Abstract
Lactic cultures were screened for their ability to accumulate γ-aminobutyric acid (GABA) in the culture medium. Lactobacillus bulgaricus CFR 2028 produced the highest yields of GABA (22.7 mM) from the substrate monosodium glutamate (MSG). Instrumental characterization by high-performance liquid chromatography and structural characterization by mass spectrometry confirmed GABA production by L. bulgaricus CFR 2028. The differential production of GABA in complex and minimal media indicated that minimal medium (modified tryptone, yeast extract and glucose; TYG) supported the highest production of GABA (37 mM), thereby signifying that the acidic pH alone could have a significant bearing on the yield of GABA. This observation opens up avenues for the optimization of the medium components for yield maximization. The biotransformation kinetics of MSG was examined in batch experiments by varying initial MSG concentration (1–5 %). The Monod model was fitted to determine the kinetic parameters under the MSG uninhibited domain, and the MSG inhibited domain was represented well by Briggs–Haldane model.
Similar content being viewed by others
Introduction
Probiotics are beneficial microorganisms that have been proven to contribute to human health by easing malnutrition and infection and modulating the host’s gastrointestinal and immune system. Probiotics have recently been recognized in the field of neuromicrobiology for their ability to synthesize neuroactive compounds that affect the host's gastrointestinal and psychological health. Probiotics, upon ingestion, enter an interactive environment comprising microbiological, immunological and neurophysiological components. They are known to produce an array of neurochemicals for which receptor-based targets on immune and neuronal elements have been well-characterized (Lyte 2011). One important neurochemical, γ-aminobutyric acid (GABA), has attracted greater attention in recent times because GABA along with glutamate known to modulate the overall excitability of the central nervous system (CNS); consequently, these molecules are believed to be involved in all functions of the CNS and in all CNS disease conditions. GABA is formed in vivo from glutamate by the action of glutamate decarboxylase (GAD; EC 4.1.1.15). In addition to its neurotransmitter function, GABA also plays physiological roles, such as inducing insulin secretion from the pancreas (Adeghate and Ponery 2002), effectively preventing diabetic conditions (Hagiwara et al. 2004), treating inhibitory motor disorders (Cho et al. 2007) and reducing blood pressure (Inoue et al. 2003). Due to these health benefits, the production of GABA through biotechnological approaches has gained considerable importance. Thus, GABA production both in biotechnological and pharmaceutical fields has been studied. Biotransformation of glutamate to GABA by probiotics may be a more selective method due to mild reaction conditions, high catalytic efficiency, safety, lower cost and environmental compatibility (Zhang et al. 2011). As a consequence, the use of probiotics with GAD activity in GABA production is thus being intensively investigated (Renye and Somkuti 2012). The production of GABA has been reported by Lactobacillus buchneri, isolated from kimchi (Cho et al. 2007), Lactococcus lactis subsp. lactis (Lu et al. 2009), Lactobacillus brevis (Zhang et al. 2010), Lactobacillus plantarum (Di Cagno et al. 2009) and probiotic NCL912 (Li et al. 2008).
Microbial production of GABA using lactic cultures is well documented (Cho et al. 2007; Lu et al. 2009; Zhang et al. 2011) but, only a few studies have focussed on the dynamics of the reaction rate (Zhang et al. 2011). A variety of kinetic substrate utilization and inhibition models are available to describe microbial growth and product formation. Among these, the Monod and Haldane models are well utilized, and although they are basically based on the specific growth rate, they can also be related to the specific rate of substrate consumption (Agarry and Solomon 2008).
The aim of our study was to examine the efficacy of lactic cultures isolated from traditional fermented foods and the faecal matter of healthy breast-fed infants and of cultures from the culture collection centres for GABA production. In addition, there is a scarcity of detailed studies on the evaluation of different fermentative media for GABA production. Such studies are warranted from the point of development of an economically viable process. A secondary arm of our investigation was to study the dynamics of the reaction rate when altered by changes in initial monosodium glutamate (MSG) concentration during the biotransformation of MSG to GABA. Our main interest was to identify the inhibitory effect of MSG level on GABA yields and to correlate the response of the maximum specific rate of MSG consumption as a function of its initial concentration. These predictions may be considered as the prior requirement for understanding any biochemical system.
