Abstract
In order to increase the molecular tools and markers needed to detect genetic diversity of rice, Random Amplified Polymorphic DNA (RAPD), Inter Simple Sequence Repeat (ISSR) and Simple Sequence Repeat (SSR) markers were used to reveal genetic diversity in 22 upland and lowland rice cultivars from Bangladesh. DNA from fresh leaves of young plants was extracted and subjected to the DNA polymorphism studies using the above mentioned markers. Among ten RAPD primers tested, four showed high polymorphism (90.90 %) with an average of 8.25 bands per primer with band size of approximately 220–1531 bp. Four ISSR primers out of ten produced high polymorphism (90.47 %) with an average of 10.5 bands per primer with band size of approximately 222–1750 bp. Among seven SSR primers tested, three amplified polymorphic bands with an average number of 4.0 bands per primer (ranging from approximately 233–950 bp) and 100 % polymorphism. Based on combined dataset generated by RAPD, ISSR and SSR markers, the highest similarity was showed between BRRI dhan 27 and BRRI dhan 29 (similarity coefficient value 0.89) and the lowest similarity was recorded between BR-3 and BRRI dhan 19 (similarity coefficient value 0.25). The UPGMA dendrogram based on genetic similarity grouped the cultivars into several clusters. Results obtained from PCA analysis were similar with UPGMA dendrogram. The evaluation of genetic similarity and cluster analysis provides useful guides for assisting plant breeders in selecting suitable genetically diverse parents for the future breeding programs.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Rice (Oryza sativa L.), a member of the plant family Poaceae, is one of the most important staple food crops supporting the world population especially in Asian countries including Bangladesh. It is grown in about 75 % of total cropped area for rice cultivation in Bangladesh. A large number of local cultivars are used as germplasmand about 5000 rice accessions are available in the Germplasm Bank of Bangladesh Rice Research Institute. However, all these varieties are not grown in Bangladesh. Developmental activities and exploitive land-use pattern are destroying natural habitats, and furthermore, modern cultivars (e.g. HYVs in rice) are replacing indigenous cultivars and land races, resulting in a substantial reduction of genetic diversity. Selection of plant varieties based on morphological characters is not very reliable because major characters of interest possess low heritability and are genetically complex. For this reason, the most successful method is the use of molecular markers for detecting genetic diversity of rice. Using DNA markers identified by DNA polymorphism study among the tested genotypes is an efficient approach in detecting genetic diversity and is widely used in Biological research. Several molecular markers viz. RFLP, RAPD, SSRs, AFLP and SNPs are presently available to assess the variability and diversity at molecular level [18]. In the last few decades, new DNA molecular markers, based on the PCR technique, such as random amplified polymorphic DNA or RAPD [34] and inter simple sequence repeats or ISSRs [38], among others, have become excellent tools for plant breeders [10, 11, 23]. However, RAPD has some limitations, such as dependence on the genetic background, low reproducibility, and level of polymorphism obtained [10, 13, 38]. In contrast to RAPD amplification, the ISSR markers are also highly polymorphic and their use is cost effective, requiring no prior information of the sequence [3]. In cereals, ISSR markers have been used to study genetic diversity and phylogenetic relationships [10, 20], for gene mapping [22], for gene tagging in molecular assisted selection [1, 21], and for DNA fingerprinting [7]. On the other hand, simple sequence repeats (SSR) or microsatellites are simple, tandemly repeated, di to tetra-nucleotide sequence motifs flanked by unique sequences [14]. SSR markers have been extensively used to identify genetic variation among Oryza species [29], analyze genetic structure within the cultivated rice [12], evaluate genetic diversity of wild rice [32] and among cultivars of cultivated rice [16, 17, 36, 37].
The objective of the present study was to assess genetic diversity among twenty two rice cultivars using RAPD, ISSR and SSR markers. The information on genetic relationship based on genetic similarity and clustering data among these rice cultivars may provide useful information for selecting appropriate parental lines in breeding program for improving rice varieties.
Materials and methods
Plant materials
Twenty two upland and lowland rice cultivars, obtained from Bangladesh Rice Research Institute, Regional Office, Rajshahi, were used as a source material in this experiment (Table 1). Fresh leaves of young plants were collected randomly from each cultivar of rice.
