Abstract
Proso millet (Panicum miliaceum L.) is one of the seven commonly cultivated millets. It is regarded as a climate-smart, ancient, and gluten-free and therefore, is healthy to humans and the environment. The exceptional nutritional properties of the grain resulted in a gradual surge in its demand in the human food market especially for people with diabetes and celiac disease. It is essential to continue the genetic improvement of proso millet to meet its ever-increasing demand. Genetic improvement of proso millet in the United States, however, is impeded by the narrow genetic base in the germplasm and lack of extensive research on its genetics and breeding. There are lots of reports on ‘omics’ (genomics, transcriptomics, proteomics, metabolomics, and phenomics) of many common crops and the technologies are being extensively used in their genetic improvement. However, such studies are scarce in proso millet. The objective of this review article is to summarize available ‘omics’ reports of proso millet and discuss their relevance for its genetic improvement. Relatively more genomics and transcriptomics reports of proso millet are available but only two proteomics and metabolomics reports focusing on grain composition and no phenomics report are available. As more efficient, fast, and cheaper ‘omics’ technologies are available, it is imperative that global proso and other millets breeders and geneticists collaborate strongly for successful utilization of ‘milletomics’ for developing of noble proso millet varieties for future need of this climate-smart superfood grain.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Millets are small-seeded annual cereal grown for food, feed, and forage. Proso millet (Panicum miliaceum L.) is one of the seven commonly cultivated millets. Other six are pearl millet (Pennisetum glaucum (L.)R.Br.), finger millet (Eleusine coracana Gaertn.), kodo millet (Paspalum scrobiculatum L.), foxtail millet (Setaria italica (L.) P. Beauvois), little millet (Panicum sumatrense Roth ex Roem. & Schult.) and barnyard millet (Echinochloa esculenta (A.Braun) H.Scholz) [11]. Proso millet, one of the earliest domesticated crops, was reported to be originated in 10,000 BC in semiarid regions of northern China [26, 40]. It is a shallow-rooted, short-season crop with high water-use efficiency, making it suitable for cultivation in hot and dry environments in the world [18, 21, 41]. It has the shortest growing season (60–90 days) among all cereals which makes it tolerant to drought stress [17, 26]. It can produce significantly higher yields on marginal lands with low fertility and minimum agricultural input compared to many major crops [11]. Proso millet is also known as common millet in the USA, broomcorn millet in China, barri in India, mijo in Spain, panic millet in France, and gijang in Korea [11]. It is cultivated widely in China, Mongolia, Republic of Korea, Nepal, Pakistan, Afghanistan, Russia, Belarus, Romania, Turkey, and a few countries in Africa and the Middle East [11, 53, 66]. In the USA, it is primarily cultivated in west-central Great Plains of the USA (primarily Colorado, Nebraska, and South Dakota). It is extensively grown in the region as a rotational crop in the winter wheat-based dryland cropping system after sunflower and corn owing to its short growing season and high water-use efficiency [1, 42, 45].
The primary use of proso millet grain is as bird feed and animal feed in the United States and Europe [30, 41, 42]. However, it constitutes a significant portion of the human diet in many countries including China, India, several countries in Africa, and the former USSR owing to its high nutritive and health-promoting attributes [56]. Besides major nutrients (carbohydrate, protein, and fat), proso millet is rich in crude fiber, minerals, phenolic compounds, and vitamins such as B1 (thiamin) and B3 (niacin) [30]. Low glycemic index and gluten-free property make this crop an apt choice for utilization in food, which is suitable for people with health conditions such as diabetes mellitus, cardiovascular disease, and celiac disease [43, 56].
Omics
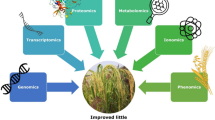
Today whole genome sequences are available for numerous crops including both major (e.g. rice, maize, soybean, and wheat) and minor crops (e.g. millets). The challenge of proper exploitation of the vast repository of genomic information led to the beginning of the “era of omics”. Omics refers to several branches of biotechnology with the suffix ‘omics’ [15].The branches of ‘omics’ are genomics, transcriptomics, proteomics, metabolomics, and phenomics. The common goal of these branches is to elucidate the underlying molecular mechanisms of various biological processes and to establish a relationship between genomic information of an organism and its phenotype. These various branches of ‘omics’ are becoming increasingly useful to today's and future plant breeders who are challenged with the herculean task of developing climate-resilient, high-yielding varieties in a short amount of time (Fig. 1). The threat to the balance between food supply and food demand by rapidly increasing world population is making the job even harder for plant breeders [12]. Advances made in genetics and omics have revolutionized plant breeding by unraveling functions of genetic factors controlling complex traits and designing molecular tools for the selection of superior genotypes [15].
