Abstract
Timely prediction of cardiovascular diseases with the help of a computer-aided diagnosis system minimizes the mortality rate of cardiac disease patients. Cardiac arrhythmia detection is one of the most challenging tasks, because the variations of electrocardiogram(ECG) signal are very small, which cannot be detected by human eyes. In this study, an 11-layer deep convolutional neural network model is proposed for classification of the MIT-BIH arrhythmia database into five classes according to the ANSI–AAMI standards. In this CNN model, we designed a complete end-to-end structure of the classification method and applied without the denoising process of the database. The major advantage of the new methodology proposed is that the number of classifications will reduce and also the need to detect, and segment the QRS complexes, obviated. This MIT-BIH database has been artificially oversampled to handle the minority classes, class imbalance problem using SMOTE technique. This new CNN model was trained on the augmented ECG database and tested on the real dataset. The experimental results portray that the developed CNN model has better performance in terms of precision, recall, F-score, and overall accuracy as compared to the work mentioned in the literatures. These results also indicate that the best performance accuracy of 98.30% is obtained in the 70:30 train-test data set.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Worldwide millions of people are suffering from cardiovascular diseases [1]. According to the reports of World Health Organization, 30% of the world’s deaths are due to cardiovascular diseases [2]. The major cause of the cardiac deaths is cardiac arrhythmia. Cardiac arrhythmia is a condition, where the functioning of the heart is abnormal [3, 4].
Due to arrhythmia, heart rhythm will be either too fast or too slow or irregular from regular rhythm of the heart. Because of arrhythmia, the heart cannot send enough blood to the body parts. Therefore the blood cannot flow in right proportion, due to which the functioning of heart, brain is affected and other parts of the body. Most arrhythmias are harmful to the heart. There are two type of arrhythmia non-life-threatening and life-threatening. Therefore, continuous monitoring of the activity of heartbeats is inevitable. So, it is very important to ascertain the arrhythmia for proper medical care by detecting heart diseases [5,6,7]. The best equipment for monitoring and identifying arrhythmia is an electrocardiogram(ECG).
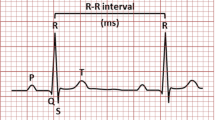
An electrocardiogram is a noninvasive and inexpensive diagnostic device, which is used for monitoring electrical activity of the heart and is also widely applied in several applications [8]. ECG is represented in the form of electrical signals, which shows the electrical activity of the heart over the period and consists of the important physiological data, which is widely used for analysis of heart-beat function. The heart’s electric field is generated by placing the electrode on different parts of the body [9]. ECG signals will construct a sequence of three waves: P wave, Q-R-S waves (known as QRS complex), and T wave, which are repeated periodically in time, so it is a periodic signal as shown in Fig. 1.
Electrocardiogram signal [4]
The ECG signal classification is widely categorized into four phases: data pre-processing, heartbeat peak segmentation [10], feature extraction, and classifiers [11, 12]. The data pre-processing phase emphasizes on noise removal from signal acquisition like power line interference and base-line wander noises [13,14,15]. Heartbeat peak segmentation phase(i.e. the finding of R peaks and Q-R-S complex in the signal) has been used approximately for the last three decades. In this part, heartbeat frequency information has been used and the signal is split into single or multiple periods of heartbeats [16, 17].
The third phase is feature extraction, which plays an important role throughout the classification process and different techniques have been introduced and validated. So in this section, the different manners of feature extractions have been categorized into two parts: the first one is: manual based feature extraction and the second one is learning-based feature extraction. In this paper learning-based feature extraction is used. For manual based methods, features are extracted from frequency domain such as wavelet transform [18,19,20], cosine transform [21], and Fourier transform [22, 23] as well as time domain, using the morphological features of the signal that can be directly extracted from the ECG data. The mostly used morphological features are R–R interval, P–R interval, P duration, Q–R–S duration, T duration, and Q–T intervals [24, 25]. Literature [26] has also explored some higher-order statistical features.
The traditional classical features, which were extracted from simple handcraft, cannot provide abundant and inner information. Learning-based feature extraction, also known as fine features and also deeper. Some dimension reduction techniques such as principal component analysis and independent component analysis, have been explored deeper features from the ECG signals [27, 28]. In [29] author applied an S-transform method for feature extraction after that proposes a learning method, based on supervised regularization-based robust subspace for learning low-dimensional vital feature from the subspace of original features. In [30] authors explored a different kind of features, such as the perspective of visual level feature combinations and view level feature combinations. The best result was obtained when all the visual features were combined.
