) family: structure, function and regulation. Biochem J 343:281
Harding C, Pompei F, Burmistrov D, Welch HG, Abebe R, Wilson R (2015) Breast cancer screening, incidence, and mortality across US counties. JAMA Intern Med 175:1483–1489
Hiscox S, Baruah B, Smith C, Bellerby R, Goddard L, Jordan N, Poghosyan Z, Nicholson RI, Barrett-Lee P, Gee J (2012) Overexpression of CD44 accompanies acquired tamoxifen resistance in MCF7 cells and augments their sensitivity to the stromal factors, heregulin and hyaluronan. BMC Cancer 12:458
Hussien R, Brooks GA (2010) Mitochondrial and plasma membrane lactate transporter and lactate dehydrogenase isoform expression in breast cancer cell lines. Physiol Gen 43:255–264
Jézéquel P, Campone M, Gouraud W, Guérin-Charbonnel C, Leux C, Ricolleau G, Campion L (2012) bc-GenExMiner: an easy-to-use online platform for gene prognostic analyses in breast cancer. Breast Cancer Res Treat 131:765–775
Jézéquel P, Frénel J-S, Campion L, Guérin-Charbonnel C, Gouraud W, Ricolleau G, Campone M (2013) bc-GenExMiner 3.0: new mining module computes breast cancer gene expression correlation analyses. Database 2013
Kim GR, Ku YJ, Cho SG, Kim SJ, Min BS (2017) Associations between gene expression profiles of invasive breast cancer and Breast Imaging Reporting and Data System MRI lexicon. Ann Surg Treat Res 93:18–26
Lacroix M, Haibe-Kains B, Hennuy B, Laes J-F, Lallemand F, Gonze I, Cardoso F, Piccart M, Leclercq G, Sotiriou C (2004) Gene regulation by phorbol 12-myristate 13-acetate in MCF-7 and MDA-MB-231, two breast cancer cell lines exhibiting highly different phenotypes. Oncol Rep 12:701–707
Liu Z, Zhang X-S, Zhang S (2014) Breast tumor subgroups reveal diverse clinical prognostic power. Sci Rep 4:4002
Luz M, Perez M, Azzalis L, Sousa L, Adami F, Fonseca F, Alves B (2017) Evaluation of MCT1, MCT4 and CD147 genes in peripheral blood cells of breast cancer patients and their potential use as diagnostic and prognostic markers. Int J Mol Sci 18:170
Mishra D, Banerjee D (2019) Lactate dehydrogenases as metabolic links between tumor and stroma in the tumor microenvironment. Cancers 11:750
Moslemi M, Sohrabi E, Azadi N, Zekri A, Afkhami H, Khaledi M, Razmara E (2020) Expression analysis of EEPD1 and MUS81 genes in breast Cancer. Bio J Sci Tech Res 29:22556–22564
Moslemi M, Moradi Y, Dehghanbanadaki H, Afkhami H, Khaledi M, Sedighimehr N, Fathi J, Sohrabi E (2021) The association between ATM variants and risk of breast cancer: a systematic review and meta-analysis. BMC Cancer 21:1–12
Pérez-Escuredo J, Van Hee VF, Sboarina M, Falces J, Payen VL, Pellerin L, Sonveaux P (2016) Monocarboxylate transporters in the brain and in cancer. Biochim Biophys Acta BBA Mol Cell Res 1863:2481–2497
Pfaffl MW, Horgan GW, Dempfle L (2002) Relative expression software tool (REST©) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res 30:e36–e36
Pinheiro C, Longatto-Filho A, Scapulatempo C, Ferreira L, Martins S, Pellerin L, Rodrigues M, Alves VA, Schmitt F, Baltazar F (2008) Increased expression of monocarboxylate transporters 1, 2, and 4 in colorectal carcinomas. Virchows Arch 452:139–146
Pinheiro C, Albergaria A, Paredes J, Sousa B, Dufloth R, Vieira D, Schmitt F, Baltazar F (2010) Monocarboxylate transporter 1 is up-regulated in basal-like breast carcinoma. Histopathology 56:860–867
Pinheiro C, Longatto-Filho A, Azevedo-Silva J, Casal M, Schmitt FC, Baltazar F (2012) Role of monocarboxylate transporters in human cancers: state of the art. J Bioenerg Biomembr 44:127–139
Rezaie E, Pour AB, Amani J, Hosseini HM (2019) Bioinformatics predictions, expression, purification and structural analysis of the PE38KDEL-scfv immunotoxin against EPHA2 receptor. Int J Pept Res Ther 26(2):979–996
