Abstract
The complete sequencing of Mycobacterium avium subspecies paratuberculosis (MAP) strain K-10 genome has shown the existence of over 4,000 genes. This opens up many opportunities to study the interaction of MAP with its hosts and the environment. Understanding the immune response directed at MAP antigens at different stages of infection, would be enhanced by the characterization of putative antigens. In this context, our comprehensive in silico analysis of MAP1138c, a putative protein demonstrates its sequential, physicochemical, structural and functional homology with Rv1411c (LprG) lipoprotein. The InterPro Scan studies have also shown that MAP1138c protein is a member of LppX/LprAFG family of lipoprotein involved in cell wall biogenesis and pathogenesis of Mycobacterium species. The structure assessment tools reveal that the theoretical structure of MAP1138c protein generated by SWISS-MODEL server shows homology with the crystal structure of Rv1411c (LprG) lipoprotein with respect to the global, local and stereochemical properties. Additionally, the structure-based ligand interaction studies using AutoDock Vina 1.1.2 shows that the triacylated glycoprotein (Ac1PIM2) also interacts with the hydrophobic pockets in the 3D theoretical structure of MAP1138c protein. Similar interactions of Rv1411c (LprG)-Ac1PIM2 leads to Toll-like Receptor 2 (TLR-2) mediated evasion of immune responses within host macrophages in tuberculosis infection. Hence, these results support our hypothesis that the MAP1138c protein is probably involved in immune evasion within host macrophages leading to virulence and infection in ruminants and human, respectively.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Johne’s disease is of foremost concern to the dairy industry worldwide (Nielsen and Toft 2009). Mycobacterium avium subspecies paratuberculosis (MAP) is the causal agent of this alarming disease of the ruminants and is associated with the Mycobacterium avium complex (MAC). MAC includes mild environmental bacteria as well as unscrupulous pathogen of humans (Turenne et al. 2007). Like any other Mycobacterial species, MAP is also specialized in the blockage of phagosome-lysosome fusion, thereby sustaining its survival (Kuehnel et al. 2001). Furthermore, they provide anti-apoptotic signals lengthening the life span of their host cell. The interaction of MAP with macrophages are mediated through variety of receptors, as well as Toll-like Receptors (TLRs) (Ferwerda et al. 2007; Koets et al. 2010). MAP can amplify interleukin (IL)-10, an immunomodulatory cytokine that represses killing of MAP by macrophages. It also triggers Th1-type immune responses which are mainly preferred to combat intracellular infections by inhibition of IL-12 (Weiss et al. 2002, 2005). Macrophages are one of the important cells of the innate immune system, but also provide signals to induce adaptive immune responses. MAP can interfere with antigen presentation, hence consequent adaptive responses (Weiss and Souza 2008). Correspondingly, the host immune response to Mycobacterial sp., namely Mycobacterium tuberculosis (M tb) is generally mediated by a TLR-2 in macrophages (Yoshida et al. 2009; Jo et al. 2007; Stenger and Modlin 2002). Mtb non-acylated lipoprotein LprG (Rv1411c) has a TLR-2 agonist action that is dependent on its association with triacylated glycolipids binding specifically with the hydrophobic pocket of Mtb LprG lipoprotein protein. The detection of a glycolipid carrier function has important implications on the role of LprG in Mycobacterial physiology (Kuehnel et al. 2001; Gehring et al. 2004). Continued exposure (>16 h) of human macrophages to LprG (acylated agonist of TLR2) results in noticeable inhibition of MHC-II Ag processing. Moreover, this inhibition depends on TLR-2. MHC-II Ag processing inhibition by mycobacterial lipoproteins may permit M. tuberculosis within infected macrophages to evade the innate immune response of the host that results in virulence and latency (Weiss et al. 2002; Drage et al. 2010). On the contrary, we know little about the evasion and survival mechanism of MAP within the host macrophages, a process common to all mycobacterial infections. Recently, it was proposed by Hassan et al. (2014) that MAP1138c a putative protein shares a structural and functional homology with Rv1411c lipoprotein. Therefore, a proper functional and structural characterization of the putative antigenic proteins will be useful in understanding the evasion and survival mechanism of MAP within host macrophages.
In this regard, the present comprehensive comparative proteomic study of MAP1138c and Rv1411c lipoproteins shows that the putative protein MAP1138c shares both structural and functional homology with Rv1411c or LprG lipoprotein. The structure-based interaction studies of the theoretical 3D structure of MAP1138c protein and triacylated glycolipid shows that the Ac1PIM2 (triacylated glycolipids) binds specifically to the hydrophobic pockets of MAP1138c protein in a similar way as it binds to the hydrophobic pockets of the crystal structure of Rv1411c or LprG lipoprotein. Thus, we propose that MAP1138c-Ac1PIM2 binary complex acts as an agonist for TLR-2 receptor and eventually leads to TLR-2 dependent inhibition of antigen processing within host macrophages and eventually reduced recognition by CD4+ T cells. This mechanism of action may lead to immune elusion and latency of MAP within host macrophages.