Proso millet is regarded as climate-smart, ancient, and gluten-free and therefore, is healthy to humans and the environment. FAO has identified proso millet as one of the future smart crops of the twenty-first century [35]. Research areas of ‘omics’ of most of the common crops are being rapidly evolving and applied for genetic improvements of the crops to meet the challenges of crops (food, fuel, and fiber) production of the century. ‘Milletomics’ (‘omics’ of millets) are also being developed slowly and applied for their genetic improvement. There are several excellent review papers on ‘omics’ of major crops and a few such review articles on millets also do exist. Proso millet ‘omics’ studies are very few. There is no single review paper on the current status of ‘omics’ in proso millet and their potential in genetic improvement of this crop. The objective of this review article is to summarize publicly available ‘omics’ studies of proso millet and discuss their relevance for its genetic improvement.
Genomics of proso millet
Genomics involves large-scale analyses of structural and functional properties of the genome paving way for understating of evolutionary and functional aspects of an organism [12]. Early genomics included studies on genome size, physical and genetic mapping. The advent of next-generation sequencing (NGS) ushered a new era of omics by endowing the area of genomics with the ability to sequence, assemble, and analyze genomes of various plant species. The availability of whole genome sequences in several major crops leads to identifying agronomically and economically important genes which revolutionized breeding methods for genetic improvement of the crops [5].
Whole genome sequence is a crucial milestone in genome-wide molecular studies as it explores the complex molecular structure of the genome of complex organisms and serves as a major resource for the improvement of important crops [2, 47]. Until last year, the whole genome sequences of three millets were available and these are foxtail millet [67], pearl millet [64], and finger millet [22]. Proso millet whole genome sequence was published last year [74]. A high-quality, chromosome-scale genome assembly was developed using NGS tools. The authors could identify two sets of homologous chromosomes that fused ~ 5.6 million years ago that resulted in present-day tetraploid proso millet. They identified 55,930 protein-coding and 339 microRNA genes [74]. The genome sequence will serve as an invaluable resource for molecular breeding of proso millet. Thousands of SNP markers can be seamlessly developed from the genome sequences, which can be used in genomic selection (GS) of superior proso millet cultivars [56].
Genomic resources such as genome sequences and molecular markers are essential to establish genetic linkage map, which is the first step of molecular breeding for improving crops [2]. Molecular markers have been extensively used in studies related to genetic diversity, taxonomic, and population structure studies of crops, including rice, wheat, maize, soybean, sorghum, and barley [7, 16, 44, 58]. Randomly amplified polymorphic DNA (RAPD), restriction fragment length polymorphism (RFLP), and simple sequence repeats (SSR) markers are regularly used in genetic diversity and quantitative trait locus (QTL) studies in finger millet [2]. SSRs and SNPs were also widely used in genomic studies in foxtail millet [27]. In pearl millet, a wide range of molecular markers, including RFLP, amplified fragment length polymorphism (AFLP), RAPD, expressed sequence tags-derived simple sequence repeats (EST-SSRs), single-stranded conformation polymorphism-SNP, and SNPs have been developed and used in genetic diversity studies, QTL analysis, and maker-assisted selection [38, 62]. However, very limited molecular markers (mostly RAPD, SSR, and GBS-SNP) are publicly available in proso millet as it is largely an underresearched crop [20, 50].
SSRs are preferred for genetic studies owing to their abundance, even distribution in the genome, multi-allelic, codominant and polymorphic attributes, easy scoring, and high reproducibility. SSRs in proso millet were studied using the genomic sequences from other plants as its genomic resources were very limited. Hu et al. [24] used SSR markers from rice, wheat, oat, and barley to assess the genetic diversity of 118 Chinese proso millet accessions. Cho et al. [10] developed the first set of SSR markers from proso millet genome. They developed 25 polymorphic SSR markers from 50 genetically diverse proso millet accessions. Hunt et al. [25] used 16 proso millet-specific SSR markers to study the dissemination of the species from its center of domestication to other regions of the world. A total of 548 SSR markers of switchgrass, genetically the closest to proso millet, were used for genetic diversity of proso millet germplasm [49].They were able to group 95 diverse genotypes from 25 different countries using the SSR markers. High throughput sequencing was used by Liu et al. [39] to develop 500 proso-millet specific SSR-primer pairs, of which 67 were successfully used to investigate the genetic diversity in 88 Chinese accessions [39].
