Abstract
Fourteen novel miniature inverted-repeat transposable element (MITE) families are found in the Florida carpenter ant genome, Camponotus floridanus. They constitute approximately 0.63 % of the entire genome. Analysis of their insertion time showed that most members of these MITEs were inserted into their host genome in less than 8 million years ago. In addition, the association between MITEs and the noncoding regions of genes in C. floridanus is random. Interestingly, an autonomous partner (named CfTEC) responsible for the amplification of these MITEs was also found in C. floridanus. Meanwhile, we present evidence, based on searches of publicly available databases, that this autonomous element was widespread in animals. Moreover, structure and phylogenetic analyses supported that TECs might represent a novel cade of transposons intermediate between the classic CACTA transposon and TRCs. Finally, their transposition mechanism and impact on host genome evolution were also discussed.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Generally, transposable elements could be grouped into two major classes: the class I or RNA transposons that transpose via RNA intermediates and can reach high copy number, and the class II or DNA transposons that move without transposition intermediates and are commonly found at a relative low copy number. DNA transposons are classified into sixteen different superfamilies: Tc1/mariner, hAT, Mutator, Merlin, Transib, P, Piggybac, PHIS, CMC, Sola, Novosib, Rehavkus, Kolobok, Zator, Ginger, and Academ (Yuan and Wessler 2011; Han et al. 2014).
In the last decades, a novel group of deletion derivatives of DNA transposon was firstly discovered in maize, named as “MITEs” for miniature inverted-repeat transposable elements (Bureau and Wessler 1992). In particular, they are structurally reminiscent of DNA non-autonomous elements with terminal inverted repeats (TIRs), target site duplication (TSD), small size (generally less than 600 bp) and lack of coding capacity (Feschotte and Pritham 2007). However, characteristics of high copy number and structural homogeneity distinguish them from other non-autonomous DNA transposons. It is now inferred that MITEs are non-autonomous DNA transposons that derived from a subset of autonomous DNA transposons based on their structure (TIRs or TSD) similarity (Feschotte et al. 2002). Therefore, MITEs could be classified into different superfamilies based on their association with autonomous elements. Although MITEs belonging to many DNA superfamilies have been reported, MITEs associated with the CACTA transposon was so far only described in plants (Lu et al. 2012; Benjak et al. 2009).
MITEs can occupy significant portions of eukaryotic genomes, and can preferentially insert into host genes, potentially influencing gene expression (Lu et al. 2012; Han et al. 2010). In addition, it also showed that massive proliferation of MITEs may be associated with more repetitive genomes in both the plant and the animal kingdoms (Tu 2000). As MITEs played important roles in host genome evolution, several programs have been developed exclusively to find MITEs: TRANSPO (Santiago et al. 2002), FINDMITE (Tu 2001), MUST (Chen et al. 2009) and MITE-Hunter (Han and Wessler 2010). Compared with these MITE discovery programs, MITE-Hunter can search large genomic data sets and is significantly more accurate.
In the present study, MITE-Hunter was firstly applied to search the Florida carpenter ant Camponotus floridanus genome. Fourteen novel MITEs associated with the CACTA transpsons are found in the Florida carpenter ant genome. Further analyses demonstrated that the autonomous partner of these MITEs might represent a novel cade of transposons intermediate between the classic CACTA transposon and TRCs.
Materials and methods
Identification and copy number calculation of the novel MITEs in the genome of Florida carpenter ant and their distribution in other sequenced ants
The assembled genome sequences (Version 3.3) of the Florida carpenter ant was downloaded from Hymenoptera Genome Database (http://hymenopteragenome.org/, Elsik et al. 2016). Then, the MITE-hunter program (Han and Wessler 2010) was used to search the Florida carpenter ant genome sequence for candidate MITEs. All elements from “Step_8” and “Step_8.singlet” files generated by MITE-hunter program were obtained. To manually check or classify a MITE sequence, the multiple sequence alignment (MSA) files of the transposon were opened by the software Bioedit (Hall 1999) to check the TIR and TSD structures. According to the common features of known MITEs (Feschotte and Pritham 2007), the following four kind elements were considered as the false positive of MITEs: (1) the putative compound transposons; (2) transposons without TIRs or TSD; (3) transposons longer than 1000 bp; (4) transposons having <10 full-length members. Then, clusters of all obtained candidate MITEs with 80 % similarity within each family were created using a custom Perl script.