Materials and methods
Chemicals
Monosodium glutamate was procured from a local supermarket. GABA was procured from Sigma-Aldrich (Saint Louis MO). All other chemicals used were of analytical grade unless otherwise stated.
Microorganisms
Lactobacillus bulgaricus CFR 2028, L. salivarius CFR 2158, L. amylovorus B-4437, L. helveticus B-4526, Lactococcus cremoris B-634 and Enterococcus faecium CFR 3002 were procured from the CFTRI culture collection centre, India; S1-2, S4-3, S4-2, S7-1, S1-4, S5-3 and S3-2 were isolated from Sandaki, a radish- and mustard-based fermented product; Lactobacillus casei NCIM 2586 was obtained from the National Collection of Industrial Microorganisms (NCIM) Pune, India; TI3T, TI3Iw, T1-2, T7-1, Enterococcus faecium and T5-1 were isolated from Takarishta, a buttermilk-based fermented product; Lactobacillus rhamnosus GG ATCC 53103 and Streptococcus thermophilus ATCC 19258 were obtained from American Type Culture Collection (ATCC; Manassas, VA); Tu6A, Tu3A, Tu4 and Tu6B were isolated from Tushambu, a fermented product based on the husk of black gram and barley; 15, M, 3, Lactobacillus fermentum CFR 2195 were isolated from faecal matter of healthy infants; Lactobacillus plantarum MTCC 1407 and Lactococcus lactis MTCC 1484 were procured from Microbial Type Culture Collection (MTCC), Chandigarh, India; Lactobacillus casei Lund was procured from the Swedish International Development Cooperation Agency (SIDA); K0A, K2A2, K7, K0A1, K23 and K1 1 were isolated from Kanjika, a rice-based fermented product; Enterococcus hirae CFR 3001 was isolated from fermented cucumber.
Preparation of inoculum and fermentation
The active cultures were prepared by transferring the culture from glycerol stock into MRS broth, followed by incubation at 37 °C for 24 h and two subcultures prior to use. Fermentation medium (MRS broth supplemented with 1 % MSG) was inoculated with 16-h-old culture (10 % v/v) and incubated at 37 °C for 48 h.
Preliminary screening: qualitative analysis of GABA by thin-layer chromatography
The culture broth was centrifuged at 8,000 rpm for 15 min at 4 °C. The supernatant (6 μl) was spotted onto thin-layer chromatography (TLC) plates [Silicagel 60 F254 plates (20 × 20 cm); Merck KGaA, Darmstadt, Germany). The plates were developed in acetic acid:1-butanol:distilled water (1:4:5) and subsequently sprayed with 0.2 % ninhydrin solution (Cho et al. 2007).
Quantitative analysis using high-performance liquid chromatography
Supernatant (10 μl) was derivatized with ortho-phthaladehyde–mercaptoethanol [OPA-MCE (20 μl)] reagent and boric acid buffer (0.4 M, pH 10.4; 100 μl). Derivatization was carried out at 30 ± 2 °C for 5 min, and the reaction mixture was filtered through microfilters (pore size 45 μm; Millipore, Bedford, MA) and analyzed by high-performance liquid chromatography (HPLC) (model LC-20 AD; Schimadzu Corp., Kyoto, Japan) equipped with a UV detector (SPA-20A). The sample aliquots (20 μl) were injected using a HPLC injector syringe (Hamilton, Bonaduz, Switzerland ) and the analysis was carried out at room temperature (30 ± 2 °C) using an analytical column (Hypersil ODS; Thermo Fisher Scientific, Waltham, MA). The flow rate of the mobile phase [sodium acetate (1.64 g) and triethylamine (200 μl) in 1,000 ml 20 % (v/v) acetonitrile] was 0.8 ml/min, and the wavelength for detection was 338 nm (Li et al. 2008).