Extraction of genomic DNA
CTAB (Cethyltrimethylammonium bromide) buffer was used for extraction of genomic DNA following the method of Doyle and Doyle [9]. DNA concentration and purity were determined by measuring the absorbance of diluted DNA solution at 260 nm by a Spectrophotometer (T80 UV/VIS Spectrometer, PG Instruments Limited, UK). Purified DNA samples were checked by using 0.8 % agarose gel on gel casting system (Bangalore GeNei gel electrophoresis system, Bangalore, India) with 0.5 X TBE buffer. Following the protocol of Borsch et al. [4] those with high band intensity and less smear were selected for polymerase chain reaction.
Primer screening
A set of eight decamer primers (Oligo Product Information, Bio Basic Inc., Canada) were used for RAPD; a set of eight primers (Sigma Aldrich, St. Louis, USA) were tested for ISSR and a set of seven primers (Sigma Aldrich, St. Louis, USA) were screened for SSR. The details of the primers are shown in Table 2.
RAPD- PCR reaction
The RAPD amplifications were carried out in 25 μl reaction mixtures containing 75 ng/μl of template DNA 1 μl, 10X reaction buffer 2.50 μl, 20 mM MgCl2 2.50 μl, 10 mM dNTPs (Oligo Product Information, Bio Basic Inc., Canada) 0.50 μl, 10 μM primer 2 μl, 1 Unit Taq DNA polymerase (Bangalore GeNei Private Ltd., Bangalore, India) 0.25 μl and deionized water 16.25 μl. The RAPD amplifications occurred under the conditions as: initial denaturation/activation at 94 °C for 4 min, 40 cycles of 0.40 min denaturation at 94 °C, 0.30 min at 37 °C, 1.30 min extension at 72 °C and a final extension at 72 °C for 7 min.
ISSR- PCR reaction
The ISSR amplifications were carried out in 25 μl reaction mixtures containing 75 ng/μl of template DNA 1 μl, 10X reaction buffer 2.50 μl, 20 mM MgCl2 2.50 μl, 10 mM dNTPs (Oligo Product Information, Bio Basic Inc., Canada) 0.50 μl, 10 μM primer 0.40 μl, 1 Unit Taq DNA polymerase (Bangalore GeNei Private Ltd., Bangalore, India) 0.25 μl and deionized water 17.75 μl. The ISSR amplifications occurred under the following conditions as: initial denaturation/activation at 94 °C for 4 min, 40 cycles of 0.40 min denaturation at 94 °C, 0.30 min at annealing temperature (depending on the primer), 1.30 min extension at 72 °C and a final extension at 72 °C for 7 min.
SSR- PCR reaction
The SSR amplifications were carried out in 25 μl reaction mixtures containing 75 ng/μl of template DNA 1 μl, 10X reaction buffer 2.50 μl, 20 mM MgCl2 2.50 μl, 10 mM dNTPs (Oligo Product Information, Bio Basic Inc., Canada) 0.50 μl, 10 μM primer 0.40 μl, 1 Unit Taq DNA polymerase (Bangalore GeNei Private Ltd., Bangalore, India) 0.25 μl and deionized water 17.75 μl. The SSR amplifications occurred under the conditions as: initial denaturation/activation at 94 °C for 4 min, 40 cycles of 0.40 min denaturation at 94 °C, 0.30 min at annealing temperature (depending on the primer), 1 min extension at 72 °C and a final elongation at 72 °C for 5 min.
PCR reactions were carried out on a thermal cycler (Nyx Technik Inc., model A6, USA). Amplified products from each sample were separated electrophoretically on 1.4 % agarose gel (Bangalore GeNei Private Ltd., Bangalore, India) containing ethidium bromide in 0.5X TBE buffer at 120 V for 1½ hour.
Data analysis
DNA bands were observed on gel documentation system (Alpha InnoTech Inc., Santa Clara, California, USA). Each amplified band was considered as a unit character regardless of its intensity and scored in terms of a binary code, based on presence (1) and absence (0) of bands. Data matrices for each of the marker have been constructed based on presence/absence of bands. A combined data matrix has also been produced. The computer package NTSYSpc Version 2.1 statistical package [30] was used to construct a dendrogram and PCA (Principal Component Analysis). At first, the data matrices were used to calculate genetic similarity based on Jaccard’s similarity coefficient [15]. These similarity matrices have been used for PCA and for constructing dendrograms using Unweighted Pair Group Method with Arithmetic averages (UPGMA) [33].