Rezaie E, Amani J, Pour AB, Hosseini HM (2020) A new scfv-based recombinant immunotoxin against EPHA2-overexpressing breast cancer cells; high in vitro anti-cancer potenc. Eur J Pharmacol 870:172912
Schwartz CE, Stevenson RE (2007) The MCT8 thyroid hormone transporter and Allan–Herndon–Dudley syndrome. Best Pract Res Clin Endocrinol 21:307–321
Si W, Li Y, Han Y, Zhang F, Wang Y, Li Y, Linghu RX, Zhang X, Yang J (2015) Epidemiological and clinicopathological trends of breast cancer in Chinese patients during 1993 to 2013: a retrospective study. Medicine 94(26):e820
Sonveaux P, Végran F, Schroeder T, Wergin MC, Verrax J, Rabbani ZN, De Saedeleer CJ, Kennedy KM, Diepart C, Jordan BF (2008) Targeting lactate-fueled respiration selectively kills hypoxic tumor cells in mice. J Clin Investig 118:3930–3942
Stein C, Colditz G (2004) Modifiable risk factors for cancer. Br J Cancer 90:299–303
Van Der Deure WM, Peeters RP, Visser TJ (2010) Molecular aspects of thyroid hormone transporters, including MCT8, MCT10, and OATPs, and the effects of genetic variation in these transporters. J Mol Endocrinol 44:1
Végran F, Boidot R, Michiels C, Sonveaux P, Feron O (2011) Lactate influx through the endothelial cell monocarboxylate transporter MCT1 supports an NF-κB/IL-8 pathway that drives tumor angiogenesis. Cancer Res 71:2550–2560
Weigelt B, Peterse JL, Van’t Veer LJ (2005) Breast cancer metastasis: markers and models. Nat Rev Cancer 5:591–602
Wolman I (2014) Berek and Novak’s gynecology, 15th edn. Springer, New York
Wong DJ, Hurvitz SA (2014) Recent advances in the development of anti-HER2 antibodies and antibody-drug conjugates. Ann Transl Med 2(12):122
Yersal O, Barutca S (2014) Biological subtypes of breast cancer: prognostic and therapeutic implications. World J Clin Oncol 5:412
Zheng A, Kallio A, Harkonen P (2007) Tamoxifen-induced rapid death of MCF-7 breast cancer cells is mediated via extracellularly signal-regulated kinase signaling and can be abrogated by estrogen. Endocrinology 148:2764–2777
Zhu S, Goldschmidt-Clermont PJ, Dong C (2005) Inactivation of monocarboxylate transporter MCT3 by DNA methylation in atherosclerosis. Circulation 112:1353–1361
Acknowledgements
This study is financially supported by Iran University of Medical Sciences, Tehran, Iran, in 2018. All parts of the work were carried out in the ‘Genetic laboratory of Ali Asghar Children’s’ and Rasoul Akram Hospitals, Tehran, Iran. The authors of the current study would like to express their deep thanks to all staff of these hospitals.
Funding
This study is financially supported by Iran University of Medical Sciences, Tehran, Iran.
Author information
Authors and Affiliations
Contributions
ES, NN, AZ, MK and JF are major contributors in writing the manuscript and design, MM, EH, MK, HA participated in writing the manuscript, MK, JF, HA and MM participated in writing the manuscript and data interpretation, NN, ES and A.Z participated in design, participated in manuscript writing, and conducted statistics. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
Ehsan Sohrabi, Masoumeh Moslemi, Ehsan Rezaie, Nahid Nafissi, Mansoor Khaledi, Hamed Afkhami, Javad Fathi and Ali Zekri declare that they have no conflict of interest.
Ethics approval and consent to participate
The study was approved by the Ethics Committee of Iran University of Medical Sciences, Tehran, Iran with code number IR.IUMS.FMD.REC.1397.159.
Consent for publication
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Sohrabi, E., Moslemi, M., Rezaie, E. et al. The tissue expression of MCT3, MCT8, and MCT9 genes in women with breast cancer. Genes Genom 43, 1065–1077 (2021). https://doi.org/10.1007/s13258-021-01116-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13258-021-01116-w