Materials and Methods
Sequence retrieval
The accession numbers for MAP1138c and Rv1411c proteins were acquired from UniProtKB (http://www.uniprot.org/). FASTA sequences of both MAP1138c and Rv1411c (LprG) proteins studied were obtained from protein database (www.ncbi.nih.gov/protein). The protein sequences in FASTA format were used to compare the physicochemical, structural and functional properties of MAP1138c and Rv1411c (LprG) proteins.
Sequence analysis
Gapped-Basic Local Search Tool (BLAST) proposed by Altschul and Madden (1997) was used by the SWISS-MODEL server to identify homologs of the query protein (MAP1138c) against the ExPDB template library of the Swiss-Model server.
Phosphorylation sites evaluation
Comparative evaluation of the serine and threonine phosphorylation locations in the orthologous proteins (MAP1138c and Rv1411c (LprG)) was executed using NetPhosBac 1.0 server (Miller et al. 2009) (http://www.cbs.dtu.dk/services/NetPhosBac 1.0).
Secretory nature profiling
The existence of signal sequence and signal peptide cleavage site in the orthologous proteins (MAP1138c and Rv1411c (LprG)) was evaluated and compared using SignalP 4.1 server (Petersen et al. 2011) (http://www.cbs.dtu.dk/services/SignalP).
Globular/order and disorder regions prediction
The globular structures/order and disorder regions between MAP1138c and Rv1411c (LprG) protein was compared using GlobPlot (Linding et al. 2003) (http://globplot.embl.de/cgiDict.py).
Hydropathy analysis
The Hydropathy plot of DNASTAR was used to predict and compare the flexible region (Karplus and Schulz 1985), surface probability (Emini et al. 1985) and hydrophilicity (Kyte and Doolittle 1982) and the antigenic index (Jameson and Wolf 1988) of MAP1138c and Rv1411c (LprG) proteins directly from its primary amino acid sequence.
Sequence feature annotation
Domain assignment
The member databases of InterPro (Zdobnov and Apweiler 2001) helps in the identification of protein domains as well as the determination of protein function. InterPro Scan sequence analysis and classification tool were used to detect and compare protein domains and functional sites of the orthologous proteins (MAP1138c and Rv1411c (LprG)).
Secondary, disorder and transmembrane prediction
PSIPRED (Jones 1999) a secondary structure prediction server based on two feed-forward neural networks will be used to compare the secondary structure, disorder region and transmembrane segment of the orthologous proteins (MAP1138c and Rv1411c (LprG)).
Homology modeling and model quality estimation
Model building
Raw model structure of MAP1138c protein was built by means of manual protein modeling server “SWISS-MODEL Workspace” (Biasini et al. 2014; Arnold et al. 2006; Bordoli et al. 2009; Kiefer et al. 2009; Kopp and Schwede 2006; Guex et al. 2009). The N-terminal secretion signal sequence of MAP1138c protein was removed before submitting to the server for model building. The hypothetical model of MAP1138c protein was made based on the results of target template sequence alignment using Promod-II (Guex and Peitsch 1997) and Modeller (Sali and Blundell 1993).
Model quality estimation
Model goodness and reliability of the model MAP1138c protein structure was evaluated by means of QMEAN score (Benkert et al. 2008; Benkert et al. 2009a, b, 2011). The estimated model reliability ranges between 0–1. The absolute model quality was measured by QMEAN Z-score. The QMEAN score obtained by the theoretical model was evaluated against the scores of high-resolution crystal structures of the same size, and a Z-score of the contributing components of QMEAN was determined. The standard Z-score of the high-resolution structure is zero. Finally, obtained model was validated at the SWISS-MODEL server using PROCHECK validation tool. PROCHECK aims to evaluate the similarity of the geometry of the amino acid residues in the predicted 3D structure of MAP1138c protein, when compared to stereochemical parameters obtained from well-refined, high-resolution structures (Laskowski et al. 1993). The analysis of the super-secondary structural motifs in model MAP1138c protein structure was performed using PROMOTIF (Hutchinson and Thornton 1996).
Ligand-binding domain (LBD) analysis
The prediction of ligand-binding sites/domains of MAP1138c protein structure was performed using Fpocket 1.0 (Guilloux et al. 2009) at http://fpocket.sourceforge.net/run_online.html.
Ligand preparation
Structure of triacylated glycolipid (Ac1PIM2) was drawn using Marvin Sketch and energy minimization of the ligand was accomplished using MMFF94 force field. Energy minimization was performed to help the docking program for identifying the bioactive conformer from the local minima.