In these techniques, the principal information may be preserved by projecting a low-dimensional space [31]. Some artificial neural network methods influence the great capacity of the networks, having many layers and several parameters, trained iteratively for extracting the course to learn features hierarchically. The features extracted from ANN are more beneficial than traditional handcrafted features. The traditional CNN has explored significant power for extracting features. Additionally, two methods: feature extraction and classification are integrated into CNN based classifiers [32, 33]. In traditional methods, both are separated. Hence this paper is based on CNN. In the last phase, various ECG signal classification techniques that are generic and fully automatic are used. The classifiers that are widely used for correct class prediction are support vector machine [34,35,36], Random-forest [37, 38], ANNs, and deep-learning [39,40,41].
Recently deep learning techniques have been applied [45] successfully for the diagnosis of ECG heartbeats and diseases. The deep-CNN [43], deep-neural network [33], and deep-belief network [44], have become very popular in ECG arrhythmia classifications. In this paper, a new CNN model has been proposed by making considerable modifications to the CNN models in previous works [46, 57, 61]. Similarly the change in number of convolutional layer, convolutional kernel size, stride, number of neurons in fully connected layers and also modify the learning rate of the CNN model can be done. In [46], the CNN model was trained and tested on the oversampled ECG database, so the result may be biased because validation was performed on the oversampled test dataset. In this paper, validation has been performed on real dataset, while the CNN model has been trained with the oversampled dataset.
Therefore in this paper, we introduced a new model, based on the CNN model for classification of ECG heartbeats into five groups, in order to significantly enhance the efficiency and effectiveness of the classification. The proposed model is highly preferable for the implementation of real-time monitoring of ECG for mobile devices. The overall proposed model consists of four steps, first normalization, second segmentation, third data-augmentation, and finally classification of the ECG signals. Since CNN requird large number of data samples for training the model that can reliably detect the multiclass classification problem. So in this study, for data augmentation technique is used which partially fulfill both requirements as class imbalence problem and number of data samples. Augmentation technique also enhance the performance of the CNN model. Once the network is trained the diagnosis of ECG signal is fast. The rest of the paper is organized as follows: Second section describes the methodology of ECG signal classification. The database used and results are described in section three. Section four discribe the discussion. Finally, the conclusion of this article is presented in section five.
Methodology
The MIT-BIH Arrhythmia database has been taken to analyse the performance of the proposed CNN model. The proposed model involves four main steps such as database normalization, heartbeat segmentation, class imbalance process, and classification. The block diagram of the proposed model is presented in Fig. 2.
ECG signal normalization
Normalization means scaling the signal at an identical level. All ECG signal values have been normalized by applying the Z-score normalization method. Generally, a Z-score is a value of standard deviation from the mean value of the given samples. This method resolves the problem of amplitude scaling and vanishing the offset effects. The Z-score normalized signal values from dataset are obtained by the following formula [47].
In MIT-BIH arrhythmia database, 48 subjects records are presented. each ECG signal record files are normalized individually. First, calculate the mean and standard deviation of each record files and then normalized the each sample value of ECG record file according to equation one.
Heart-beat segmentation
The MIT-BIH arrhythmia database continuously records the signals. To obtain individual heartbeats, ECG signals are segmented and stored using the annotation file information given by the cardiologists. In the segmentation process, firstly find all the peak values from the given annotation file information and then based on the peak values, entire ECG signal sets are divided into segments. The length of each segment is 360 samples long and is centered on the peak locations [48] that are 179 samples taken before the R-peak value and 180 samples have been taken after the R-peak value in each segment according to the given annotation files information.
Imbalance process
The MIT-BIH ECG arrhythmia database is highly a non-uniform distribution of heartbeats in the different classes. There are approximately 80% (According to ANSI-AAMI) hearts beats, that belong to one class and the remaining 20% of the heartbeats belong to the other four classes, so this dataset is referred to as a class imbalance problem because the number of samples in the majority class is a hundred folds higher than that of the minority class samples.
The approaches used for handling the class imbalance problems are as follows:
Algorithm level
Data level
Hybrid of algorithm and data.