Results
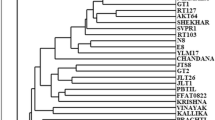
Based on the accurate amplified band profiles and the produced polymorphic patterns of DNA fingerprinting, four primers were chosen both for RAPD and ISSR and three for SSR (Table 3). The list of selected primers, their reproducibility, size of fragments and percentage of polymorphism are shown in Table 3. Profile for RAPD, ISSR and SSR are shown in Fig. 1. Dendrograms for RAPD, ISSR, SSR and combined datasets are shown in Fig. 2. Three dimensional plots for PCA for RAPD, ISSR, SSR and combined datasets are shown in Fig. 3.
Amplified products obtained from different markers. a RAPD profile obtained with MT370578; b ISSR profile obtained with UBC825and c SSR profile obtained with primer CMCCA145. The lanes represent, M: 100 bp DNA marker, 1–22: rice cultivars listed as in Table 1
Polymorphism of RAPD markers and cluster analysis
The number of scorable fragments produced per primer ranged from 7 to 10 and size of the products ranged from approximately 220–1531 bp. Among the four primers, MT370571 and MT370573 showed 100 % polymorphism and ranges of fragments were between 220 and 1531 bp and 260–1200 bp respectively (Table 3). RAPD marker profile produced by the primer MT370578 is shown in Fig. 1a. The similarity coefficients of RAPD markers ranged from 0.10 to 0.94. Among the 22 rice cultivars, BRRI dhan-27 and BRRI dhan-29 showed the highest similarity index (0.94), and BR-16 and IR-64 showed the lowest similarity index (0.10). The dendrogram (Fig. 2a) grouped rice cultivars into two main clusters. First cluster was divided into two main subclusters. The first subcluster was consisted of BR-1, BR-15, BR-14, BR-11, BR-16, BR-17, BR-19, BR-18, BRRI dhan-35, BRRI dhan-36, BRRI dhan-45, BRRI dhan-47, BR-8, BR-9, BR-26, BRRI dhan-27, BRRI dhan-29 and BRRI dhan-28 while the second subcluster was consisted of BR-3 and BRRI dhan-50. The second cluster consisted of BR-7 and IR-64 (Fig. 2a). The PCA yielded eigenvalues of each principle component axes of coordination of cultivars with the first three axes (percentage of PCA1, PCA2, PCA3 recorded 50.6295, 10.4703 and 5.2868 respectively) totally accounted for 66.3866 (Fig. 3a).
Polymorphism of ISSR markers and cluster analysis
The sizes of amplified products ranged from 222 to 1750 bp (Table 3) and in average 90.47 % polymorphism were observed. ISSR marker profile produced by UBC825 in agarose gel is shown in Fig. 1b. In ISSR analysis, the similarity coefficients observed for the rice cultivars rangied between 0.94 and 0.30. Among the 22 rice cultivars, similarity index was highest between BRRI dhan-27 and BRRI dhan-29 (0.94) and it was lowest between BR-3 and BR-19 (0.30). The cluster analysis of ISSR markers grouped the rice cultivars into four distinct clusters (Fig. 2b). The first cluster included seventeen cultivars of rice with two subsequent sub-clusters. These seventeen cultivars were BR-1, BR-9, BR-16, BR-11, BR-18, BR-14, BR-7, BR-8, BR-26, BRRI dhan-27, BRRI dhan-29, BRRI dhan-35, BRRI dhan-28, BRRI dhan-36, BRRI dhan-47, BRRI dhan-45 and IR-64. The second cluster was consisted of only one cultivar BR-15, The third and fourth clusters were constituted of BR-3 and BR-17 and BR-19 and BRRI dhan-50, respectively. The PCA yielded eigenvalues of each principle component axes of coordination of varieties with the first three axes (percentage of PCA1, PCA2, PCA3 recorded 64.7295, 6.9856 and 5.5923 respectively) totally accounted for 77.3073 (Fig. 3b).