Molecular docking
The PDB of the 3D theoretical structure of MAP1138c protein generated by SWISS-MODEL server was used for docking using AutoDock Vina 1.1.2 (Trott and Olson 2010). Fpocket web server was employed to detect the binding pockets in MAP1138c protein structure. The docking procedure for the interaction studies of MAP1138c (protein) and Ac1PIM2 (ligand) was fixed to rigid conditions, and the dock grid was set encircling the protein–LBD of MAP1138c protein. Exhaustiveness was set to 20 with all other parameters set to default standards. The docking and simulation studies was performed using the system configuration namely; Windows® seven operating system (OS) with 8 GB of RAM and Intel® Core™ i7 processor. The binary complex (Ac1PIM2-MAP1138c) categorized by their binding energy values, was examined for geometry and docking. After docking, the preeminent binary complex model was selected on the basis of lowest binding energy. Whereas the most appropriate complex (Ac1PIM2-MAP1138c) conformation was chosen on the basis of hydrogen bond interactions between the protein and ligand near the ligand-binding site. The lowermost energy poses specify the maximum binding affinity since high energy produces the unstable conformations. For visualization purpose, AutoDock Vina was used to generate docking poses, which was loaded directly to PyMol (Seelier and Bert 2010). PyMol was used further to produce the images of the protein ligands complex models.
Results
Gapped-BLAST analysis
A gapped-BLAST was performed for MAP1138c protein sequence to obtain its homologs that provided significant functional and structural motifs for the query protein. The gapped-BLAST analysis of the query sequence leads to the identification of a possible template with known X-Ray structure (PDB ID: 3MH8A) having 70 % identity and 83 % positive match (Supplemental Fig. 1). The template was found to be the crystal structure of LprG from H37Rv strain of Mycobacterium tuberculosis.
Analysis of phosphorylation potential of orthologous proteins
The phosphorylation of serine and threonine residues plays a key role in the regulation of host-pathogen interactions and cell signaling. The potency of the phosphorylation reaction is based upon the phosphorylation potential. The amino acids (serine and threonine) phosphorylation potential was calculated using NetPhosBac 1.0 server. The amino acids are having a phosphorylation potential greater than a threshold value are considered effective site of phosphorylation (Miller et al. 2009). Gain of phosphorylation site was observed for MAP1138c protein, where new eight potent phosphorylation sites (one threonine and seven serine residues) were found when compared to Rv1411c (LprG) protein. Whereas, Rv1411c (LprG) protein shows only four serine residues having phosphorylation potential greater than the threshold value (0.5). The predicted phosphorylation site and their position are tabulated in Table 1.
Analysis of signal sequences of orthologous proteins
Estimation of the signal sequence in a protein is important to categorize the protein as secretory or non-secretory protein (Petersen et al. 2011). In this study, MAP1138c and Rv1411c (LprG) proteins were used as query proteins to detect appropriate signal sequence. The results of SignalP 4.1 server for each of the orthologous proteins (Rv1411c (LprG) and MAP1138c) showed that these are preproteins and are secretory by nature due to the presence of secretory signal sequences in the N-terminal region of preprotein (Supplemental Fig. 2a, b). The length of the signal peptide for Rv1411c (LprG) and MAP1138c proteins were found to be 30 amino acids each. The presence of twin-arginine at N-terminal region of the signal peptide for both Rv1411c (LprG) and MAP1138c proteins relates to their translocation across the membrane via Tat translocases. It can also be stated that after translocation there is a possibility of cleavage of the preprotein by SPase I (Paetzel et al. 2002).The mature peptide for MAP1138c and Rv1411c (LprG) proteins were found to be 31–238 and 31–235, respectively. Thus, we conclude that MAP1138c and Rv1411c (LprG) proteins are preproteins and are secretory by nature.
Analysis of the globular and disorder domains in orthologous proteins
Globular and disorder domains in a protein plays significant roles in protein function (Linding et al. 2003). Thereby, a change in the profile of order and disorder region in a protein may lead to a change in the functionality of the protein. MAP1138c and Rv1411c (LprG) proteins of MAP and H37Rv exhibit similar profile of the globular domain. Figure 1a, b graphically represents the comparative GlobPlot profiles of both MAP1138c and Rv1411c (LprG) proteins. The GlobPlot profile of both MAP1138c and Rv1411c (LprG) show absence of low complexity, transmembrane and coil & coil region, thereby highlighting its role as a secretory globular protein.
Analysis of the hydropathy plot of orthologous proteins
Figure 2a, b shows the result of the hydropathy analysis of both MAP1138c and Rv1411c (LprG) proteins using DNASTAR software (Madison, WI, USA) which reveals regions of high antigenic index (potential antigenic determinants). In silico analysis of MAP1138c protein revealed the presence of high antigenic index regions (above zero-reticle): (31–60, 78–85, 90–95, 100–106, 120–134, 140–146, 155–180, 200–207, 210–220 and 230–238 aa). Similarly, for Rv1411c (LprG) protein (31–60, 78–92, 98–105, 118–134, 138–144, 152–175, 198–206, 210–218 and 222–236 aa) amino acids stretches with high antigenic index were observed. The comparative antigenic profiling of MAP1138c and Rv1411c (LprG) proteins were promising as the antigenic contours for both the proteins all along the sequence were nearly similar. Moreover, the flexible regions, surface probability and hydrophilicity plot of both the protein were also identical.