In this paper, data level approach is applied, namely Synthetic minority oversampling technique (SMOTE) [49].
Synthetic minority oversampling technique (SMOTE)
It is a most widely used oversampling method introduced by Chawla in 2002 [49]. This method generates new synthetic minority samples without duplicating the samples. The generation of synthetic data is done by the following equation.
where \(x^i\) represents the instance of minority class under consideration, \(x^j\) is a randomly selected instance from the K-nearest neighbours of minority instance \(x^i\). \(\delta\) is a vector in which every element is a random number from [0, 1]; it is used to generate a synthetic instance in reference to two original instances \(x^i\) and \(x^j\) respectively, which are also known as primary reference instance and assistant reference instance respectively. xsynthetic is generated along the line between \(x^{i}\) and \(x^j\) and direction of line joining towards \(x^i\). It also minimizes the risk of over-fitting.
In this way, synthetically created balancing set is formed that contains an equal number of instances in training dataset of each class. According to AAMI standard, ECG signal database is divided into five classes’ namely normal, supraventricular, ventricular, fusion, and unknown class. The total samples of class N are not oversampled as they are available in enough quantity. The classes (S, V, F, & Q) are oversampled to increase the number of samples in order to match the total number of samples of class N [50]. Table 1 describes the total number of heart beats before and after synthetic data generation in training dataset.
Convolutional neural network model
The property of CNN architecture is that it automatically emphasizes and extracts the most suitable features from the input data by applying convolution operation. It keeps more concentration on the local features and its location among the other features. Otherwise, on the same feature maps on CNN, the weights of neurons are the same resulting in network learning in parallel, which markedly saves the learning time [51]. The proposed architecture of CNN model for heartbeat classification is shown in Fig. 3.
In this paper, four convolutional layers, the first, third, fifth, and seventh are convolved along with filter size 27, 14, 3, and 1 according to the Eq. 3.
where Y represents the output vector, x represents the signal values, f denote the filters, and N denotes the number of elements.
After each convolutional layer, a second layer max-polling is used, which aims to diminish the size of the feature map. The Brute force technique is used in this task for the obtained parameter of filter (kernel) size in the layers 1, 3, 5, and 7. The rectifier linear unit (ReLU) [52] activation function is used for layer first, third, fifth, seventh, ninth, and tenth. There are three fully connected layers ninth, tenth, and eleventh consisting of 148, 50, and 5 outputs neurons, respectively. The final layer (11) use the soft-max function to classify each output class separately (as N, S, V, F, & Q). In this model, CNN was trained using Adam optimizer technique. The training parameters are set as follows: learning rate is \(1e-3\), and regularization is 0.01 respectively. The CNN model uses a batch size of 34. The loss parameter is calculated using the categorical cross-entropy function with SMOTE technique used on the database and weighted categorical cross-entropy function without SMOTE technique used on the database. The CNN model has been trained and tested iteratively. The iteration count has been taken are 30. The testing is also performed after every iteration. The proposed CNN model architecture is summarized in Table 2.
Results
In this work, we have taken ECG signals from the open-source Physio-Bank ATM MIT-BIH Arrhythmia datasets [53]. The Arrhythmia database was composed by gathering data 48 ECG recordings from 47 subjects in which 22 women databases aged between 23 to 89 years old and 25 men database aged between 32 to 89 years old. The dataset specifies 1,09,449 labelled cardiac beats of 15 different heartbeat classes. In accordance with ANSI/AAMI EC 57: 1998 standard, four records 102, 104, 107, 217 contain paced beats. These records are removed from the evaluation database. Further, the heartbeats are categorized into 15 original classes of the MIT-BIH arrhythmia database are rearranged into five categories according to ANSI/AAMI standard.
Performance of heartbeats detection according to ANSI-AAMI standards is compared. Observational results show that the CNN model for heartbeats classification is effective and also analyses the computational complexity. The proposed CNN model is implemented in Win-Python 3.6.6 using Keras libraries. All observations are performed on Dell workstation with dual Intel Xeon E5-2600, 2.4 GHz processor, and 64GB RAM. The average minimum CPU time is 296 and maximum is 489 seconds per epochs in order to complete the training and testing of the ECG database.