Next, their respective consensus sequences were used as queries to blast against the Florida carpenter ant genome for estimating copy number of each MITE family. Their copy numbers were calculated based on the following criteria: (1) all fragments showed more than 80 % identity and 50 % coverage to their consensus sequences; (2) fragments were defined as a single insertion when they were separated by <50 bp. Meanwhile, copies having nucleotide identity >80 % and coverage >80 % of the consensus sequences were defined as intact members. Meanwhile, when we analyzed the TIRs of MITEs in the Florida carpenter ant genome, we found that many MITEs were flanked by the TIRs across their whole sequence. It meant that their copies will be calculated twice (forward and reverse plus) when the consensus sequence was used to masked their host genome. To reduce this redundancy, only the best hit (higher coverage and similarity) were chosen as its copy.
Finally, we determined the present of related MITEs with the above novel MITEs in other seven sequenced ants including the Argentine ant Linepithema humile, the red fire ant Solenopsis invicta, the Fungus growing Ant Acromyrmex echinatior, the Jerdon’s jumping ant Harpegnathos saltator, the leaf cutter ant Atta cephalotes, the red harvester ant Pogonomyrmex barbatus and the clonal raider ant Cerapachys biroi. Genomes of these ants were downloaded from Hymenoptera Genome Database or NCBI.
Sequence analysis
Their consensus sequences were reconstructed using the software DAMBE (Xia and Xie 2001). Then, their secondary structure was analyzed using UNAFOLD (http://unafold.rna.albany.edu/). To identify paralogous empty sites, the MITEs 100 bp flanking sequences were used as a query to blast against the host genome.
For all MITEs, their potential autonomous partners were identified using extensive analysis. Briefly, we searched the large DNA fragments that shared similar or same terminal sequences with those of MITEs. All DNA fragments ranging from 1000 bp to 10 kb flanked by the termini of MITEs were obtained. Then, their potential open reading frames (ORF) were predicted using getorf in EMBOSS-6.3.1 package (Rice et al. 2000).
The obtained transposase sequences were used as initial queries (tBLASTn using default parameters) to find these elements elements in other genomes available at the NCBI, including nucleotide collection (nr/nt), genome survey sequences (GSS), expressed sequence tag (EST), high throughput genomic sequences (HTGS), and the whole-genome shotgun (WGS) databases. This element was considered in a species if hits showed significant similarity (E-values less than 10−5) to the query and contained the specific motif (E-X4-L-X7-K-X-DNL-X2-H-X8-F-X6-7-CRF/YC) of the query shown in Fig. 3. We used these sequences along with the sequences of known CACTA transposons from other species to create an alignment using MUSCLE (Edgar 2004). Alignments were imported to the software Genedoc (Nicholas et al. 1997) for shading of conserved residues. Further analysis with MUSCLE (Edgar 2004) and the neighbor-joining method, using the Transposase_21 domain and excluding positions with gaps, resulted in the phylogenetic tree shown in Fig. 4b. The confidence values were evaluated by bootstrap analysis using 1000 samplings. Phylogenetic trees were built using MEGA 4 (Tamura et al. 2007).
Age analyses
Because the DNA elements have evolved neutrally since their insertion, the sequence divergence between copies and the consensus sequence can be used to determine the age of the elements. We used MUSCLE (Edgar 2004) for aligning all full-length copies from one family and its consensus sequence and used the Kimura two parameter method to estimate the nucleotide substitution (K). Then, the insertion time of each element was estimated by the formula T = K/2r (Li 1997), where T corresponds to the insertion time in millions of years, K corresponds to the number of nucleotide substitutions per site, and r corresponds to the neutral mutation rate of the species lineage. In this case we chose a rate of 0.54 × 10−8 synonymous substitutions per site per year as reported for 95 orthologous protein-coding sequences from three ant species C. floridanus, Solenopsis invicta and Pogonomyrmex barbatus (Zhou and Cahan 2012).