Polymorphism of SSR markers and cluster analysis
The 22 rice cultivars were fingerprinted using seven SSR primers out of which three generated 100 % polymorphism and the sizes of alleles ranged from 233 to 950 bp (Table 3). A representative agarose gel for primer CMCCA145 is shown in Fig. 1c. The number of alleles detected by a single marker ranged from 3 to 5 with an average of 4.0. The similarity coefficients of SSR markers ranged from 0.0 to 1.0. The highest similarity index (1.0) was found between several cultivars such as BR- 8 and BR-9; BR-14 and BR-15; BRRI dhan-36, BRRI dhan-45 and BRRI dhan-47; BRRI dhan-28, BRRI dhan-29 and BRRI dhan-50. However, these were distributed in several sub-cluster of the dendrogram. In the dendrogram, the 22 rice cultivars were divided into 2 clusters (Fig. 2c). The first cluster was constituted with three rice cultivars namely BR-1, BR-3 and BR-16. The second cluster which was consisted of the rest of the 22 cultivars, was further divided into two distinct subclusters. The PCA yielded eigenvalues of each principle component axes of coordination of varieties with the first three axes (percentage of PCA1, PCA2, PCA3 recorded 51.8799, 12.2734 and 9.4953 respectively) totally accounted for 73.6485 (Fig. 3c).
Combined analysis with RAPD, ISSR and SSR data
The combined similarity coefficients of 22 rice cultivars based on RAPD, ISSR and SSR markers ranged from 0.25 to 0.89. BRRI dhan-27 and BRRI dhan-29 showed the highest similarity index (0.89), while the lowest similarity index (0.25) was found between BR-3 and BR-19. Cluster analysis performed with combined data of the markers generated a dendrogram that grouped the genotypes into several clusters (Fig. 2d). The first cluster was consisted of seventeen cultivars. The first cluster was constituted with BR-1, BR-11, BR-16, BR-8, BR-9, BR-18, BR-14, BR-15, BR-17, BR-26, BRRI dhan-27, BRRI dhan-29, BRRI dhan-28, BRRI dhan-35, BRRI dhan-36, BRRI dhan-45 and BRRI dhan-47. This cluster was divided into two distinct subclusters, each of which were subsequently divided into two subclusters. The PCA yielded eigenvalues of each principle component axes of coordination of varieties with the first three axes (percentage of PCA1, PCA2, PCA3 recorded 57.6700, 7.4895 and 4.4610 respectively) totally accounted for 69.6205 (Fig. 3d).
Comparison of RAPD, ISSR and SSR markers
All the three markers were highly polymorphic among the rice cultivars. Of these, SSR showed 100 % polymorphism (Table 4). RAPD and ISSR are comparable in respect to polymorphism (90.90 % and 90.47 % for RAPD and ISSR, respectively). However, it is notable that SSR produced fewer polymorphic bands than both RAPD and ISSR (Table 4). Comparison of different similarity matrices revealed that ISSR and combined dataset were the most correlated (r = 0.80673) followed by RAPD and combined dataset (r = 0.75267).
Discussion
In the present investigation, the average number of fragments amplified by RAPD primers among the genotypes was 8.25 with a range of 7 to 10 where average of polymorphic and monomorphic bands were 7.5 and 0.75 respectively. Kanawapee et al. [19] also evaluated 20 RAPD and 20 SSR markers and a total of 161 RAPD alleles were produced which revealed 68.94 % polymorphism. Other authors also reported higher level of polymorphism for rice i.e. 80 % among 42 Indian elite varieties estimated by using 40 RAPD primers [8], 72.9 % among 7 Egyptian genotypes using 8 RAPD primers [31] and 89.4 % among 40 cultivars of Pakistani rice using 25 RAPD primers [27]. Rahman et al. [28] detected 53.85 % of polymorphism in rice of Bangladesh. Such variation in the number of fragments produced by these arbitrary primers may be attributed to the differences in the binding sites throughout the genome of the genotypes included. ISSR primers generated 7 to 14 bands with average of 10.5 of which 9.5 was with polymorphic bands and 1.0 was with monomorphic bands. The total percentage of polymorphism was recorded as 90.47. Earlier, generation of 3 to 6 fragments in drought tolerant rice [35] and 8 fragments in Assam rice [2D:\Programs\ProductionJournal\AppData\Local\Microsoft\Windows\Temporary Internet Files\Content.IE5\index.html - 7#7] by ISSR primers were reported. Nagaoka and Ogihara [24] have reported that the ISSR primers produced several times more information than RAPD markers in wheat. Bhuyan et