Electrospray tandem mass spectroscopy
Electrospray tandem mass spectroscopy (ESI-MS) analysis was carried out on a Waters 2695 separations module consisting of a Micromass® Q-TofUltimaTM, Waters 2996 HPLC system and Waters 2996 photodiode array detector (Waters Corp., Milford, MA). Data were collected and processed on a personal computer with Mass Lynx software ver. 4.0 (Micromass, a diversion of Waters Corp.). The ESI-MS analysis of GABA was performed using underivatized samples. Positive ion mode ESI-MS was acquired, with the needle voltage set at 3.50 kV, cone voltage at 100 V, source temperature at 120 °C and a desolvation temperature of 300 °C. The ESI-MS was performed using argon as the cone gas (50 l/h) and nitrogen as the desolvation gas (500 l/h). Mass spectra of standard GABA and samples were recorded.
Differential production of GABA in complex and minimal media
Five different MRS media were evaluated. The compositions of these media in grams per litre were as follows:
-
MRS1: Protease peptone, 10; beef extract, 10; yeast extract, 5; dextrose, 20; polysorbate-80, 1; ammonium citrate, 2; sodium acetate, 5; MSG, 10; initial pH 6 (Ratanaburee et al. 2011).
-
Modified MRS: Yeast extract, 6; dextrose, 25; polysorbate-80, 1; tryptone, 6; MgSO4·7H20, 0.2; MnSO4·4H20, 0.05; MSG, 10; initial pH 5 (Li et al. 2008).
-
MRS5: Protease peptone, 10; beef extract, 10; yeast extract, 5; dextrose, 20; polysorbate-80, 1; ammonium citrate, 2; sodium acetate, 5; MSG, 50; initial pH 6.5 (Cho et al. 2007).
-
GYP: Proteose peptone, 5; yeast extract, 10; dextrose, 10; sodium acetate, 2; MgSO4·7H2O, 0.2; MnSO4·4H2O, 0.01, FeSO4·7H20, 0.01; NaCl, 0.01; MSG, 10; initial pH 6.8 (Choi et al. 2006).
-
TYG: Yeast extract, 5; dextrose, 10; tryptone, 5, MSG, 10; initial pH 7 (Nomura et al. 1999)
These different media (50 ml) in 250-ml Erlenmeyer flasks were inoculated with 16-h old culture [4–5 optical density (OD) units] of L. bulgaricus CRF 2028 and the flasks incubated at 37 °C for 48 h. The experiments were carried out in duplicate. Changes in pH were recorded, and biomass and GABA after 48 h of incubation were quantified.
Effect of substrate concentration on GABA production and time-course study
The effect of substrate concentration on the growth of L. bulgaricus CFR 2028 and GABA production as a function of fermentation time was studied. TYG medium (100 ml in 500-ml Erlenmeyer flasks) supplemented with 1, 2, 3, 4 or 5 % MSG was inoculated with a 16-h-old culture (4–5 OD units) of L. bulgaricus CRF 2028. The initial pH of the media was adjusted to 7.0. The flasks were incubated at 37 °C, and samples were drawn at regular intervals. The experiments were carried out in duplicate. Changes in pH were recorded, and biomass, residual sugar and GABA as a function of fermentation time were quantified.
Residual glucose estimation
Residual glucose concentration in the supernatant was estimated using the GOD-POD kit (BEACON Diagnostics Pvt Ltd., Navasari, Gujarat, India). The sample (10 μl) was incubated with 1 ml working reagent at 37 °C for 10 min. Absorbance of the incubated samples were read at 505 nm against the standard. Residual glucose concentration in the samples was estimated using Eq. 1:
Biomass estimation
Growth of L. bulgaricus CFR 2028, in terms of OD was determined by measuring the culture turbidity at 600 nm during the course of fermentation (Robbe-Saule et al. 2006).
pH measurement
The pH of the samples drawn at regular time intervals was measured using a digital pH meter equipped with a glass electrode over a range 0.0–14.0 (Analab Scientific Instruments Pvt Ltd, Gujarat, India).
Modelling or theoretical aspects
Monod’s model: MSG uninhibited domain
The differential method of analysis was used to calculate MSG consumption rate, rs (from MSG concentration [S] versus time data). The relationship between rs and [S], may be evaluated by the simple and practical Monod model. The model assumes MSG to be a non-inhibitory compound and its non-inhibitory kinetic form is presented in Eq. 2 (Sukumar 2009).