Relationship of MITEs to genes
The distribution of MITEs relative to predicted genes in the WGS assembly was analyzed for all scaffolds. One file (gff file, v 3.3) for positions of predicted genes in scaffolds was downloaded from Hymenoptera Genome Database. Another file is the lengths of scaffolds calculated using Perl script. Then, Perl script was written to investigate the information of MITEs close to or in predicted genes. To determine whether the relationship of MITEs and genes are due to random, a computer simulation similar to previous studies was performed (Naito et al. 2006; Han et al. 2010).
Results
Identification and characterization of novel MITEs in the genome of Florida carpenter ant
MITE-hunter (Han and Wessler 2010), a program designed to discover MITEs as well as other short non-autonomous “cut-and-paste” Class 2 transposons in genomic data sets, was firstly applied to search the Florida carpenter ant genome with default parameters. Using this bioinformatics tool, 443 putative candidate MITEs were retrieved from “Step_8” and “Step_8.singlet” files generated by MITE-hunter program. Then, we filtered out pseudo-MITEs from predicted ones by the methods described in experimental procedures. After curation and classification, the filtered library had a total of 125 putative MITEs (data not shown). When checking the TIR and TSD structures of the above MITEs, we found a 2 bp direct repeat suggestive of TSD flanking the inverted repeat in all fourteen families and they were flanked by CACHS (the H represents A, T or C; S represents C or G) termini (Table 1; Fig. 1 and Table S1). These characteristics support classification as CACTA DNA transposons. Generally, the classic CACTA DNA transposons were flanked by the highly conserved CACTA motif in the first and last 5 bp of the TIRs and it can create a 3 bp TSD during excision (Buchmann et al. 2014). Although a novel CACTA transposon named as TRC recently discovered in animal and fungi genomes was also flanked by 2 bp TSD, it was presence a CCC core sequence in the first three bases in their TIRs (DeMarco et al. 2006). Therefore, we speculated that the autonomous partners of 14 MITE families identified in this study might represent a novel clade of the CACTA transposons. Meanwhile, these 14 MITE families formed the starting point for the below analysis.
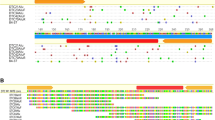
Determination of TSD of the Florida carpenter ant MITEs. a Multiple alignments of copies of CfMITE1. TSD was shown using red color. b Two examples (CfMITE1 and CfMITE2) of alignments of the flanking sequences of MITE insertions with a paralogous sequences found within the same genome but devoid of the transposons
14 MITE families range in size from 106 to 685 bp (Table 1). Then, a homology search was used to determine number of copies for each MITE family in the Florida carpenter ant genome. With this approach, we identified 3555 MITEs in total, which masked a total of ~1.51 Mb (0.63 % of the assembly) of the Florida carpenter ant genome (Table 1). Meanwhile, the information about insertion sites of the MITEs into scaffold was shown in Table S2. Further analysis indicated that about 70 % (2475/3555) copies of 14 MITE families identified in the Florida carpenter ant genome were full-length copies (Table 1), suggesting that they have been amplified from a single or few founder elements in their host genome. Meanwhile, we also noticed that copy number varies across different MITE families but in some cases is high (up to 888 copies; Table 1). This result implied that some MITEs families were specific proliferation in the Florida carpenter ant genome.
The average AT content for each MITE family varied from 63.9 to 82 % (Table 1).
Meanwhile, only CfMITE8 has less than the average AT content (with a A + T frequency of 66 %) of the Florida carpenter ant genome (Bonasio et al. 2010), suggesting the Florida carpenter ant MITEs are AT rich. Meanwhile, these MITEs form stable secondary structure (Figure S1).
Finally, we also found the present of related MITEs with the above novel MITEs in other seven sequenced ants (Table S1).