al. [2] reported 201 polymorphic fragments which showed 98 % polymorphism in Assam rice using ISSR primers. It may be due to highly polymorphic, abundant nature of the microsatellites due to slippage in DNA replication. Galván et al. [11] also concluded that ISSR would be a better tool than RAPD for phylogenetic studies. Compared to RAPD analysis, higher level of polymorphism was observed with SSR markers among twenty two rice cultivars. The average number of fragments amplified by SSR primers among the cultivars was 4.0 with a range of 3 to 5 and 100 % polymorphism was observed. Thus, it can be said that SSR revealed more polymorphism than RAPD and ISSR despite producing fewer bands. Zeng et al. [37] and Bounphanousay et al. [5] reported in average 4.3 bands per locus and 3.1 bands per locus respectively. In contrast, Brondani et al. [6] determined 6–22 bands per locus in average 14.6 for Brazilian rice. In the present study, the polymorphic bands detected in rice varieties will be very useful in future rice research. The molecular markers identified by the present laboratory procedure are expected to accelerate the detection of drought tolerant, saline tolerant or disease tolerant genotypes more efficiently as compared to those identified by field screening which is a very difficult procedure with less accuracy. Pakniyat and Tavakol [26] found markers related to drought tolerance in bread wheat genotypes using these markers. Moreover, Nazari and Pakniyat [25] found markers associated with drought tolerance in wild and cultivated barley genotypes using RAPD markers.
Cluster analysis was carried out on four sets of marker profiling data based on RAPD, ISSR, SSR and combination of RAPD, ISSR and SSR. The results based on all the four combinations of DNA marker profiles broadly grouped the 22 cultivars into several clusters where the clusters were subsequently divided into further sub clusters. However, the formation of subclusters within the cluster varied between RAPD, ISSR and SSR. The highest similarity was noticed between BRRI dhan-27 and BRRI dhan-29 in the combined cluster. This was also found in RAPD and ISSR based dendrogram while their position in the SSR based dendrogram was different. The highest similarity coefficient varied among different markers but the highest value was recorded as 0.94 between BRRI dhan-27 and BRRI dhan-29 in ISSR marker system. The dendrogram obtained from the UPGMA method was supported by the PCA analysis. There was close relationship among some of the rice cultivars used in this study, presumably they might have been collected from similar locations or these cultivars may have been derived from the same pedigree. Based on the findings, it can be concluded that some distantly related varieties like BR-1 and IR-64 can be utilized for breeding better varieties in future development programme. Despite being the most commercially important crop in Bangladesh, the research findings on genetic analysis of different rice cultivars using molecular markers are scarce. Molecular markers are scattered throughout the genome and their association with various agronomic traits is influenced by the cultivator under selection pressure induced by domestication. Exploration and evaluation of diversity among these landraces would be of great significance for in situ conservation and rice breeding programs.
References
Akagi H, Yokozeki Y, Inagaki A, Nakamura A, Fujimura T. A co-dominant DNA marker closely linked to the rice nuclear restorer gene, Rf-1, identified with inter-SSR fingerprinting. Genome. 1996;39:1205–9.
Bhuyan N, Borah BK, Sarma RN. Genetic diversity analysis in traditional lowland rice (Oryza sativa L.) of Assam using RAPD and ISSR markers. Curr Sci. 2007;93:967–72.
Bornet B, Muller C, Paulus F, Branchard M. Highly informative nature of inter simple sequence repeat (ISSR) sequences amplified using tri- and tetra-nucleotide primers from DNA of cauliflower (Brassica oleracea var. ‘botrytus’ L.). Genome. 2002;45:890–6.
Borsch T, Hilu KW, Quandt D, Wilde V, Neinhuis C, Barthlott W. Noncoding plastid trnT-trnF sequences reveal a well resolved phylogeny of basal angiosperms. J Evol Biol. 2003;16:558–76.
Bounphanousay C, Jaisil P, McNally KL, Sanitchon J, Hamilton NRS. Variation of microsatellite markers in a collection of Lao’s black glutinous rice (Oryza sativa L.). Asian J Plant Sci. 2008;7:140–8.