Thus, by plotting experimentally measured values of \( -\frac{1}{\mathrm{rs}} vs\frac{1}{\left[\mathrm{S}\right]} \), the kinetic parameters K s [saturation constant (mM)] and rs,max [maximum specific rate of MSG consumption (mM/h)] can be obtained.
Briggs–Haldane model: MSG inhibited domain
The Briggs–Haldane’s model was evaluated to present the dynamics of MSG inhibitory effects due to the mathematical simplicity of this model and its wide acceptance for representing the kinetics of inhibitory substrates. Haldane’s inhibitory kinetics is represented as follows:
where K I is the Briggs–Haldane inhibition constant (mM). By fitting the experimental data into the above simplified equation and by plotting \( -\frac{\left[\mathrm{S}\right]}{\mathrm{rs}} \) versus [S], the kinetic parameters can be estimated (Sukumar 2009).
Results and discussion
Qualitative analysis using TLC
Our screening and selection of potent GABA-producing lactic acid bacteria (LAB) strains were based on the qualitative analysis of GABA, carried out using TLC. All of the tested strains were found to be positive for GABA production (Fig. 1). However, there was a distinct variation in the intensity of developed spots, indicating that the production of GABA was strain-dependent.
Thin-layer chromatogram of gamma-aminobutyric acid (GABA) production. Lanes: 1 GABA standard (3 mg/ml) 2 Enterococcus faecium 3 E. hirae CFR 3001, E. faecium, 4 CFR 3002, 5 Streptococcus thermophilus ATCC 19258, 6 Lactobacillus bulgaricus CFR 2028, 7 L. casei Lund, 8 monosodium glutamate (MSG) standard (3 mg/ml)
Quantitative analysis using HPLC
The result of the TLC analysis was further verified by analyzing the supernatant using HPLC. The result of quantitative analysis of GABA produced by the lactic cultures screened is depicted in Fig. 2. The HPLC chromatogram of supernatants of all GABA-producing strains exhibited a peak with a retention time that matched that of the GABA standard (11.7 min). The concentration of GABA in the supernatant varied from 0.091 to 22.79 mM. Among all strains screened, L. bulgaricus CFR 2028 produced the highest amount of GABA (22.79 mM) after 48 h of fermentation. The results of the screening trials indicated that GABA production by L. salivarius CFR 2158, E. faecium CFR 3002, S1-2 and L. amylovorus B-4437 was also considerable, ranging from 9 mM to 17 mM. The culture chosen for further analysis, L. bulgaricus CFR 2028, exhibited a higher biotransformation potential for GABA production than other GABA-producing reported in earlier studies (Yokoyama et al. 2002; Park and Oh 2007; Ali et al. 2009).
Electrospray tandem mass spectroscopy
The structural characterization of GABA was carried out by MS analysis. All measurements were performed in methanol. Under specified conditions GABA appeared as [M+H]+ (hydrogen adduct of compound ‘M’). The mass spectra of the GABA standard showed a distinct peak at an m/z value of 104.99. The presence of a distinct peak at an m/z of 104.92 (Fig. 3) in the underivatized supernatant of L. bulgaricus CFR 2028 confirmed the production of GABA.
Differential production of GABA in complex and minimal media
Maximum GABA production by L. bulgaricus CFR 2028 was observed in TYG medium. Under these conditions of nutrient stress in TYG medium, it is possible that the cells switched to survival mode, catabolizing the available MSG and other nutrients for GABA production and growth, respectively. GABA production was found to be slightly lower in complex media when compared to that in TYG medium. One possible reason for this result could be that the regulatory factors which govern and favour growth may not favour the efficient expression of GAD (Castanie-Cornet and Foster 2001). MRS, a selective complex medium intended for the cultivation of Lactobacillus, supported maximum growth. The production of GABA was highest in TYG medium (38 mM) and least in modified MRS medium (6 mM). The amount of GABA produced was 20, 18 and 9 mM in MRS1, GYP and MRS5 medium, respectively.