The performance of CNN model was evaluated in terms of accuracy (Acc), Recall (Se), Precision (P), and F-score. These parameters are determined by the value of true positive (TP), false positive(FP), false negative (FN), true negative (TN), and the whole instances presented in the database (\(\sum\)) are as follows:
In this study, first, the real data is divided into different sets of training and testing dataset. In the first section, 50% of real data was oversampled using SMOTE technique; then this oversampled data was used for training of proposed CNN model. The trained CNN model has then used on 50% real data for testing phase, which that the model had never seen in the learning phase. Similarly, in the second, third, fourth, and fifth section 60%, 70%, 80%, and 90% real data were oversampled using SMOTE technique. Then this oversampled data were applied for the training of proposed classification CNN model. The trained CNN model used the rest 40%, 30%, 20%, and 10% real data respectively for testing of the model, which had never seen in the learning phase. Figs. 4, 5 shows that the testing accuracy and loss performance graphs used learning rate of 0.001 of the proposed CNN model during 30 iterations for five class’s heartbeat classification.
Table 3 tabulates the performance of the CNN model classification on various test datasets such as 50%, 40%, 30%, 20%, and 10% respectively. This table specify the average precision, average recall, average F-score and overall accuracy that were calculated for each class analysis based on confusion matrix evaluation according to Eqs. 4 to 7. The overall best testing accuracy of the CNN model for five class classification was 98.30%, which was obtained by using training test ratio 70:30. The training and test dataset were randomly selected while adjusting the hyper parameters of CNN model. The statistical performance shows that the best results are obtained in the model.
The confusion matrix of the 11-layer CNN classifier model has obtained the best overall accuracy using training test ratio 70:30 and learning rate is 0.001, as shown in the Table 4. It maps the right and wrong identification of heartbeats.
Figure 6 shows the average CPU time is taken to train per heartbeat signals (segment) of the proposed CNN model and to use in Ref. [46, 57].
Discussion
Table 5 summarizes the performance comparison works published in various ECG classification using machine learning and deep learning techniques. Those techniques have obtained highest overall classification performance using the MIT-BIH arrhythmia database. The performance of our 11-layer CNN model was comparable with the best obtained existing work performance, thus validation of the effectiveness of our proposed CNN model in classifying the MIT-BIH arrhythmia database. Kutulu et al. [54], have developed a multistage automatic classification system. In this system, 150 features were extracted with the help of higher-order statistics, wavelet packet decomposition, morphological features, and Fourier transform by applying a combination of 5 K-NN classifiers. This work has achieved an accuracy of 93.49% using 3345 beats of training and 2542 beats of testing set.
In [55], a hybrid approach using segmented and fixed interval wave shape; that switches between those two features resulted in the best multi-way classification performance. In the work [24], four types of heartbeats were classified based on non-normalized and normalized R–R intervals. The classification of the heart beats has been done with the help of the linear discriminate classifiers and the morphological features were extracted by linear prediction modelling and wavelet analysis. In [59], Hermit function coefficients and R-R intervals have been used as a feature vectors. The block based neural network trained with particle swarm optimization technique was used for optimizing the structure and weights of the neural networks.
In [57], author proposed a 7-layer, 1-Dimensional CNN model for the classification of ECG beats. In this model, the input segment length was 128 samples centered on the R-peak. The learning rate was initially set to 0.001 and has been changed in accordance with the mean square errors. For fair comparision with [57], we have also been trained and tested our model without using SMOTE method, and results are shown in appendix, which gives better overall accuracy. In [61], kachuee et al. proposed a model based on deep CNN for heartbeat classification. Specifically, the model was trained with residual connections for detecting the heartbeats. In [59], a Principal Component Analysis network has been used by the author for extracting the features based on a raw ECG signal. Targeting at the improvement of classification accuracy, Linear SVM was applied, and then the accuracy achieved is 97.77%. In [46], Acharya et al. has proposed a 9-layer deep CNN model for classifying the heartbeats using synthetic data generation for oversampling of minority classes, while we have used 11-layer deep CNN model using SMOTE method. In this model [46], the convolutional layers have the kernel size 3, 4, and 4 respectively, while our model used kernel size 27, 14, 3, and 1 respectively. Heartbeat segment size was 260 samples long, while our’s sement size was 360 samples long. The three fully connected layers in [46] have consisted the number of output neurons 30, 20, and 5 respectively, while our model consist of 148, 50, and 5, respectively. In [46], leaky-Relu activation functions with learning rate \(3\times1e-3\) was used while in our model used Relu activation functions with learning rate \(1e-3\). The performance results of this paper may be optimistically biased since both the training and validation sets were oversampled using synthetic data generation.