ProtScan domain and functional analysis of orthologous proteins
ProtScan Domain analysis showed that both MAP1138c and Rv1411c (LprG) proteins share a common domain DUF1396 ranging from 40–231 amino acids. This domain comprises of various lipoproteins and as a group; they are known as the LppX/LprAFG family from Mycobacterium species. The members of this family are involved in virulence and localization of complex lipids to the outer membrane of MTB (Bigi et al. 1997). According to MEMSAT3 (Jones et al. 1994), tool both proteins share a similar profile of transmembrane segment at their N-Terminal region. Figure 3a, b shows the presence of a transmembrane segment at the N-terminal region of both MAP1138c and Rv1411c (LprG) proteins. The transmembrane segment of MAP138c protein consists of an outside loop (1–9 a.a), inside helix cap (10–13 a.a), central transmembrane helix segment (14–22 a.a) and outside helix cap (23–26 a.a). Similarly, the transmembrane segment of Rv1411c protein constitute of an outside loop (1–10 a.a), inside Helix cap (11–14 a.a), central transmembrane helix segment (15–23 a.a) and outside helix cap (24–27 a.a) for Rv1411c (LprG) protein. The N-terminal region of MAP1138c protein corresponds to the signal sequence as predicted by SignalP 4.1. Therefore, the presence of central transmembrane helix at the C-terminal region of the signal sequence of the protein is important for anchoring the protein to the outer membrane of the bacteria during secretion across the membrane.
Template search and homology modeling
A theoretical model of MAP1138c protein was generated using the automated protein-modelling SWISS-MODEL server. The coordinates of crystal structure of Rv1411c (LprG) protein from MTB (PDB ID: 3MH8A) was used as a template. Figure 4 shows the resultant 3D model of MAP1138c protein showing the secondary structure conformations of the same. The superimposition of the crystal structure of 3MH8A with the resultant 3D model of MAP1138c protein depicts secondary structural homology between the predicted 3D model of MAP1138c protein and the crystal structure of Rv1411c (LprG) (Supplemental Fig. 3). Pairwise alignment of MAP1138c (query) and Rv1411c (target) protein sequence displayed 70 % sequence identity and 83 % of sequence similarity (Supplemental Fig. 1).
Model quality evaluation
The model goodness and reliability were evaluated by measuring QMEAN score (Qualitative Model Energy Analysis). Furthermore, a Z-Score (standard score) relative to the scores attained for high-resolution experimental structure of similar size is specified. Absolute quality of the model is measured with the help of Z-Score. The Z-Score of the individual component of QMEAN and QMEAN Z-score of the predicted model is also illustrated (Fig. 5a, b). The global QMEAN score 0.9 of the predicted model reflect upon the reliability of the model (Fig. 5a).
The QMEAN Z-score value for the predicted structure of MAP1138c protein is 1.33 as the score of all except torsion energy is close to the ideal value. The overall score of QMEAN shows a deviation of approximately one standard deviation from the ideal value that is zero (Fig. 5b). This deviation is considered fair, and the structure can be validated for further studies. The local error estimate of each residue of the modeled protein was estimated and represented using a color ranging from red (unreliable region) to blue (reliable region) (Supplemental Fig. 4). We may distinguish the reliability of generated model since the blue color region occupies a significant portion of the protein sequence and nothing in the potentially unreliable region (red).
Model structure assessment
As expected, the predicted structure of MAP1138c protein, shares secondary structural features with Rv1411c (LprG) protein. The PROMOTIF analysis shows that the MAP1138c protein consists of a single domain of an alpha/beta fold along with a β-sheet. This region consists of eleven anti-parallel strands on one side and six helices (3 [α-helix] + 3 [310 helix]) on the opposite side. Additionally, nine Hairpins, seventeen B-Turns, one G-Turn and five Bulges were featured in the PROMOTIF analysis of the model protein. Between the β-sheet and α-helices lies a large cavity. The entrance to the cavity lies near the beta strands (β 3 to β 6) of modeled MAP1138c protein. The lower part of the molecule has a narrow cavity between a single α/β fold and β 10 and β 11. Similar profile of secondary, super secondary and cavity structure of Rv1411c (LprG) protein is demonstrated in the crystal structure of LprG lipoprotein by Drage et al. (2010). The amino acids are lining the central cavity and portal lies in the hydrophobic stretches of MAP1138c (LprG) protein predicted and represented by the hydrophobicity surface plot (Supplemental Fig. 5). The percentage of strand, alpha helix and 3–10 helices in the predicted model were 13.7, 47.2 and 5.1, respectively. This result confirms the globular nature of MAP1138c protein owing to its higher strand percentage.
The geometry of the predicted model was evaluated using PROCHECK. The Ramachandran plots of generated model structure, as evaluated by PROCHECK, showed overall good quality. For MAP1138c protein structure 85.8 % residues resulted in the core regions, 12.4 % in the generally allowed regions, 0.6 % in the additionally tolerable region and 1.2 % in the forbidden regions (only 9 residues labeled out of 195) of the Ramachandran plot (Fig. 6).