Effect of substrate concentration on GABA production and time-course study
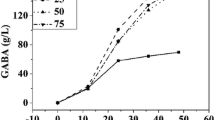
The growth of L. bulgaricus CFR 2028 increased with increasing MSG concentration; consequently, maximum growth was observed in TYG medium supplemented with 5 % MSG. There was a near-complete utilization of glucose by 48, 60, 84, 84 and 96 h of fermentation in the TYG medium supplemented with 1, 2, 3, 4 and 5 % MSG, respectively, supporting the growth accordingly. A slight increase in pH at the end of the fermentation process in TYG medium with an increase in MSG concentration (1 to 5 %) was observed, corresponding to a value of 4.03 ± 0.06, 4.57 ± 0.02, 4.56 ± 0.00, 4.66 ± 0.00 and 4.73 ± 0.01, respectively. GABA production was found to increase with increasing MSG concentration (12.34 ± 0.28 and 46.64 ± 1.35 mM) at 1 and 2 % MSG, respectively. However, it was found to decrease with an increase of >2 % (30.1 ± 1.86, 22.40 ± 3.36, 16.17 ± 1.10 mM GABA at 3–5 % MSG, respectively). L. bulgaricus CFR 2028 produced the highest amount of GABA (47 mM) at an initial concentration of 2 % MSG. It was also observed that the initial pH (7.0) of the TYG medium supplemented with 2 % MSG decreased to pH 5.4 after the addition of the 10 % (v/v) aliquot of 16-h-old inoculum prepared in MRS broth (Fig. 4). The maximum yield (Yp/s) obtained was 0.0449 mM GABA/mM MSG, with a conversion of 39.43 % and volumetric productivity of 0.48 mM/h. At the end of the fermentation period, residual MSG in TYG medium supplemented with 1, 2, 3, 4 and 5 % MSG was found to be 12.10 ± 0.94, 14.88 ± 2.96, 32.85 ± 0.63, 40.58 ± 3.04, 57.44 ± 1.87 mM, respectively.
Monod’s model: MSG uninhibited domain
The production of GABA at an initial MSG concentration of ≤2 % indicated that the system may follow the classical Monod model (Samantha et al. 2008). The suitability of the Monod's equation to fit the data was evaluated using the experimental data (rs and [S]) within the MSG uninhibited region. Simulated work led to a reasonable straight line using the equations shown below for the plot \( -\frac{1}{\mathrm{rs}} vs\frac{1}{\left[\mathrm{S}\right]} \) with a slope and intercept of \( \frac{\mathrm{Ks}}{\mathrm{rs}, \max } \) and \( \frac{1}{\mathrm{rs}, \max } \), respectively.
Thus it can be concluded that the reaction engineering performance of the present system on the MSG uninhibited domain can effectively be described by the Monod model. The average numerical values for rs,max and K s were found to be 27.52 mM/h and 86.53 mM, respectively. The rate equation for the MSG uninhibited domain may, therefore, be written as
Briggs-Haldane model: MSG inhibited domain
The second order polynomial equation for the simplified Briggs–Haldane model was used to determine kinetic parameters for MSG concentrations ranging from 3 to 5 %. The following equations were obtained for MSG concentrations of 3, 4 and 5 %, respectively.
The kinetic parameters of the Briggs–Haldane model were determined using the experimental data for deriving the above equations. The predicted profiles of –rs versus [S] are compared with the experimental in data (Fig. 5). Monod’s model agreed well with the experimental results at 2 % MSG with a standard deviation (SD) of 0.09, while the Briggs–Haldane model fitted the data with a SD of 0.40, 0.11 and 0.10 for 3, 4 and 5 % MSG, respectively.
The variation in kinetic parameters with initial MSG concentration is presented in Table 1. As seen from Table 1, the kinetic parameters show an explicit trend of variation. The rs,max decreased and K s increased with an increased initial MSG concentration. This increase in K s values can be correlated to the decrease in the affinity for L. bulgaricus CFR 2028 for MSG in the MSG inhibited domain (Agarry and Solomon 2008). It is thus apparent that the inhibition became prominent as the initial MSG concentration increased. Thus, we conclude that the rs,max varies with initial MSG concentration and is a strong function of initial MSG concentration. The overall rate equation under the MSG inhibited domain may therefore be written as
The overall magnitude of K I in the present case is greater than the K s value, indicating that the overall system dynamics cannot be explained by the Briggs–Haldane model alone. This necessitates the addition of two biological switches which will be active in the present system. An integrated equation capable of describing the dynamics of overall MSG consumption rate may therefore be compounded as follows (Samantha et al. 2008):
where, β and € are dynamic biological switches. β = 1 and € = 0 when the MSG concentration is ≤ 2 %; β = 0 and € = 1 when the concentration of MSG is ≥ 2 %. The critical MSG concentration for our study was identified to be 2 %.