In the methodical literature, the works focus on the classification of heartbeats into five classes. A total number of 44 files in the MIT-BIH database are used for the purpose of performance evaluation. From Table 5, it can be seen that we have obtained the overall best accuracy of 98.30% for recognition of 5-classes heartbeat. Our proposed model has given better results than the works summarized in the above table. We have also implemented categorical cross-entropy loss and weighted categorical cross-entropy loss [62] for imbalenced dataset without using SMOTE method. The results are shown in the “Appendix” section in Tables 6 and 7. In Table 6, the results are poor compared to Table 3 results, but in Table 7 the overall accuracy in little bit improved compared to Table 3, while recall and F-score is not good. Since in healthcare point of view recall plays an important role in correctly identifying disease. Thus compared to Tables 6 and 7, Table 3 shows better recall and F-score of the heartbeat detection in the MIT-BIH arrhythmia database using SMOTE method. This proposed CNN model is easy to use and less complex because in this model, data has been fed without performing the de-noising process and feature extraction, selection, classification were combined into end-to-end CNN model.
Conclusion
The objective of this paper is to implement a deep learning CNN model that which automatically classifies the ECG heartbeat according to the AAMI standards. The performance of the novel CNN model that correctly classifies heartbeat classes was found to be improved after generating the synthetic data using SMOTE technique and reducing the minority class imbalance problem. This model is fully automatic, hence there is no requirement of an additional system like feature extraction, feature selection, and classification. Once the model is trained, it can be applied to predict ECG signals for arrhythmia patients. By performing experiments on the arrhythmia database (MIT-BIH database), our model shows greater effectiveness and efficiency for ECG signal classification. Due to less computational complexity, the proposed CNN model can be used for classifying long term ECG signals and detecting disease events in real time. It is very much effective for wearable devices.
References
Mehra R (2007) Global public health problem of sudden cardiac death. J Electrocardiol 40(6):S118–S122
World Health Organization (2017) Noncommunicable diseases: progress monitor 2017
Hadhoud MM, Eladawy MI, Farag A (2006) “Computer aided diagnosis of cardiac arrhythmias.” Computer Engineering and Systems, The 2006 International Conference on. IEEE
Singh S et al (2018) Classification of ECG arrhythmia using recurrent neural networks. Proced Comput Sci 132:1290–1297
De Chazal P, O’Dwyer M, Reilly RB (2004) Automatic classification of heartbeats using ECG morphology and heartbeat interval features. IEEE Trans Biomed Eng 51(7):1196–1206
Alonso-Atienza F et al (2014) Detection of life-threatening arrhythmias using feature selection and support vector machines. IEEE Trans Biomed Eng 61(3):832–840
Khadra L, Al-Fahoum AS, Al-Nashash H (1997) Detection of life-threatening cardiac arrhythmias using the wavelet transformation. Med Biol Eng Comput 35(6):626–632
Sörnmo L, Laguna P (2005) Bioelectrical signal processing in cardiac and neurological applications, vol 8. Academic Press, Amsterdam
Melillo P et al (2015) “Wearable technology and ECG processing for fall risk assessment, prevention and detection.” Engineering in Medicine and Biology Society (EMBC), 2015 37th Annual International Conference of the IEEE. IEEE
Jain S et al (2017) QRS detection using adaptive filters: a comparative study. ISA Trans 66:362–375
Adam G, Witold P (2011) ECG signal processing, classification and interpretation: a comprehensive framework of computational intelligence. Springer, London
Singh BN, Tiwari AK (2006) Optimal selection of wavelet basis function applied to ECG signal denoising. Digital Signal Proc 16(3):275–287
Sameni R et al (2007) A nonlinear Bayesian filtering framework for ECG denoising. IEEE Trans Biomed Eng 54(12):2172–2185
Pasolli E, Melgani F (2015) Genetic algorithm-based method for mitigating label noise issue in ECG signal classification. Biomed Signal Proc Control 19:130–136