Timing of amplification and identification of genes associated with MITEs
To gain further insight into the timing of amplification of these MITEs, we perform a calculation of the time elapsed since the amplification of MITEs in the Florida carpenter ant genome. The estimated ages of the 2475 intact MITE members varied from 0 to 14.26 million years, with most of them (~98 %; 2420/2475) being less than 8 million years old (Fig. 2 and Table S3). In addition, copies of CfMITE4 shared a high level of sequence similarity with each other and thus had the shortest estimated insertion time (ranging from 0.19 to 2.87 million years) when compared with other families (Fig. 2 and Table S3).
MITEs were often found near genes and they might play an important role in the gene biological functions and regulation (Oki et al. 2008; Han et al. 2010; Tu 2001). Therefore, we examined whether MITEs described in the Florida carpenter ant genome were also preferentially in the non-coding regions of the genes or not. One MITE was considered as close to a gene if the distance of this MITE to the coding sequence of this gene was shorter than 500 bp (Lu et al. 2012). Our results showed that 34 % (838/2475) of MITEs in the Florida carpenter ant were found near or in genes (Table 2). Of these sequences, approximately 730 were found in gene regions (exons and introns) and 108 in 5′ and 3′ flanking regions of the closest genes. In gene regions, most MITEs (25.7 %) were located in introns. Meanwhile, about 3.8 % of MITEs were located in exons. This finding was consistent with previous studies, which demonstrated that insertions of MITEs in exons were generally deleterious (Hirochika 2001). In order to examine whether biased insertions of these MITEs were due to chance, a computer simulation (a negative control) described by recent studies was performed (Naito et al. 2006; Han et al. 2010). It was found that MITEs seemed to be not preferentially associated with genes in the Florida carpenter ant as a result of similar insertion frequencies with those in control (34 versus 37 %) (Table 2).
Autonomous partners of MITEs
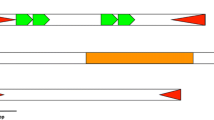
The real origin of MITEs remains controversial. At present, the prevalent ideal is that MITEs may be derived from DNA transposons (Feschotte et al. 2002). It had been proposed that DNA transposons gave rise to MITEs by a two step model. Small defective elements originated by internal deletions from an autonomous transposon and were subsequently amplified to generate a high number of highly similar elements by the transposase encoded by the autonomous (Feschotte et al. 2002). Therefore, we identified the potential autonomous elements of MITEs discovered in the Florida carpenter ant using extensive analysis as described in experimental procedures. Surprisingly, potential autonomous elements responsible for the spread of CfMITE9 were found (Fig. 3a). This autonomous element was named as CfTEC for C. floridanus Transposable Element related to CACTA. CfMITE9 and CfTEC were almost flanked by the identical TIRs (CACTGTAAAAAAA and CACTGTAAAAAA, respectively). Besides, CfMITE9 show extensive sequence similarity with the related master element. For example, 99 bp of the 5′ end and 109 bp of the 3′ end of CfMITE9 showed high similarity (from 76.1 to 84.8 %) with those of CfTEC. However, the internal sequence of CfMITE9, which is 388 bp long and is highly conserved in all copies, did not have similarity to CfTEC. This result suggested that CfMITE9 could capture, mobilize, and amplify foreign or host genomic sequences. The overall size of CfTEC was 8870 bp (Fig. 3a). Such a size was not expected for a so-called miniature transposon. Interestingly, two intact ORF (one with similarity to Tnp2, another with unknown function), which did not have premature stop codon was clearly identified for CfTEC (Fig. 3b). It suggested that it might be a source of functional protein and was still active in the Florida carpenter ant. Meanwhile, we found one CACTA transposon (named ENS1_Cis; http://www.girinst.org/protected/repbase_extract.php?access=ENS1_Cis&format=EMBL) from Ciona savignyi deposited in Repbase also appears to duplicate 2 bp TSD (especially for TA). Further analysis demonstrated that transposase of ENS1_Cis also encodes two ORFs and these two ORFs show high sequence similarity with those of CfTEC (Fig. 3b), suggesting it also should belong to a member of TEC.