Conclusion
The results of our preliminary screening indicte that the LAB tested could serve as a potential model to study GABA production. Instrumental characterization by HPLC confirmed the production of GABA by the tested organisms and revealed that L. bulgaricus CFR 2028 produced the highest yield. Further, structural characterization by MS confirmed the production of GABA by L. bulgaricus CFR 2028. Upon evaluating complex and minimal media, we found that the highest production of GABA by L. bulgaricus CFR 2028 was obtained in the TYG medium, clearly indicating that the composition of the medium had a significant effect on the yield of GABA. This result opens up avenues for the optimization of the medium components for maximization of yields. A better growth rate and low GABA production were observed in MRS medium and a low growth rate and high GABA production were obtained in TYG supplemented with 1 % MSG. The use of TYG medium could have implications on the economics of process development.
The inhibitory effect of MSG on the specific rate of substrate consumption was quite appreciable above a critical level of 2 %. Two distinct domains were revealed by studying the effect of initial MSG levels in TYG medium on the dynamics of GABA yields. One domain is characterized by the MSG uninhibited domain (<2 % MSG) where the dynamic response was described well by the classical Monod model. The second domain is a situation of MSG inhibition, which was successfully simulated by the Briggs–Haldane model. A unified biotransformation rate for the entire region (from 4.06 to 27.52 mM/h) covering both the MSG uninhibited and inhibited domains was proposed and which will be useful in bioreactor design for this system.
Abbreviations
- GABA:
-
Gamma-aminobutyric acid
- K I :
-
Briggs–Haldane inhibition constant (mM)
- K s :
-
Saturation constant (mM)
- MCE:
-
Mercaptoethanol
- [M+H]+ :
-
Hydrogen adduct of compound ‘M’
- MSG:
-
Monosodium Glutamate
- OPA:
-
Ortho-phthaladehyde
- rs:
-
Rate of MSG consumption (mM/h)
- rs,max:
-
Maximum specific rate of MSG consumption (mM/h)
- TEA:
-
Triethylamine
References
Adeghate E, Ponery AS (2002) GABA in the endocrine pancreas: cellular localization and function in normal and diabetic rats. Tissue Cell 34:1–6
Agarry SE, Solomon BO (2008) Kinetics of batch microbial degradation of phenols by indigenous Pseudomonas fluorescens. Int J Environ Sci Tech 5(2):223–232
Ali FWO, Saari N, Baker FA, Abdulamir AS, Mohammed AS, Manap YA, Zulkifli AH (2009) Novel, practical and cheap source for isolating beneficial γ-Aminobutyric acid-producing Leuconostoc NC5 bacteria. Res J Med Sci 3(4):146–153
Castanie-Cornet MP, Foster JW (2001) Escherichia coli acid resistance: cAMP receptor protein and a 20 bp cis-acting sequence control pH and stationary phase expression of the gadA and gadBC glutamate decarboxylase genes. Microbiology 147:709–715
Cho YR, Chang JY, Chang HC (2007) Production of γ-aminobutyric acid (GABA) by Lactobacillus bunchneri: ioslated from Kimchi and its neuroprotective effect on neuronal cells. J Microbiol Biotechnol 17(1):104–109
Choi SI, Lee JW, Park SM, Lee MY, Ji GE, Park MS, Heo TR (2006) Improvement of gamma-aminobutyric acid (GABA) production using cell entrapment of Lactobacillus brevis GABA 057. J Microbiol Biotechnol 16:562–568
Di Cagno R, Mazzacane F, Rizzello CG, De Angelis M, Giuliani G, Meloni M, De Servi B, Gobbetti M (2009) Synthesis of gamma-aminobutyric acid (GABA) by Lactobacillus plantarum DSM19463: functional grape must beverage and dermatological applications. Appl Microbiol Biotechnol 86:731–741