Brondani C, Caldeira KDS, Borba TCO, Rangel PN, de Morais OP, de Castro EM, Rangel PDN, Mendonça JA, Brondani RV. Genetic variability analysis of elite upland rice genotypes with SSR markers. Crop Breed Appl Biotechnol. 2006;6:9–17.
Carvalho A, Matos M, Lima-Brito J, Guedes-Pinto H, Benito C. DNA fingerprint of F1 interspecific hybrids from the Triticeae tribe using ISSRs. Euphytica. 2005;143:93–9.
Davierwala AP, Chowdari KV, Kumar S, Reddy APK, Ranjekar PK, Gupta VS. Use of three different marker systems to estimate genetic diversity of Indian elite rice varieties. Genetica. 2000;108:269–84.
Doyle JJ, Doyle JL. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 1987;19:11–5.
Fernández E, Figueiras M, Benito C. The use of ISSR and RAPD markers for detecting DNA polymorphism, genotype identification and genetic diversity among barley cultivars with known origin. Theor Appl Genet. 2002;104:845–51.
Galván MZ, Bornet B, Balatti PA, Branchard M. Inter simple sequence repeat (ISSR) markers as a tool for the assessment of both genetic diversity and gene pool origin in common bean (Phaseolus vulgaris L.). Euphytica. 2003;132:297–301.
Garris AJ, Tai TH, Coburn J, Kresovich S, McCouch S. Genetic structure and diversity in Oryza sativa L. Genetics. 2005;169:1631–8.
Godwin ID, Aitken EA, Smith LW. Application of inter simple sequence repeat (ISSR) markers to plant genetics. Electrophoresis. 1997;18:1524–8.
Hamada H, Petrino MG, Kakunaga T. A novel repeated element with Z-DNA-forming potential is widely found in evolutionarily diverse eukaryotic genomes. Proc Natl Acad Sci USA. 1982;79:6465–9.
Jaccard P Nouvelles rescherches sur la distribution florale. Bull Soc Vaudoise Sci Nat. 1908;44:223–70.
Jain S, Jain RK, McCouch SR. Genetic analysis of Indian aromatic and quality rice (Oryza sativa L.) germplasm using panels of fluorescently-labeled microsatellite markers. Theor Appl Genet. 2004;109:965–77.
Jayamani P, Negrão S, Martins M, Oliveira MM. Genetic relatedness of Portuguese rice accessions from diverse origins as assessed by microsatellite markers. Crop Sci. 2007;47:879–84.
Joshi SP, Gupta VS, Aggarwal RK, Ranjekar PK, Brar DS. Genetic diversity and phylogenetic relationship as revealed by inter-simple sequence repeat (ISSR) polymorphism in the genus Oryza. Theor Appl Genet. 2000;100:1311–20.
Kanawapee N, Sanitchon J, Srihaban P, Theerakulpisut P. Genetic diversity analysis of rice cultivars (Oryza sativa L.) differing in salinity tolerance based on RAPD and SSR markers. Electronic J Biotechnol. 2011;14(6):2–2.
Kantety RV, Zeng X, Bennetzen JL, Zehr BE. Assessment of genetic diversity in dent and popcorn (Zea mays L.) inbred lines using inter-simple sequence repeat (ISSR) amplification. Mol Breed. 1995;1:365–73.
Kaushik A, Saini N, Jain S, Rana P, Singh RK, Jain RK. Genetic analysis of a CSR10 (indica) × Taraori Basmati F3 population segregating for salt tolerance using ISSR markers. Euphytica. 2003;134:231–8.
Kojima T, Nagaoka T, Noda K, Ogihara Y. Genetic linkage map of ISSR and RAPD markers in Einkorn wheat in relation to that of RFLP markers. Theor Appl Genet. 1998;96:37–45.
Lima-Brito J, Carvalho A, Martin A, Heslop-Harrison JS, Guedes-Pinto H. Morphological, yield, cytological and molecular characterization of a breed wheat × tritordeum F1 hybrid. J Genet. 2006;85:123–31.
Nagaoka T, Ogihara Y. Applicability of inter-simple sequence repeat polymorphisms in wheat for use as DNA markers in comparison to RFLP and RAPD markers. Theor Appl Genet. 1997;94:597–602.