Single nucleotide polymorphism (SNP) markers are considered ideal for genetic studies, high-density genetic maps, and genomic selection because of their abundance in the genome [56, 70]. A limited number, even though not adequate, SNP markers are available for proso millet. Rajput and his coworkers identified 833 GBS-SNPs using 93 recombinant inbred lines (RILs) and developed the first genetic linkage map in proso millet.They established eighteen major linkage groups (LG), which were considered as the 18 chromosomes of proso millet. They were also able to identify QTLs for eight morpho-agronomic traits including lodging and grain shattering [51]. Six SNPs of the waxy (wx-L) locus were identified using sequences of PCR amplicons of 132 proso millet accessions and the allelic variation of the waxy gene was reported [68]. While conducting genome-wide population genetic studies in three millets viz., kodo millet, little millet, and proso millet, Johnson, and his coworkers identified 1882 SNPs for proso millet [29]. Population genetic studies indicated that those SNPs could accurately reflect the genome-wide genetic variation.
Transcriptomics of proso millet
Transcriptomics is the study of transcriptome, total collection of messenger RNA (mRNAs) in a cell or tissue of an organism [54]. It involves comprehensive analysis and quantification of the genome-wide changes of transcripts [33]. There are several reports on transcriptome studies in foxtail millet [60, 71], finger millet [22, 48], pearl millet [14, 32, 61, 64], and barnyard millet [28]. There are a limited number of reports on transcriptome research in proso millet. Yue et al. (2016) employed Illumina sequencing to sequence and assembled proso millet transcriptome. The sequence reads obtained from the sequencing of two genotypes namely, Yumi No. 2 and Yumi No.3, were assembled into unigenes. 62,543 contigs could be assigned to 315 categories of gene ontology (GO). 15,514 unigenes were mapped to 202 Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways using KEGG analysis. Yue et al. were also able to identify 292 differentially expressed genes. They also identified ~ 32,000 SSR markers and ~ 400,000 SNP markers which may be utilized in molecular breeding and population genetic studies [72]. Hou et al. (2017) used Illumina sequencing to generate read sequences and developed a de novo transcriptome assembly of proso millet. They identified 25,341 gene sequences and 4,724 SSR loci in the transcriptome. They also identified 14 polymorphic SSR primers exhibiting a total of 43 alleles. The percentage of polymorphic markers (6.1%) showed that 288 polymorphic genic SSR markers can be utilized for genetic assay [23]. More recently, Zhang et al. (2019) reported a comparative study of leaf transcriptomes of proso millet to understand the underlying molecular underpinning of drought tolerance mechanisms. They constructed a cDNA library using leaf mRNA from the drought-tolerant (Neimi 5) and drought-sensitive (Jinshu 6) cultivars and subsequently sequenced using Hiseq 4000 sequencing platform. They reported 833 and 2,166 differentially expressed genes (DEGs) in Jinshu 6 and Neimi 5, respectively in response to PEG-induced drought stress [73]. More research aimed at understanding the functional aspects of the genes involved in various other key regulatory processes in proso millet is warranted.
Proteomics of proso millet
‘Proteome’ is the entire set of proteins expressed in an entire organism or a specific organ or tissue or cell at any given stage or condition. ‘Proteomics’ refers to a large-scale structural and functional analysis of ‘proteome’ for understanding the underlying regulatory mechanism of the expressed proteins [8]. Since proteins act directly on biochemical processes, proteomics becomes vital to understand biological functions [19]. The most commonly used tools used in structural proteomics are two-dimensional electrophoresis (2-DE) and mass spectrometry (MS)-based techniques such as MALDI-TOF [6]. Most of the proteome is proportional to the transcriptome. However, levels and diversity of proteome are not always directly in proportion to the transcriptome due to post-translational modification. Although extensive proteomics reports are available in most of the major crop plants such as rice, corn, soybean, potato, wheat, proteomics of proso and other millets are very scanty [9, 52].