Roonizi EK, Sassi R (2016) A signal decomposition model-based Bayesian framework for ECG components separation. IEEE Trans Signal Proc 64(3):665–674
da Silva Luz EJ et al (2016) ECG-based heartbeat classification for arrhythmia detection: a survey. Comput Methods Programs Biomed 127:144–164
Pinheiro E, Postolache O, Girão P (2011) “Method for segmentation of cardiac signals based on four parameter sine fitting.” EUROCON-International Conference on Computer as a Tool (EUROCON), 2011 IEEE. IEEE
Ceylan R, Özbay Y (2007) Comparison of FCM, PCA and WT techniques for classification ECG arrhythmias using artificial neural network. Expert Syst Appl 33(2):286–295
Ye C, Coimbra MT, Vijaya Kumar BVK (2010) Arrhythmia detection and classification using morphological and dynamic features of ECG signals. Engineering in Medicine and Biology Society (EMBC), 2010 Annual International Conference of the IEEE. IEEE
Kutlu Y, Kuntalp D (2012) Feature extraction for ECG heartbeats using higher order statistics of WPD coefficients. Comput Methods Progr Biomed 105(3):257–267
Shinde AA, Kanjalkar P (2011) The comparison of different transform based methods for ECG data compression. Signal processing, communication, computing and networking technologies (ICSCCN), 2011 International Conference on IEEE
Gu Z, Zhang N, Qian S (2013) Analyzing electrocardiogram signals with multiscale short-time fourier transforms. U.S. Patent No. 8,494,622. 23 Jul
Uslu E, Bilgin G (2012) Exploiting locality based Fourier transform for ECG signal diagnosis. Applied Electronics (AE), 2012 International Conference on. IEEE
Lin CC, Yang CM (2014) Heartbeat classification using normalized RR intervals and morphological features. Math Probl Eng. https://doi.org/10.1155/2014/712474
Ye Can, Vijaya Kumar BVK, Coimbra MT (2012) Heartbeat classification using morphological and dynamic features of ECG signals. IEEE Trans Biomed Eng 59(10):2930–2941
Martis RJ et al (2013) Cardiac decision making using higher order spectra. Biomed Signal Proc Control 8(2):193–203
Chawla MPS (2009) A comparative analysis of principal component and independent component techniques for electrocardiograms. Neural Comput Appl 18(6):539–556
Imah EM et al (2011) A comparative study on Daubechies Wavelet Transformation, Kernel PCA and PCA as feature extractors for arrhythmia detection using SVM.” TENCON 2011-2011 IEEE Region 10 Conference. IEEE
Lu W et al (2018) Wearable computing for internet of things: a discriminant approach for human activity recognition. IEEE Internet of Things J 6(2):2749–2759
Jing P et al (2018) A framework of joint low-rank and sparse regression for image memorability prediction. IEEE Trans Circuits Syst Video Technol 29(5):1296–1309
Wang JS et al (2013) ECG arrhythmia classification using a probabilistic neural network with a feature reduction method. Neurocomputing 116:38–45
Kiranyaz S et al (2015) Convolutional Neural Networks for patient-specific ECG classification. Engineering in Medicine and Biology Society (EMBC), 2015 37th Annual International Conference of the IEEE. IEEE
Pourbabaee B, Roshtkhari MJ, Khorasani K (2017) Deep convolutional neural networks and learning ecg features for screening paroxysmal atrial fibrillation patients. IEEE Trans Syst Man Cybern 99:1–10
Khalaf AF, Owis MI, Yassine IA (2015) A novel technique for cardiac arrhythmia classification using spectral correlation and support vector machines. Expert Syst Appl 42(21):8361–8368
Varatharajan R, Manogaran G, Priyan MK (2018) A big data classification approach using LDA with an enhanced SVM method for ECG signals in cloud computing. Multimed Tools Appl 77(8):10195–10215
Raj S, Ray KC (2017) ECG signal analysis using DCT-based DOST and PSO optimized SVM. IEEE Trans Instrum Meas 66(3):470–478
Alickovic E, Subasi A (2016) Medical decision support system for diagnosis of heart arrhythmia using DWT and random forests classifier. J Med Syst 40(4):108
Kropf M, Hayn D, Schreier G (2017) ECG classification based on time and frequency domain features using random forests. Computing 44:1
Xiong Z et al (2018) ECG signal classification for the detection of cardiac arrhythmias using a convolutional recurrent neural network. Physiol Meas 39(9):094006