Characteristics of the autonomous partner of CfMITEs. a CFMITE9 and their master CfTEC. Gray rectangles are homologous regions between CfTEC and CFMITE9. Percentages of identity were calculated using Bioedit. Numbers on the edges of the gray rectangles are the coordinates of the region of homology limits. b Schematic structure of CfTEC from C. floridanus and ENS1_Cis from Ciona savignyi. The arrows indicated their transcriptional direction. c Multiple alignments of the Transposase_21 domains of proteins of TECs, the classic CACTA transposons and TRCs. Boxes with Roman numbers I to III indicate conserved motifs of the Transposase_21 domain in all organisms. Boxes with N indicate the NLPP conserved motifs of the Transposase_21 domain shown by TEC and the classic CACTA transposons. Meanwhile, specific conserved residues (E-X4-L-X7-K-X-DNL-X2-H-X8-F-X6-7-CRF/YC) observed in TEC group were shown using red color
Next, we determine whether CfTEC could be found in other organisms. To test this possibility, a BLASTP and TBLASTN search against the nr and WGS database at GenBank was performed using the amino acid sequences encoded by CfTEC transposase as a query. Interestingly, these searches produced hits indicating high similarity between the deduced CfTEC translated sequence and translated sequences from genomes of widespread animals (Fig. 4a and Table S4). In addition, one significant hit (with E-values 6e−37) was also obtained from Parabasalids Trypanosoma vivax. However, no such elements were found in plant and fungi genomes. Multiple alignments of transposases of TEC with other known CACTA transposons showed that three different conserved motifs could be discerned in their Transposase_21 region, marked I-III in the aligned proteins (Fig. 3c). In most of transposons, a first highly conserved motif I/L/V-X1-I/L/V/F-X1-I/L/V/F-X2-D-X14-19-I/M/L/V of Transposase_21 region was present (Fig. 3c, Box I). However, some components of the signature strings are shared by only two groups. For example, the P residue 3–6 aa downstream of the D residue was shared by the TEC and classical CACTA groups, but not by TRC group. In contrast, the S or T residue 7–9 aa upstream of the I/M/L/V residue was shared by the TEC and TRC groups, but not by classical CACTA group. Interestingly, we identified a NLPP motif 4 bp downstream of the first conserved motif in the Transposase_21 domain only shown by TEC and classical CACTA groups (Fig. 3c, Box N), suggesting this motif might have an important role and was essential for activity of transposition. For the second and third conserved domains (Fig. 3c, Box II and III), they were more conserved in TRC and classical CACTA groups when compared with those of the TEC group. For example, four additional conserved residues (G, P, F/Y/I, L/M) were only shared by TRC and classical CACTA groups. Although these three conserved motifs were present in the Transposase_21 domain of all these three groups of CACTA transposons, specific conserved residues (E-X4-L-X7-K-X-DNL-X2-H-X8-F-X6-7-CRF/YC) were also observed in TEC groups and all of them were located in the first and third motifs of TEC Transposase_21 domain (Fig. 3c, shown by red letters). Meanwhile, these specific residues were also used as a measurement for determining whether TEC transposons were present in other organisms or not when we used the homology-based strategy as described in experimental procedures.
a Taxonomic distribution of the TEC transposons. Number in parentheses on the right indicates the number of species in each taxa have TEC transposons. b Phylogeny for the Transposase_21 domains of CACTA-like transposases. TECs, the classic CACTA transposons and TRCs were indicated using red, green and purple, respectively. c Multiple alignments of TIR sequences from TECs, the classic CACTA transposons and TRCs as well as CfMITEs. The regions with high and medium levels of identity among the sequences are shown as black and gray columns, respectively
A phylogenetic tree (Fig. 4b) was created from the three conserved regions (I–III) shown in the alignment of Fig. 3c. It showed that these CACTA elements could clearly be classified into three groups: TEC, TRC and the classic CACTA transposons. Next, we investigated the similarity of TIRs of transposons from the above three groups. It is interesting to note that the plant CACTA transposons and TECs shared almost perfect terminal nucleotide sequences, 5′-CACTG/ANAA/GAAAW-3′ (the N represents A, T, C or G; W represents A or T), of TIRs (Fig. 4c). However, no such level of similarity is found in TRC TIR sequences, which display at most 5 coincident bases.