In proso millet, possibly the first proteomics report was seed protein analysis of four Korean cultivars using 2-D electrophoresis and mass fingerprinting [55]. The objective was to map proso millet seed proteins and identify the functional characteristics of the identified proteins. Of 1152 differentially expressed proteins on 2-D gel, 26 were identified by MALDI-TOF/MS. Two proteins were upregulated in all cultivars, 13 were upregulated and 11 were downregulated in two cultivars. Most of the upregulated proteins were associated with starch metabolism, transcription, and pathogenesis whereas, downregulated proteins were involved in glycolysis, stress responses, and transduction. The authors reported that the differential expression of the proteins may be cultivar specific. In another comparative proteomics analysis between ancient bread and reference cereals (proso millet, foxtail millet, barley, and wheat), the researchers identified proso millet and barley as an ingredient of ancient bread excavated in Subeixi cemetery (500–300 BC) in Xinjiang, China [59].
However, comparatively more proteomics studies were reported in other millets. Four proteomic studies were reported in foxtail millet, which included two on response to abiotic stresses—salinity and drought [46, 65] and two on seed proteome [37, 69]. In pearl millet, two reports were proteome analyses related to downy mildew disease [3, 4]. There was only one proteomic report on the jasmonic acid signaling pathway in finger millet [57].
Metabolomics of proso millet
‘Metabolomes’ is defined as the complete set of low molecular weight compounds present in a biological sample. Metabolomics is the study of metabolites of an organism, organ, tissue, or cell under a specific stage and external or internal condition [13]. Metabolome profile indicates the physiological condition of the cell or tissue or the whole organism, which indirectly provides functional aspects of the metabolites. Many a times, transcriptome or proteome do not provide the biochemical basis of some biological functions especially stress tolerance in plants. Therefore, metabolomics becomes necessary to understand the functional basis of plant genomes [63]. Metabolomics provides a bridge between genotype and phenotype. Reports on metabolomics of millets are very scanty.
The first proso millet metabolomics was studied using gas chromatography–time-of-flight mass spectrometry (GC-TOFMS) for grain quality assessment [31]. They studied primary metabolites and phenolics in matured seeds of three Korean proso millet varieties. They identified a total of 48 metabolites including 43 primary metabolites and five phenolic acids. The seed metabolomes could separate the three varieties using principal component analysis (PCA) and partial least-squares discriminate analysis (PLS-DA). Relatively in a more recent report, ‘metabolome’ of conventionally and organically grown grains of two proso millet varieties were analyzed [36]. They did not see any difference in metabolome in the millet grains grown conventionally or organically except levels of glucose and fructose in organic millets. However, there were significant differences between the varieties. In the case of other millets, the only metabolomics study was on foxtail millet by Li et al. [34] where they studied primary and secondary metabolites in foxtail millet and inheritance patterns of the metabolome in the millet hybrids.
Conclusion
A comprehensive study of all ‘omics’ of millet has great potential for genetic improvement of millets, climate-smart cereal crops. However, the field of ‘omics’ in millet is not very rich. There are a decent number of studies of genomics, transcriptomics in pearl millet, foxtail millet and finger millet. Similar reports in proso millet and other minor millets such as kodo millet, barnyard millet, and Japanese millet are very few. Even lesser number of reports on proteomics and metabolomics are available for pearl millet and foxtail millet. But very few (1–3) proteomics and metabolomics studies were reported in proso millet. Not a single published report on phenomics is available in any millet yet. Genome sequence of most of the millets (proso millet, pearl millet, foxtail millet, finger millet) are now available. Genome sequence of other three minor millets (kodo millet, barnyard millet, and Japanese millet) will soon be available with increasingly fast and efficient methodologies at decreasingly cheap cost. Similarly, more efficient, fast, and cheaper ‘omics’ technologies are getting available. Therefore, it is expected that the global millet scientists will be able to take advantage of such technological advancement to advance the ‘omics’ research in millets. In the near future, the millet breeders and geneticists will use the fruits of such ‘omics’ research for genetic improvement of all these millets. However, a strong collaboration among the millet geneticists and breeders across the globe is unquestionably will be necessary for the successful application of the ‘omics’ in millet genetic improvement and to see noble millet varieties in the seed industry in future.
References
Agdag M, Nelson L, Baltensperger D, Lyon D, Kachman S. Row spacing affects grain yield and other agronomic characters of proso millet. Commun Soil Sci Plant Anal. 2001;32:2021–32.
Antony Ceasar S, Maharajan T, Ajeesh Krishna TP, Ramakrishnan M, Victor Roch G, Satish L, et al. Finger millet [Eleusine coracana (L.) Gaertn.] improvement: Current status and future interventions of whole genome sequence. Front Plant Sci. 2018;9.