The stereochemical parameters like Chi-chi2, Main-chain and Side-chain parameters were evaluated and declared fit for further investigation. Correspondingly, bond lengths (B.L), bond angles (B.A) and planar groups were 99.8, 98.8 and 89.5 % respectively were within limits. Whereas 0.2, 1.2 and 10.5 % of B.L, B.A and planar groups respectively of the total residues were highlighted above limits. The summary of PROCHECK analysis by SWISS-MODEL server for the theoretical 3D model of MAP1138c protein is pictorially represented (Supplemental Fig. 6).
Analysis of ligand binding sites
Fpocket software identified hydrophobic binding pockets represented by green dot in the cavity between the beta sheet and the helical region of MAP1138c protein (Supplemental Fig. 7). Table 2 shows a list of pocket identified by fpocket in MAP1138c protein and these pockets are ranked according to score obtained.
Protein–ligand docking studies
Figure 7 displays the minimum energy of binding (−9.08e+006 kcal/mol) docked state of Ac1PIM2-MAP1138c protein. The ligand molecule Ac1PIM2 (triacylated glycolipid) is anchored within the hydrophobic pocket, or cavity formed by hydrophobic amino acid residues forming the cavity between the alpha and the beta strands of MA1138c protein structure. The docked structure of the binary complex (Ac1PIM2-MAP1138c) is stabilized by two intermolecular hydrogen bonds is pictorially represented (Supplemental Fig. 8). The oxygen (O6) of the mannose attached to 6-OH position of inositol in Ac1PIM2 with the NH of VAL58 amino acid residue of MAP1138c protein. The hydrogen bond distance between these groups is 3.214 Å. Another hydrogen bond was formed between oxygen (O1) of the mannose attached to 6-OH of inositol in PIM2 with the NH of amino acid VAL133 of MAP1138c protein at a distance of 3.322 Å. Four distinct conformational clusters were found using an RMSD (Root Mean Square Deviation) tolerance of 2.0 A. Table 3 shows the clustering histogram showing the lowest/mean binding energy of the binary complex (MAP1138c-Ac1PIM2) and the number of conformation in each cluster with their RMSD value.
Discussion
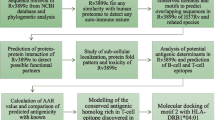
We carried out a comprehensive comparative proteomic, structural and functional analysis of MAP1138c and Rv1411c (LprG) proteins. From our analysis of gapped BLAST, it appears that MAP1138c protein shares a 70 % sequential homology with the crystal structure of Rv1411c (LprG) lipoprotein (Supplemental Fig. 1). A comparative analysis of the ProtParam parameters namely GRAVY, instability index and aliphatic index by Hassan et al. (2014) shows that both MAP1138c and Rv1411c (LprG) proteins share similar physicochemical properties. Both MAP1138c and Rv1411c (LprG) proteins shares a common profile of order and disorder region as predicted by GlobPlot server. These comparison shows that the high percentage of sequence identity between the orthologous protein (MAP1138c and Rv1411c) leads to similarities in their physicochemical and structural properties.
In addition, as evident from the comparative NetPhosBac analysis, MAP1138c protein exhibits additional phosphorylation sites than Rv1411c (LprG) proteins. As specified before, phosphorylation plays a significant role in signaling-transduction process (Paetzel et al. 2002). Therefore, it can be inferred that MAP1138c protein could also interact actively with other proteins leading to signal transduction and cell signaling due to the presence of potential phosphorylation sites in the protein sequence. We also predicted the presence of a signal peptide processing sequence (MLGMQTRRRLSAVFASLTLATALIAGCSSG) having twin Arginine in the N-terminal region of the signal peptide of MAP1138c protein. The presence of twin-arginine suggests that this transcript is a preprotein, and a mature protein is secreted via the Tat pathway and cleaved by SPase I peptidases. The presence of secretory signal sequence indeed could be essential for virulence and pathogenesis of MAP. In our SignalP 4.1 prediction studies, we also found Rv1411c (LprG) protein having secretory property via Tat pathway. We can, therefore, hypothesize that MAP1138c is a preprotein and the mature protein upon cleavage by specific peptidases is secreted out into the host, by SignalP 4.1.
The comparative antigenic profile of MAP1138c and Rv1411c (LprG) proteins are promising since the antigenic contours, flexible regions, surface probability and hydrophilicity for both the proteins all along the sequence were nearly similar. The profile created by antigenic index of DNASTAR (Madison, WI, USA) software for MAP1138c and Rv1411c (LprG) proteins can be used to map the antigenic domains of these proteins and assess the antigenic potential of these peptides in eliciting an innate and differential immune response.