Discussion
Diverse deletion derivatives from the CACTA transposons were first identified in insect
Up to now, a large array of MITE families have been described from many insects, including the silkworm Bombyx mori (Han et al. 2010), the yellow fever mosquito Aedes aegypti (Tu 2000), the fly Drosophila willistoni (Holyoake and Kidwell 2003), the African malaria mosquito Anopheles gambiae (Tu 2001) and the triatomine bug Rhodnius prolixus (Zhang et al. 2013). MITEs identified in these insects have been grouped into known superfamilies based on their association with structure characteristics of these superfamilies, majorly including Tc1/mariner, hAT, piggyBac, Ginger and Sola. To our best knowledge, no MITEs belonging to the CACTA transposons has been reported. The main reason why CACTA elements have remained undiscovered in insects for so long is that the CACTA transposons were firstly identified in the plants (Capy et al. 1998) and almost all studies focused on the studying plant CACTA transposons. In this study, 14 novel families of mobile elements were discovered in the Florida carpenter ant genome. Characteristics of short length and no coding ability suggested that they were members of MITEs. One typical feature present in these MITEs is the presence of a CACTG/A core, indicating that they might be more closely related to the CACTA superfamily of DNA elements. However, another typical feature of these MITEs is the presence of flanking 2 bp direct repeats resulting from the target site duplication, suggesting that autonomous elements responsible for the transposition of these MITEs might belong to be a novel group of the CACTA transposon. Indeed, this hypothesis was supported by the further analyses (please see the following part for the detail information).
TECs-a clade of transposons intermediate between the classic CACTA transposon and TRCs
MITEs are non-autonomous DNA elements that might originate from a subset of autonomous DNA elements (Feschotte and Pritham 2007). In this study, we present evidence that a family of MITEs from the Florida carpenter ant genome, CfMITE9, derives from a larger element, CfTEC, which has coding capacity for a putative CACTA transposase. Many significant hits were obtained from animal genomes when the coding sequence of CfTEC was used as a query to search against the databases in NCBI. Generally, DNA transposons can be classified into different superfamilies or families based on their transposase or different structure (TIRs or TSD) in its flanking sequences (Yuan and Wessler 2011). According to our results, we concluded that TECs might represent a cade of transposons intermediate between the classic CACTA transposon and TRCs based on the following three points: (1) TECs show a similar core sequence (CACTA/G) and terminal sequences (5′-CACTG/ANAA/GAAAW-3′) at the both ends with those of the plant CACTA transposons; (2) both TECs and TRCs are flanked by 2 bp TSD, not 3 bp TSD like that of the plant CACTA transposons; (3) phylogenetic analysis indicated that TEC, TRC and the classic CACTA transposons should belong to be three different groups of CACTA transposons.
Transposition mechanism of novel MITEs identified in the Florida carpenter ant genome
There were at least 3555 CfTEC-related MITEs in the available Florida carpenter ant genome. We also observed that some MITEs (up to several hundred copies) had experienced burst transposition in its host genome. Besides, copies of these MITE families in the Florida carpenter ant showed the high sequence identity and structural homogeneity. Meanwhile, the estimated insertion time of most of them (~98 %) was less than 8 million years old. Strikingly, CfTEC is the only longer element with coding capacity in the current ant database. Further analysis showed that CfTEC encoded two intact ORFs and might be still active in the Florida carpenter ant. Therefore, CfTEC might be responsible for the transposition of these MITEs. However, we should also notice that significant homology of TIRs between CfTEC and the classical CACTA transposons was also observed, suggesting that they had similar cleavage sites or patterns during the process of their transposition (Cui et al. 2002). Therefore, we cannot exclude the possibility that there is, elsewhere in the Florida carpenter ant genome, a functional classical CACTA transposon that could provide a source of transposase for these MITEs.