Anup CP, Melvin P, Shilpa N, Gandhi MN, Jadhav M, Ali H, et al. Proteomic analysis of elicitation of downy mildew disease resistance in pearl millet by seed priming with β-aminobutyric acid and Pseudomonas fluorescens. J Proteomics. 2015;120:58–74. https://doi.org/10.1016/j.jprot.2015.02.013.
Anup CP, Kini KR. Analysis of dynamics of proteome in resistant cultivar of pearl millet seedlings during Sclerospora graminicola infection. J Appl Biol Biotechnol. 2016;4:67–71.
Aydin G. Genome analysis of plants. In: Hakeem KR, Tombuloğlu H, Tombuloğlu G, editors. Plant omics: trends and applications. Springer: Cham; 2016. p. 1–27.
Barkla BJ, Castellanos-Cervantes T, Diaz de León JL, Matros A, Mock HP, Perez-Alfocea F, et al. Elucidation of salt stress defense and tolerance mechanisms of crop plants using proteomics-Current achievements and perspectives. Proteomics. 2013;13:1885–900.
Boyles RE, Brenton ZW, Kresovich S. Genetic and genomic resources of sorghum to connect genotype with phenotype in contrasting environments. Plant J. 2019;97:19–39.
Budzinski IGF, Regiani T, Labate MTV, Guidetti-Gonzalez S, da Silva DI, Rodrigues MJC, et al. Proteomics. In: Borém A, Fritsche-Neto R, editors. Omics in Plant Breeding. John Wiley & Sons, Inc, New York; 2014. pp. 59–79.
Cánovas FM, Dumas-Gaudot E, Recorbet G, Jorrin J, Mock HP, Rossignol M. Plant proteome analysis. Proteomics. 2004;4:285–98.
Cho Y Il, Chung JW, Lee GA, Ma KH, Dixit A, Gwag JG, et al. Development and characterization of twenty-five new polymorphic microsatellite markers in proso millet (Panicum miliaceumL.). Genes and Genomics. 2010;32:267–73.
Das S, Khound R, Santra M, Santra DK. Beyond bird feed: Proso millet for human health and environment. Agric. 2019;9. https://doi.org/10.3390/agriculture9030064
de Oliveira AC, da Maia LC, Farias DR, Marini N. Genomics. In: Borém A, Fritsche-Neto R, editors. Omics in Plant Breeding. Wiley, New York; 2014. pp. 13–31.
Diola V, de Daloso DM, Antunes WC. Metabolomics. In: Borém A, Fritsche-Neto R, editors. Omics in Plant Breeding. Wiley, New York; 2014. pp. 1–11.
Dudhate A, Shinde H, Tsugama D, Liu S, Takano T. Transcriptomic analysis reveals the differentially expressed genes and pathways involved in drought tolerance in pearl millet [pennisetum glaucum (l.) r. Br]. PLoS ONE. 2018;13:1–14.
Fritsche-Neto R, Borém A. Omics: Opening up the "black box" of the phenotype. In: Borém A, Fritsche-Neto R, editors. Omics in Plant Breeding. Wiley, New York; 2014. pp. 1–11.
Gedil M, Menkir A. An integrated molecular and conventional breeding scheme for enhancing genetic gain in maize in Africa. Front Plant Sci. 2019;10:1–17.
Goron TL, Raizada MN. Genetic diversity and genomic resources available for the small millet crops to accelerate a New Green Revolution. Front Plant Sci. 2015;6. https://doi.org/10.3389/fpls.2015.00157
Graybosch RA, Baltensperger DD. Evaluation of the waxy endosperm trait in proso millet (Panicum miliaceum). Plant Breed. 2009;128:70–3.
Gygi SP, Rochon Y, Franza BR, Aebersold R. Mb001720. Mol Cell Biol. 1999;19:1720–30.
Habiyaremye C, Matanguihan JB, D’Alpoim Guedes J, Ganjyal GM, Whiteman MR, Kidwell KK, et al. Proso millet (Panicum miliaceum L.) and its potential for cultivation in the Pacific Northwest, U.S.: A review. Front Plant Sci. 2017;7:1–17.
Henry WB, Nielsen DC, Vigil MF, Calderón FJ, West MS. Proso millet yield and residue mass following direct harvest with a stripper-header. Agron J. 2008;100:580–4.
Hittalmani S, Mahesh HB, Shirke MD, Biradar H, Uday G, Aruna YR, et al. Genome and Transcriptome sequence of Finger millet (Eleusine coracana (L.) Gaertn.) provides insights into drought tolerance and nutraceutical properties. BMC Genomics. 2017;18:1–16.