The ProtScan domain analysis for both MAP1138c and Rv1411c (LprG) proteins shows that they share a common domain DUF1396 ranging from 40–231 amino acids. DUF1396 consists of several lipoproteins belonging to LppX/LprAFG family from Mycobacterium species. The best known of these is LprG from MTB (Bigi et al. 1997). Functional characterization of LprG revealed that it halted MHC II antigen processing; a mechanism evolved by MTB to avoid the host MHC-II-restricted CD4+ T cell response within host macrophages (Gehring et al. 2004). LppX is a secreted antigen, which might be a potent target for the design of vaccine against MTB (Al-Attiyah and Mustafa 2004). While LprF is a membrane-associated lipoprotein responsible for signal transduction leading to reaction to osmotic stress (Steyn et al. 2003). In this context, we can assume that MAP1138c is a lipoprotein and possesses a signal sequence of 30 amino acids at the N-terminal region. The presence of the signal sequence direct the preprotein to the cell membrane for secretion via the Tat pathway and eventually cleaved and secreted into the host environment. Therefore, we propose a hypothesis that the MAP1138c protein may play a role in the pathogenesis of paratuberculosis infection by inhibiting the MHC-II mediated antigen processing within the host macrophages. Thus, evading the host MHC-II-restricted CD4+ T cell response, which leads to virulence of M. paratuberculosis infection in ruminants.
Based on the high pairwise sequential identity between the query and target protein, a template structure (protein from Mycobacterium tuberculosis (PDB ID: 3MH8A) was selected. Theoretical model for the MAP1138c protein (query protein) was generated using alignment protein-modelling mode of SWISS-MODEL server. Once the model structure of MAP1138c protein was made, quality assessment tools of SWISS-MODEL server were used to estimate the reliability of the model. Model quality estimation is essential in homology modeling. Hence, mean force potential and empirical force-field methods provided by SWISS-MODEL server were used to validate the quality of the protein model structure.
QMEAN provide scoring functions to estimate the quality of the protein structure model (Benkert et al. 2008, 2009a, b). The QMEAN score reflects the reliability of the whole model and it value ranges from zero (unreliable) to one (reliable). QMEAN score for MAP1138c protein model was found to be 0.92, which means that that the predicted 3D model for a given protein is reliable. QMEAN Z-score evaluates the absolute quality of the query model, and the score is standardized to standard deviation (SD) 1 and mean 0. The Z-score also predicts by much the model structure is deviated from the normalized standard deviation value obtained from the expected values for reference structures. QMEAN Z-scores are calculated for all four structural descriptors (all-atom pairwise, C β interaction, solvation and torsion angle energy) actuality part of the QMEAN score. The investigation of the Z-scores for each structural descriptors can help in detecting the structural features accountable for an observed strongly negative (red region) QMEAN Z-score. Models of good quality are supposedly to have the Z-score values of the individual structural descriptors in the favorable area (light red to blue) (Benkert et al. 2011). The QMEAN Z-score for MAP1138c protein was observed to be 1.33, i.e., <2. The result signifies that the predicted 3D structure of MAP1138c protein shows a standard deviation of approximately one from the template X-Ray structure. This variation is acceptable and allows us to use this predicted structure for further structural and functional analysis of MAP1138c protein. The predicted 3D structure of MAP138c protein falls in the “Good Structure” category since the individual score of the pseudo energies and the QMEAN have values in the light red to the blue region.
Similarly, the predicted three-dimensional structure of MAP1138c protein was evaluated for per residue error in the protein chain. Residue error was estimated and visualized using a color gradient extending from blue (reliable regions) to red (possibly unreliable regions). The local error estimate per residue for MAP1138c protein lies in the more reliable regions (blue color) (Supplemental Fig. 4). The three-dimensional structure of MAP1138c protein shares a similar profile of secondary structures with Rv1411c (LprG) protein demonstrated using PROMOTIF program. Both MAP1138c protein structure and crystal structure of Rv1411c (LprG) protein shares a common profile of hydrophobic amino acids lining the central cavity (ligand binding domain). The presence of hydrophobic amino acids in the ligand binding domain of MAP138c protein structure suggests an analogous mode of protein–ligand (triacylated glycolipids) interactions in both the protein. The structure assessment tool namely Ramachandran plots of PROCHECK analysis was used to evaluate the secondary structure profile of MAP1138c protein model. The Ramachandran plot showed overall good quality since 98.8 % residues of the predicted 3D structure of MAP1138c proteins lies favorable regions of the plot. While, only 1.2 % of the total residues lie in the disallowed regions of the plot. In the same way, stereochemical properties obtained and evaluated for MAP1138c protein model by PROCHECK further shows that the theoretical structure generated by SWISS-MODEL server is worth investigating. Fpocket predicted hydrophobic binding site of the ligand in MAP1138c protein structure complemented well with the results of the docking procedure. The docking and simulation studies revealed that the binary complex of MAP1138c protein and triacylated glycolipids were stabilized by two intermolecular hydrogen bonds. The successful docking procedure showed that Ac1PIM2 (triacylated glycolipid) could bind to the hydrophobic binding pocket of MAP1138c protein of MAP. The binding pocket of MAP1138c protein for TLR-2 agonist (Ac1PIM2) resembles the complex crystal structure of Rv1411c (LprG) lipoprotein and triacylated glycolipid (Ac1PIM2) demonstrated by Drage et al. (2010). This ligand–protein interactions study indicates that the MAP1138c protein may contribute to cell wall assembly and facilitate recognition of triacylated glycolipids by TLR-2 leading to TLR-2 mediated immune evasion within host macrophages. The current study also provides an opportunity to design drug targeting the TLR-2 agonist activity of the binary complex (Ac1PIM2-MAP1138c) apparently contributing to immune evasion and virulence. Additional, functional and biochemical experimental studies are necessary to establish the role of MAP1138c protein in the evasion of the cell-mediated response within host macrophages. Such studies will improve our current knowledge of the mechanisms of pathogenesis and latency of MAP but also will contribute to developing new therapeutic for Johne’s disease.