Nazari L, Pakniyat H. Genetic diversity of wild and cultivated barley genotypes under drought stress using RAPD markers. Biotechnol. 2008;7:745–50.
Pakniyat H, Tavakol E. RAPD markers associated with drought tolerance in bread wheat (Triticum aestivum L.). Pak J Biol Sci. 2007;10:3237–9.
Rabbani MA, Pervaiz ZH, Masood MS. Genetic diversity analysis of traditional and improved cultivars of Pakistani rice (Oryza sativa L.) using RAPD markers. Electron J Biotechnol. 2008;11:3.
Rahman SN, Islam MS, Alam MS, Nasiruddin KM. Genetic polymorphism in rice (Oryza sativa L) through RAPD analysis. Indian J Biotechnol. 2007;6:224–9.
Ren F, Lu B-R, Li S, Huang J, Zhu Y. A comparative study of genetic relationships among the AA-genome Oryza species using RAPD and SSR markers. Theor Appl Genet. 2003;108:113–20.
Rohlf FJ. NTSYS-pc: Numerical Taxonomy and multivariate analysis System. New York: Ver. 2.1. Exeter Publishing Ltd. Setauket; 2000.
Saker MM, Youssef SS, Abdallah NA, Bashandy HS, El El Sharkawy AM. Genetic analysis of some Egyptian rice genotypes using RAPD, SSR and AFLP. Afr J Biotechnol. 2005;4:882–90.
Shishido R, Kikuchi M, Nomura K, Ikehashi H. Evaluation of genetic diversity of wild rice (Oryza rufipogon Griff.) in Myanmar using simple sequence repeats (SSRs). Genet Resour Crop Evol. 2006;53:179–86.
Sneath PHA, Sokal RR. Numerical taxonomy. San Francisco, USA: W.H. Freeman and Company; 1973.
Williams JGK, Kubelik AR, Livak KJ, Rafalski JA, Tingey SV. DNA polymorphisms amplified by arbitrary primers are useful as genetic markers. Nucleic Acids Res. 1990;18:6531–5.
Youssef MA, Mansour A, Solliman SS. Molecular markers for new promising drought tolerant lines of rice under drought stress via RAPD-PCR and ISSR markers. J Amer Sci. 2010;6:355–63.
Yu SB, Xu WJ, Vijaykumar CHM, Ali J, Fu BY, Xu JL, Jiang YZ, Marghirang R, Domingo J, Aquino C, Virmani SS, Li ZK. Molecular diversity and multilocus organization of the parental lines used in the International Rice Molecular Breeding Program. Theor Appl Genet. 2003;108:131–40.
Zeng L, Kwon T-R, Liu X, Wilson C, Grieve CM, Gregorio GB. Genetic diversity analyzed by microsatellite markers among rice (Oryza sativa L.) genotypes with different adaptations to saline soils. Plant Sci. 2004;166:1275–85.
Zietkiewicz E, Rafalski A, Labuda D. Genome fingerprinting by simple sequence repeat (SSR)-anchored polymerase chain reaction amplification. Genomics. 1994;20:176–83.
Acknowledgments
The authors are gratefully acknowledged to Professor Joarder DNA and Chromosome Laboratory, Department of Genetic Engineering and Biotechnology, University of Rajshahi and USDA (Grant No. 09.111.014.10.01.002.2009-13) and Ministry of Science and Technology (MoST), Bangladesh (Grant No. BS 58 (2010-2011)), for financial support. The authors also acknowledge the support and encouragement of the Principal, Vivekananda College, Thakurpukur, West Bengal, India.
Authors’ contribution
Shah Md. Mahbub Alam, Sadia siddika, Md. Enamul Haque, Md. Asadul Islam and Biswanath Sikdar designed the workplan, performed the experiments and wrote the paper. Ashutosh Mukherjee performed statistical analysis.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Alam, S.M.M., Siddika, S., Haque, M.E. et al. Genetic diversity of some upland and lowland rice cultivars in Bangladesh using RAPD, ISSR and SSR markers. Nucleus 59, 15–23 (2016). https://doi.org/10.1007/s13237-015-0148-x
Received:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13237-015-0148-x