Hagiwara H, Seki T, Ariga T (2004) The effect of pre-germinated brown rice intake on blood glucose and PAI-1 levels in streptozotocin-induced diabetic rats. Biosci Biotech Biochem 68:444–447
Inoue K, Shirai T, Ochiai H, Kasao M, Hayakawa K, Kimura M, Sansawa H (2003) Blood-pressure-lowering effect of a novel fermented milk containing γ-aminobutyric acid (GABA) in mild hypertensives. Eur J Clin Nutr 57:490–495
Li H, Gao D, Cao Y, Xu H (2008) A high GABA producing Lactobacillus brevis isolated from Chinese traditional paocai. Ann Microbiol 58(4):649–653
Lu X, Xie C, Gu Z (2009) Optimisation of fermentation parameters for GABA enrichment by Lactococcus lactis. Czech J Food Sci 27(6):433–442
Lyte M (2011) Probiotics function mechanistically as delivery vehicles for neuroactive compounds: microbial endocrinology in the design and use of probiotics. BioEssays 33(8):574–581
Nomura M, Nakajima I, Fujita Y, Kobayashi M, Kimoto H, Suzuki I, Aso H (1999) Lactococcus lactis contains only one glutamate decarboxylase gene. Microbiology 145:1375–1380
Park KB, Oh SH (2007) Production of yogurt with enhanced levels of gamma-aminobutyric acid and valuable nutrients using lactic acid bacteria and germinated soybean extract. Biores Technol 98:1675–1679
Ratanaburee A, Kantachote D, Charernjiratrakul W, Penjamras P, Chaiyasut C (2011) Enhancement of GABA in a fermented red seaweed beverage by starter culture Lactobacillus plantarum DW12. Electronic J Biotechnol 4(3). doi:10.2225/vol14-issue3-fulltext-2
Renye JA Jr, Somkuti GA (2012) Vector-mediated chromosomal integration of the glutamate decarboxylase gene in Streptococcus thermophilus. Biotechnol Lett 34:549–555. doi:10.1007/s10529-011-0802-6
Robbe-Saule V, Jaumouillé V, Prévost MC, Guadagnini S, Talhouarne C, Mathout H, Kolb A, Norel F (2006) Crl activates transcription initiation of Rpos-regulated genes involved in the multicellular behavior of Salmonella enterica serovar Typhimurium. J Bacteriol 188(11):3983–3994
Samantha K, Chowdhury R, Bhattacharya P (2008) Effect of substrate concentration on the transient dynamics of specific cell growth during bioconversion of Cr+6 to Cr+3 using polyculture consortia. Indian J Chem Tech 15:209–215
Sukumar M (2009) Reduction of hexavalent chromium by Rhizopus oryzae. Afr J Environ Sci Technol 4(7):412–418
Yokoyama S, Hiramatsu JI, Hayakawa K (2002) Production of γ-Aminobutyric acid from alcohol distillery lees by Lactobacillus brevis IFO-12005. J Biosci Bioeng 93:95–97
Zhang Y, Goa N, Feng Y, Song L, Goa Q (2010) Biotransformation of sodium L-glutamate to GABA by L. brevis with two glutamate decarboxylase genes. The Fourth Int Conf Bioinformatics and Biomedical Engineering (iCBBE), vol 2. Chengdu, China, 1–4 June 18th, 20th 2010. doi: 10.1109/ICBBE.2010.5518023
Zhang Y, Song L, Goa Q, Yu SM, Li L, Goa NF (2011) The two-step biotransformation of monosodium glutamate to GABA by Lactobacillus brevis growing and resting cells. Appl Microbiol Biotech 94(6):1619–1627. doi:10.1007/s00253-012-3868-8
Acknowledgements
DivyaShri Gangaraju is thankful to the Council of Scientific and Industrial Research (CSIR), New Delhi, India for the award of a Junior Research Fellowship. The Director, CFTRI, is acknowledged for supporting the research work.
Conflict of interest
None.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Gangaraju, D., Murty, V.R. & Prapulla, S.G. Probiotic-mediated biotransformation of monosodium glutamate to γ-aminobutyric acid: differential production in complex and minimal media and kinetic modelling. Ann Microbiol 64, 229–237 (2014). https://doi.org/10.1007/s13213-013-0655-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13213-013-0655-4