References
Al-Attiyah R, Mustafa AS (2004) Computer-assisted prediction of HLA-DR binding and experimental analysis for human promiscuous Th1-cell peptides in the 24 kDa secreted lipoprotein (LppX) of Mycobacterium tuberculosis. Scand J Immunol 59:16–24
Altschul SF, Madden TL (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402
Arnold K, Bordoli L, Kopp J, Schwede T (2006) The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics 22:195–201
Benkert P, Tosatto SCE, Schomburg D (2008) QMEAN: A comprehensive scoring function for model quality assessment. Proteins: Struct Funct Bioinform 71:261–277
Benkert P, Kuenzli M, Schwede T (2009a) QMEAN server for protein model quality estimation. Nucleic Acids Res 37:W510–W514
Benkert P, Schwede T, Tosatto SCE (2009b) QMEANclust: estimation of protein model quality by combining a composite scoring function with structural density information. BMC Struct Biol 9:35
Benkert P, Biasini M, Schwede T (2011) Toward the estimation of the absolute quality of individual protein structure models. Bioinformatics 27:343–350
Biasini M, Bienert S, Waterhouse A, Arnold K, Studer G, Schmidt T, Kiefer F, Cassarino TG, Bertoni M, Bordoli L, Schwede T (2014) SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res 42:W252–W258
Bigi F, Espitia C, Alito A, Zumarraga M, Romano MI, Cravero S, Cataldi A (1997) A novel 27 kDa lipoprotein antigen from Mycobacterium bovis. Microbiology 143:3599–3605
Bordoli L, Kiefer F, Arnold K, Benkert P, Battey J, Schwede T (2009) Protein structure homology modelling using SWISS-MODEL workspace. Nat Protoc 4:1–13
Drage MG, Tsai HC, Pecora ND, Cheng TY, Arida AR, Shukla S, Rojas RE, Seshadri C, Moody DB, Boom WH, Sacchettini JC, Harding CV (2010) Mycobacterium tuberculosis lipoprotein LprG (Rv1411c) binds triacylated glycolipid agonists of Toll-like receptor 2. Nat Struct Mol Biol 17:1088–1095
Emini EA, Hughes JV, Perlow DS, Roger J (1985) Induction of hepatitis A virus neutralizing antibody by a virus specific synthetic peptide. J Virol 55:836–839
Ferwerda G, Kullberg BJ, de Jong DJ, Girardin SE, Langenberg DM, van Crevel R, Ottenhoff TH, Van der Meer JW, Netea MG (2007) Mycobacterium paratuberculosis is recognized by Toll-like receptors and NOD2. J Leukoc Biol 82:1011–1018
Gehring AJ, Dobos KM, Belisle JT, Harding CV, Boom WH (2004) Mycobacterium tuberculosis LprG (Rv1411c): a novel TLR-2 ligand that inhibits human macrophage class II MHC antigen processing. J Immunol 173:2660–2668
Guex N, Peitsch MC (1997) SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis 18:2714–2723
Guex N, Peitsch MC, Schwede T (2009) Automated comparative protein structure modeling with SWISS-MODEL and Swiss-PdbViewer: a historical perspective. Electrophoresis 30:S162–S173
Guilloux VL, Schmidtke P, Tuffery P (2009) Fpocket: an open source platform for ligand pocket detection. BMC Bioinform 10:168
Hassan SA, Hasnain SE, Halawani SM (2014) In silico characterization of a putative ORF-MAP1138c of Mycobacterium avium subspecies paratuberculosis (MAP) with its implications in virulence. BMC Genomics 15(Suppl 2):P14
Hutchinson EG, Thornton JM (1996) PROMOTIF—a program to identify and analyze structural motifs in proteins. Protein Sci 5:212–220
Jameson BA, Wolf H (1988) The antigenic index: a novel algorithm for predicting antigenic determinants. Comput Appl Biosci 4(1):181–186
Jo EK, Yang CS, Choi CH, Harding CV (2007) Intracellular signaling cascades regulating innate immune responses to Mycobacteria: branching out from toll-like receptors. Cell Microbiol 9:1087–1098
Jones DT (1999) Protein secondary structure prediction based on position-specific scoring matrices. J Mol Biol 292:195–202
Jones DT, Taylor WR, Thornton JM (1994) A model recognition approach to the prediction of all-helical membrane protein structure and topology. Biochemistry 33:3038–3049
Karplus PA, Schulz GE (1985) Prediction of chain flexibility in proteins. Naturwissenschaften 72:212–213