Impact on genome
MITEs related to CfTEC are highly reiterated in the Florida carpenter ant genome. They constitute approximately 0.63 % of the entire genome. These results might be consistent with the hypothesis that massive proliferation of MITEs may be associated with more repetitive genomes in both the plant and the animal kingdoms (Tu 2000). In addition, amplification of MITEs in the ant might also provide an opportunity for genetic innovation. However, MITEs related to CfTEC is probably an underestimate because genome regions of the Florida carpenter ant that cannot be assembled are enriched in repeats (Bonasio et al. 2010).
MITEs in many plants and in insects are frequently discovered in the flanking regions and introns of genes (Lu et al. 2012; Han et al. 2010). It also showed that MITEs might provide coding sequences for genes and could also regulate the expressions of host genes in which they reside (Oki et al. 2008; Naito et al. 2009). If MITEs contained a regulatory motif, they might upregulate expression of its host genes. Alternatively, MITE may downregulate expression of its host genes through MITE-derived small RNAs (Lu et al. 2012). In this study, 34 % (838/2475) of MITEs in the Florida carpenter ant were present near or in genes. Therefore, it will be very interesting to investigate whether or not, and to what extent, these MITEs contribute to the evolution of gene regulation in the Florida carpenter ant.
References
Benjak A, Boué S, Forneck A, Casacuberta JM (2009) Recent amplification and impact of MITEs on the genome of grapevine (Vitis vinifera L.). Genome Biol Evol 1:75–84
Bonasio R, Zhang G, Ye C, Mutti NS, Fang X, Qin N, Donahue G, Yang P, Li Q, Li C et al (2010) Genomic comparison of the ants Camponotus floridanus and Harpegnathos saltator. Science 329:1068–1071
Buchmann JP, Löytynoja A, Wicker T, Schulman AH (2014) nalysis of CACTA transposases reveals intron loss as major factor influencing their exon/intron structure in monocotyledonous and eudicotyledonous hosts. Mob DNA 5:24
Bureau TE, Wessler SR (1992) Tourist: a large family of small inverted repeat elements frequently associated with maize genes. Plant Cell 4:1283–1294
Capy P, Bazin C, Higuet D, Langin T (1998) Dynamics and evolution of transposable elements. In Molecular biology intelligence unit. Landes Bioscience, Austin, p 197 (North American distributor Chapman & Hall)
Chen Y, Zhou F, Li G, Xu Y (2009) MUST: a system for identification of miniature inverted-repeat transposable elements and applications to Anabaena variabilis and Haloquadratum walsbyi. Gene 436:1–7
Cui Z, Geurts AM, Liu G, Kaufman CD, Hackett PB (2002) Structure-function analysis of the inverted terminal repeats of the sleeping beauty transposon. J Mol Biol 318:1221–1235
DeMarco R, Venancio TM, Verjovski-Almeida S (2006) SmTRC1, a novel Schistosoma mansoni DNA transposon, discloses new families of animal and fungi transposons belonging to the CACTA superfamily. BMC Evol Biol 6:89
Edgar RC (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797
Elsik CG, Tayal A, Diesh CM, Unni DR, Emery ML, Nguyen HN, Hagen DE (2016) Hymenoptera genome database: integrating genome annotations in Hymenoptera mine. Nucleic Acids Res 44:D793–D800
Feschotte C, Pritham EJ (2007) DNA transposons and the evolution of eukaryotic genomes. Annu Rev Genet 41:331–368
Feschotte C, Zhang X, Wessler SR (2002) Miniature inverted-repeat transposable elements (MITEs) and their relationship with established DNA transposons. In: Craig NL, Craigie R, Gellert M, Lambowitz AM (eds) Mobile DNA II. American Society for Microbiology Press, Washington, pp 1147–1158
Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser 41:95–98
Han Y, Wessler SR (2010) MITE-Hunter: a program for discovering miniature inverted-repeat transposable elements from genomic sequences. Nucleic Acids Res 38:e199