Hou S, Sun Z, Li Y, Wang Y, Ling H, Xing G, et al. Transcriptomic analysis, genic SSR development, and genetic diversity of proso millet ( Panicum miliaceum; Poaceae). Appl Plant Sci. 2017;5:1600137.
Hu X, Wang J, Lu P, Zhang H. Assessment of genetic diversity in broomcorn millet (Panicum miliaceum L.) using SSR markers. J Genet Genomics. 2009;36:491–500. https://doi.org/10.1016/S1673-8527(08)60139-3
Hunt HV, Campana MG, Lawes MC, Park YJ, Bower MA, Howe CJ, et al. Genetic diversity and phylogeography of broomcorn millet (Panicum miliaceum L.) across Eurasia. Mol Ecol. 2011;20:4756–71.
Hunt HV, Badakshi F, Romanova O, Howe CJ, Jones MK, Heslop-Harrison JSP. Reticulate evolution in Panicum (Poaceae): the origin of tetraploid broomcorn milletP miliaceum. J Exp Bot. 2014;65:3165–75.
Jaiswal V, Gupta S, Gahlaut V, Muthamilarasan M, Bandyopadhyay T, Ramchiary N, et al. Genome-Wide Association Study of Major Agronomic Traits in Foxtail Millet (Setaria italica L.) Using ddRAD Sequencing. Sci Rep. 2019;9:1–11.
Jayakodi M, Madheswaran M, Adhimoolam K, Perumal S, Manickam D, Kandasamy T, et al. Transcriptomes of Indian barnyard millet and barnyardgrass reveal putative genes involved in drought adaptation and micronutrient accumulation. Acta Physiol Plant. 2019;41. https://doi.org/10.1007/s11738-019-2855-4
Johnson M, Deshpande S, Vetriventhan M, Upadhyaya HD, Wallace JG. Genome-wide population structure analyses of three minor millets: Kodo Millet, Little Millet, and Proso Millet. Plant Genome. 2019;12:190021. https://doi.org/10.3835/plantgenome2019.03.0021.
Kalinova J, Moudry J. Content and quality of protein in proso millet (Panicum miliaceum L.) varieties. Plant Foods Hum Nutr. 2006;61:45–9.
Kim JK, Park SY, Yeo Y, Cho HS, Kim YB, Bae H, et al. Metabolic profiling of millet (Panicum miliaceum) using gas chromatography-time-of-flight mass spectrometry (GC-TOFMS) for quality assessment. Plant Omics. 2013;6:73–8.
Kulkarni KS, Zala HN, Bosamia TC, Shukla YM, Kumar S, Fougat RS, et al. De novo transcriptome sequencing to dissect candidate genes associated with pearl millet-downy mildew (sclerospora graminicola sacc.) interaction. Front Plant Sci. 2016;7:1–16.
Lata C. Advances in omics for enhancing abiotic stress tolerance in millets. Proc Indian Natl Sci Acad. 2015;81:397–417.
Li S, Dong X, Fan G, Yang Q, Shi J, Wei W, et al. Comprehensive profiling and inheritance patterns of metabolites in foxtail millet. Front Plant Sci. 2018;871:1–16.
Li X, Siddique KHM. Future smart food- Rediscovering hiddent treasures of neglected and underutilized species for zero hunger in Asia. Bangkok: FAO; 2018.
Liang K, Liang S, Lu L, Zhu D, Zhu H, Liu P, et al. Metabolic variation and cooking qualities of millet cultivars grown both organically and conventionally. Food Res Int. 2018;106:825–33. https://doi.org/10.1016/j.foodres.2018.01.023.
Liang K, Liang S, Zhu H. Comparative proteomics analysis of the effect of selenium treatment on the quality of foxtail millet. Lwt. 2020;131:109691. https://doi.org/10.1016/j.lwt.2020.109691.
Liang Z, Gupta SK, Yeh CT, Zhang Y, Ngu DW, Kumar R, et al. Phenotypic data from inbred parents can improve genomic prediction in pearl millet hybrids. G3 Genes, Genomes, Genet. 2018;8:2513–22.
Liu M, Xu Y, He J, Zhang S, Wang Y, Wang Y, et al. Genetic diversity and population structure of broomcorn millet (Panicum miliaceum L.) cultivars and landraces in China based on microsatellite markers. Int J Mol Sci. 2016;17:1–18.