Kiefer F, Arnold K, Künzli M, Bordoli L, Schwede T (2009) The SWISS-MODEL Repository and associated resources. Nucleic Acids Res 37:D387–D392
Koets A, Santema W, Mertens H, Oostenrijk D, Keestra M, Overdijk M, Labouriau R, Franken P, Frijters A, Nielen M, Rutten V (2010) Susceptibility to paratuberculosis infection in cattle is associated with single nucleotide polymorphisms in Toll-like receptor 2 which modulate immune responses against Mycobacterium avium subspecies paratuberculosis. Prev Vet Med 93:305–315
Kopp J, Schwede T (2006) The SWISS-MODEL repository: new features and functionalities. Nucleic Acids Res 34:D315–D318
Kuehnel MP, Goethe R, Habermann A, Mueller E, Rohde M, Griffiths G, Valentin-Weigand P (2001) Characterization of the intracellular survival of Mycobacterium avium ssp. paratuberculosis: phagosomal pH and fusogenicity in J774 macrophages compared with other mycobacteria. Cell Microbiol 3:551–566
Kyte J, Doolittle RF (1982) A simple method for displaying the hydropathic character of a protein. J Mol Biol 157:105–132
Laskowski RA, MacArthur MW, Moss DS, Thornton JM (1993) PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Cryst 26:283–291
Linding R, Russell RB, Neduva V, Gibson TJ (2003) GlobPlot: exploring protein sequences for globularity and disorder. Nucleic Acids Res 31:3701–3708
Miller ML, Soufi B, Jers C, Blom N, Macek B, Mijakovic I (2009) NetPhosBac-a predictor for Ser/Thr phosphorylation sites in bacterial proteins. Proteomics 9:116–125
Nielsen SS, Toft N (2009) A review of prevalences of paratuberculosis in farmed animals in Europe. Prev Vet Med 88:1–14
Paetzel M, Karla A, Strynadka NCJ, Dalbey RE (2002) Signal peptidases. Chem Rev 102:4549–4579
Petersen TN, Brunak S, Heijne GV, Nielsen H (2011) SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods 8:785–786
Sali A, Blundell TL (1993) Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol 234:779–815
Seelier D, Bert LDG (2010) Ligand docking and binding site analysis with PyMol and Autodock/Vina. J Comput Aided Mol Des 24:417–422
Stenger S, Modlin RL (2002) Control of Mycobacterium tuberculosis through mammalian toll-like receptors. Curr Opin Immunol 14:452–457
Steyn AJ, Joseph J, Bloom BR (2003) Interaction of the sensor module of Mycobacterium tuberculosis H37Rv KdpD with members of the Lpr family. Mol Microbiol 47:1075–1089
Trott O, Olson AJ (2010) AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization and multithreading. J Comput Chem 31:455–461
Turenne CY, Wallace R Jr, Behr MA (2007) Mycobacterium avium in the postgenomic era. Clin Microbiol Rev 20:205–229
Weiss DJ, Souza CD (2008) Review paper: modulation of mononuclear phagocyte function by Mycobacterium avium subsp. Paratuberculosis. Vet Pathol 45:829–841
Weiss DJ, Evanson OA, Moritz A, Deng MQ, Abrahamsen MS (2002) Differential responses of bovine macrophages to Mycobacterium avium subsp. paratuberculosis and Mycobacterium avium subsp. avium. Infect Immun 70:5556–5561
Weiss DJ, Evanson OA, de Souza C, Abrahamsen MS (2005) A critical role of interleukin-10 in the response of bovine macrophages to infection by Mycobacterium avium subsp. Paratuberculosis. Am J Vet Res 66:721–726
Yoshida A, Inagawa H, Kohchi C, Nishizawa T, Soma G (2009) The role of toll-like receptor 2 in survival strategies of Mycobacterium tuberculosis in macrophage phagosomes. Anticancer Res 29:907–910
Zdobnov EM, Apweiler R (2001) InterProScan—an integration platform for the signature-recognition methods in InterPro. Bioinformatics 17:847–848
Acknowledgments
This work is funded by the Deanship of Scientific Research (DSR), King Abdulaziz University, Jeddah under Grant Number (830-263-D1435). The author, therefore, acknowledge with thanks DSR technical and financial support.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Hassan, S.A. In silico approach to identify the role of a putative protein MAP1138c in the virulence of Johne’s disease. Genes Genom 37, 327–338 (2015). https://doi.org/10.1007/s13258-014-0258-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13258-014-0258-x