Nurmaini S, Gani A (2018) Cardiac arrhythmias classification using deep neural networks and principle component analysis algorithm. Int J Adv Soft Comput Appl 10(2):14–32
Muthuvel K et al (2015) Classification of ECG signal using hybrid feature extraction and neural network classifier. New Delhi, power electronics and renewable energy systems. Springer, New Delhi, pp 1537–1544
Acharya UR et al (2017) Automated detection of arrhythmias using different intervals of tachycardia ECG segments with convolutional neural network. Inform Sci 405:81–90
Acharya UR et al (2017) Application of deep convolutional neural network for automated detection of myocardial infarction using ECG signals. Inform Sci 415:190–198
Wu Z, Ding X, Zhang G (2016) A novel method for classification of ECG arrhythmias using deep belief networks. Int J Comput Intell Appl 15(04):1650021
Al Rahhal MM et al (2016) Deep learning approach for active classification of electrocardiogram signals. Inform Sci 345:340–354
Acharya UR et al (2017) A deep convolutional neural network model to classify heartbeats. Comput Biol Med 89:389–396
Wikipedia contributors (2018) Feature scaling. Wikipedia, The Free Encyclopedia. Wikipedia, The Free Encyclopedia, 24 Jun 2018. https://en.wikipedia.org/w/index.php?title=Feature-scaling&oldid=847274325
Chen S et al (2017) Heartbeat classification using projected and dynamic features of ECG signal. Biomed Signal Proc Control 31:165–173
Chawla NV et al (2002) SMOTE: synthetic minority over-sampling technique. J Artif Intell Res 16:321–357
Prati RC, Batista GEAPA, Monard MC (2008) A study with class imbalance and random sampling for a decision tree learning system. IFIP International Conference on Artificial Intelligence in Theory and Practice. Springer, Boston
Yi Z et al (2014) Time series classification using multi-channels deep convolutional neural networks. International Conference on Web-Age Information Management. Springer, Cham
Krizhevsky A, Sutskever I, Hinton GE (2012) Imagenet classification with deep convolutional neural networks. Advances in neural information processing systems
Goldberger AL et al (2000) PhysioBank, PhysioToolkit, and PhysioNet: components of a new research resource for complex physiologic signals. Circulation 101(23):e215–e220
Kutlu Y, Kuntalp D (2011) A multi-stage automatic arrhythmia recognition and classification system. Comput Biol Med 41(1):37–45
De Chazal P (2013) A switching feature extraction system for ECG heartbeat classification. Computing in Cardiology Conference (CinC), 2013. IEEE, USA
Raj S, Maurya K, Ray KC (2015) A knowledge-based real time embedded platform for arrhythmia beat classification. Biomed Eng Lett 5(4):271–280
Zubair M, Kim J, Yoon C (2016) An automated ECG beat classification system using convolutional neural networks. IT Convergence and Security (ICITCS), 2016 6th International Conference on. IEEE
Li T, Zhou M (2016) ECG classification using wavelet packet entropy and random forests. Entropy 18(8):285
Shadmand S, Mashoufi B (2016) A new personalized ECG signal classification algorithm using block-based neural network and particle swarm optimization. Biomed Signal Proc Control 25:12–23
Yang W et al (2018) Automatic recognition of arrhythmia based on principal component analysis network and linear support vector machine. Comput Biol Med 101:22–32
Kachuee M, Fazeli S, Sarrafzadeh M (2018) ECG heartbeat classification: a deep transferable representation. arXiv preprint arXiv:1805.00794
Li J, Chen R (2018) Detection of premature ventricular contractions using densely connected deep convolutional neural network with spatial pyramid pooling layer.” arXiv preprint arXiv:1806.04564
Acknowledgements
This study was not funded by any agencies.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
All authors declare that they have no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Appendix
Appendix
Rights and permissions
About this article
Cite this article
Pandey, S.K., Janghel, R.R. Automatic detection of arrhythmia from imbalanced ECG database using CNN model with SMOTE. Australas Phys Eng Sci Med 42, 1129–1139 (2019). https://doi.org/10.1007/s13246-019-00815-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13246-019-00815-9