Lu H, Zhang J, Liu KB, Wu N, Li Y, Zhou K, et al. Earliest domestication of common millet (Panicum miliaceum) in East Asia extended to 10,000 years ago. Proc Natl Acad Sci U S A. 2009;106:7367–72.
Lyon DJ, Baltensperger DD. Proso millet (Panicum miliaceum) tolerance to several postemergence herbicides. Weed Tech. 1993;7:230–3.
Lyon D, Burgener P, DeBoer K, Harveson R, Hein G, Hergert G, Producing, and Marketing Proso millet in the Great Plains., et al. Publication # EC137. Lincoln, NB: Univ of Nebraska Ext Serv; 2008.
McSweeney MB, Seetharaman K, Ramdath DD, Duizer LM. Chemical and physical characteristics of proso millet (Panicum miliaceum)-based products. Cereal Chem. 2017;94:357–62.
Nadeem MA, Nawaz MA, Shahid MQ, Doğan Y, Comertpay G, Yıldız M, et al. DNA molecular markers in plant breeding: current status and recent advancements in genomic selection and genome editing. Biotechnol Biotechnol Equip. 2018;32:261–85.
Nielsen DC, Unger PW, Miller PR. Efficient water use in dryland cropping systems in the Great Plains. Agron J. 2005;97:364–72.
Pan J, Li Z, Wang Q, Garrell AK, Liu M, Guan Y, et al. Comparative proteomic investigation of drought responses in foxtail millet. BMC Plant Biol. 2018;18:1–19.
Pandian S, Ramesh M. Decoding of finger millet genome: a milestone of millet genomics. Sign Transduct Insights. 2019;8:117864341882054. https://doi.org/10.1177/1178643418820541.
Parvathi MS, Nataraja KN, Reddy YAN, Naika MBN, Gowda MVC. Transcriptome analysis of finger millet (Eleusine coracana (L.) Gaertn.) reveals unique drought responsive genes. J Genet. 2019;98:1–12. https://doi.org/10.1007/s12041-019-1087-0
Rajput SG, Plyler-Harveson T, Santra DK. Development and characterization of SSR markers in proso millet based on Switchgrass genomics Am J Plant Sci. 2014;05:175–86.
Rajput SG, Santra DK. Evaluation of genetic diversity of proso millet germplasm available in the united states using simple-sequence repeat markers. Crop Sci. 2016;56:2401–9.
Rajput SG, Santra DK, Schnable J. Mapping QTLs for morpho-agronomic traits in proso millet (Panicum miliaceum L.). 2016;36. org/https://doi.org/10.1007/s11032-016-0460-4
Reddy DS, Bhatnagar-Mathur P, Vadez V, Sharma KK. Grain legumes (Soybean, Chickpea, and Peanut): omics approaches to enhance abiotic stress tolerance. Improv Crop Resist Abiotic Stress. 2012;2:995–1032.
Reddy VG, Upadhyaya HD, Gowda CLL. Morphological characterization of world’s proso millet germplasm collection. J SAT Agric Res. 2007;3:1–4.
Rodrigues CM, Mafra VS, Machado. Transcriptomics. In: Borém A, Fritsche-Neto R, editors. Omics in Plant Breeding. Wiley, New York; 2014. pp. 33–57.
Roy SK, Kwon S-J, Yu J-H, Sarker K, Cho S-W, Moon Y-J, et al. Comparison of Protein Profiles of Proso Millet (Panicum miliaceum) Seeds of various Korean. Korean J Crop Sci. 2017;62:40–50.
Santra DK, Khound R, Das S. Proso millet (Panicum miliaceum L.) breeding: Progress, challenges and opportunities. In: Al-Khayri, Jain SM, Johnson DV, editors. Adv Plant Breed Strateg Cereal. Springer Nature, Heidelberg; 2019. 5:223–57.
Sen S, Kundu S, Dutta SK. Proteomic analysis of JAZ interacting proteins under methyl jasmonate treatment in finger millet. Plant Physiol Biochem Elsevier Masson SAS. 2016;108:79–89. https://doi.org/10.1016/j.plaphy.2016.05.033.
Shabir G, Aslam K, Khan AR, Shahid M, Manzoor H, Noreen S, et al. Rice molecular markers and genetic mapping: current status and prospects. J Integr Agric. 2017;16:1879–91. https://doi.org/10.1016/S2095-3119(16)61591-5
Shevchenko A, Yang Y, Knaust A, Thomas H, Jiang H, Lu E, et al. Proteomics identifies the composition and manufacturing recipe of the 2500-year old sourdough bread from Subeixi cemetery in China. J Proteomics. 2014;105:363–71. https://doi.org/10.1016/j.jprot.2013.11.016.