Han MJ, Shen YH, Gao YH, Chen LY, Xiang ZH, Zhang Z (2010) Burst expansion, distribution and diversification of MITEs in the silkworm genome. BMC Genom 11:520
Han MJ, Xu HE, Zhang HH, Feschotte C, Zhang Z (2014) Spy: a new group of eukaryotic DNA transposons without target site duplications. Genome Biol Evol 6:1748–1757
Hirochika H (2001) Contribution of the Tos17 retrotransposon to rice functional genomics. Curr Opin Plant Biol 4:118–122
Holyoake AJ, Kidwell MG (2003) Vege and Mar: two novel hAT MITE families from Drosophila willistoni. Mol Biol Evol 20:163–167
Li WH (1997) Molecular evolution. Sinauer Associates, Sunderland
Lu C, Chen J, Zhang Y, Hu Q, Su W, Kuang H (2012) Miniature inverted-repeat transposable elements (MITEs) have been accumulated through amplification bursts and play important roles in gene expression and species diversity in Oryza sativa. Mol Biol Evol 29:1005–1017
Naito K, Cho E, Yang G, Campbell MA, Yano K, Okumoto Y, Tanisaka T, Wessler SR (2006) Dramatic amplification of a rice transposable element during recent domestication. Proc Natl Acad Sci USA 103:17620–17625
Naito K, Zhang F, Tsukiyama T, Saito H, Hancock CN, Richardson AO, Okumoto Y, Tanisaka T, Wessler SR (2009) Unexpected consequences of a sudden and massive transposon amplification on rice gene expression. Nature 461:1130–1134
Nicholas KB, Nicholas HB, Deerfield DW (1997) GeneDoc: analysis and visualization of genetic variation. EMBNEW News 4:14
Oki N, Yano K, Okumoto Y, Tsukiyama T, Teraishi M, Tanisaka T (2008) A genome-wide view of miniature inverted-repeat transposable elements (MITEs) in rice, Oryza sativa ssp. japonica. Genes Genet Syst 83:321–329
Rice P, Longden I, Bleasby A (2000) EMBOSS: the European molecular biology open software suite. Trends Genet 16:276–277
Santiago N, Herraiz C, Goni JR, Messeguer X, Casacuberta JM (2002) Genome-wide analysis of the Emigrant family of MITEs of Arabidopsis thaliana. Mol Biol Evol 19:2285–2293
Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol 24:1596–1599
Tu Z (2000) Molecular and evolutionary analysis of two divergent subfamilies of a novel miniature inverted repeat transposable element in the yellow fever mosquito, Aedes aegypti. Mol Biol Evol 17:1313–1325
Tu Z (2001) Eight novel families of miniature inverted repeat transposable elements in the African malaria mosquito, Anopheles gambiae. Proc Natl Acad Sci USA 98:1699–1704
Xia X, Xie Z (2001) DAMBE: software package for data analysis in molecular biology and evolution. J Hered 92:371–373
Yuan YW, Wessler SR (2011) The catalytic domain of all eukaryotic cut-and-paste transposase superfamilies. Proc Natl Acad Sci USA 108:7884–7889
Zhang HH, Xu HE, Shen YH, Han MJ, Zhang Z (2013) The origin and evolution of six miniature inverted-repeat transposable elements in Bombyx mori and Rhodnius prolixus. Genome Biol Evol 5:2020–2031
Zhou Y, Cahan SH (2012) A novel family of terminal-repeat retrotransposon in miniature (TRIM) in the genome of the red harvester ant, Pogonomyrmex barbatus. PLoS One 7:e53401
Acknowledgments
This work was supported by the National Science Foundation of the People’s Republic of China (No. 31360371) and the Natural Science Foundation of Jiangxi Province (No. 20161BAB214151).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
This article does not contain any studies with human subjects or animals performed by any of the authors
Conflict of interest
SHJ declares that he has no conflict of interest. GYL declares that he has no conflict of interest. XMX declares that she has no conflict of interest.
Additional information
S.-H. Jiang and G.-Y. Li contributed equally.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Jiang, SH., Li, GY. & Xiong, XM. Novel miniature inverted-repeat transposable elements derived from novel CACTA transposons were discovered in the genome of the ant Camponotus floridanus . Genes Genom 38, 1189–1199 (2016). https://doi.org/10.1007/s13258-016-0464-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13258-016-0464-9