Shi W, Cheng J, Wen X, Wang J, Shi G, Yao J, et al. Transcriptomic studies reveal a key metabolic pathway contributing to a well-maintained photosynthetic system under drought stress in foxtail millet (Setaria italica L.). PeerJ. 2018;2018.
Shinde H, Tanaka K, Dudhate A, Tsugama D, Mine Y, Kamiya T, et al. Comparative de novo transcriptomic profiling of the salinity stress responsiveness in contrasting pearl millet lines. Environ Exp Bot. 2018;155:619–27. https://doi.org/10.1016/j.envexpbot.2018.07.008.
Srivastava RK, Singh RB, Pujarula VL, Bollam S, Pusuluri M, Chellapilla TS, et al. Genome-wide association studies and genomic selection in pearl millet: advances and prospects. Front Genet. 2020;10:1–10.
Sumner LW, Mendes P, Dixon RA. Plant metabolomics: Large-scale phytochemistry in the functional genomics era. Phytochemistry. 2003;62:817–36.
Varshney RK, Shi C, Thudi M, Mariac C, Wallace J, Qi P, et al. Pearl millet genome sequence provides a resource to improve agronomic traits in arid environments. Nat Biotechnol. 2017;35:969–76.
Veeranagamallaiah G, Jyothsnakumari G, Thippeswamy M, Chandra Obul Reddy P, Surabhi GK, Sriranganayakulu G, et al. Proteomic analysis of salt stress responses in foxtail millet (Setaria italica L. cv. Prasad) seedlings. Plant Sci. 2008;175:631–41.
Vetriventhan M, Upadhyaya HD. Diversity and trait-specific sources for productivity and nutritional traits in the global proso millet (Panicum miliaceum L.) germplasm collection. Crop J. 2018;6:451–63.
Wang J, Zhang G, Liu X, Quan Z, Cheng S, Xu X, et al. Genome sequence of foxtail millet (Setaria italica) provides insights into grass evolution and biofuel potential. Nat Biotechnol. 2012;30:549–54.
Wang R, Wang H, Liu X, Ji X, Chen L, Lu P, et al. Waxy allelic diversity in common millet (Panicum miliaceum L.) in China. Crop J. 2018;6:377–85.
Weng Q, Song X, Zhao Y, Yuan J, Dong Z, Zhao Z, et al. Proteomic profiling of foxtail millet hybrid Zhangzagu10 and its parent lines using iTRAQ-based technique. J Plant Biochem Biotechnol. 2020;2. https://doi.org/10.1007/s13562-020-00551-2
Xia W, Luo T, Zhang W, Mason AS, Huang D, Huang X, et al. Development of high-density snp markers and their application in evaluating genetic diversity and population structure in elaeis guineensis. Front Plant Sci. 2019;10:1–11.
Xu B qin, Gao X li, GaoJ feng, Li J, Yang P, Feng B li. Transcriptome profiling using RNA-seq to provide insights into foxtail millet seedling tolerance to short-term water deficit stress induced by PEG-6000. J Integr Agric. CAAS. 2019;18:2457–71. https://doi.org/10.1016/S2095-3119(19)62576-1
Yue H, Wang M, Liu S, Du X, Song W, Nie X. Transcriptome-wide identification and expression profiles of the WRKY transcription factor family in Broomcorn millet (Panicum miliaceum L.). BMC Genomics.2016;17:1–11. https://doi.org/10.1186/s12864-016-2677-3
Zhang Y, Gao X, Li J, Gong X, Yang P, Gao J, et al. Comparative analysis of proso millet (Panicum miliaceum L.) leaf transcriptomes for insight into drought tolerance mechanisms. BMC Plant Biol. 2019;19:1–17.
Zou C, Li L, Miki D, Li D, Tang Q, Xiao L, et al. The genome of broomcorn millet. Nat Commun. 2019;10. https://doi.org/10.1038/s41467-019-08409-5
Funding
The project was supported by ‘Research State Aided’ internal funds: 21-6243-1001.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Corresponding Editor : Manoj Prasad.
Rights and permissions
About this article
Cite this article
Khound, R., Santra, D.K. Omics for proso millet genetic improvement. Nucleus 63, 241–247 (2020). https://doi.org/10.1007/s13237-020-00339-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13237-020-00339-8