Abstract
Heat shock protein 90 (HSP90) is important for many organisms, including plants. Based on the whole genome information, the gene number, gene structure, evolutionary relationship, protein structure, and active site of the HSP90 gene family in Rosa chinensis and Rubus idaeus were determined, and the expression of the HSP90 gene under salt, and drought stresses in two rose varieties Wangxifeng and Sweet Avalanche were analyzed. Six and eight HSP90 genes were identified from R. chinensis and Ru. idaeus, respectively. Phylogenetic analysis revealed that the analyzed genes were divided into two Groups and four subgroups (Classes 1a, 1b, 2a, and 2b). Although members within the same classes displayed highly similar gene structures, while the gene structures and conserved domains of Group 1 (Class 1a and 1b) and the Group 2 (Class 2a and 2b) are different. Tandem and segmental duplication genes were found in Ru. idaeus, but not in R. chinensis, perhaps explaining the difference in HSP90 gene quantity in the two analyzed species. Analysis of cis-acting elements revealed abundant abiotic stress, photolight-response, and hormone-response elements in R. chinensis HSP90s. qRT-PCR analysis suggested that RcHSP90-1–1, RcHSP90-5–1 and RcHSP90-6–1 in Sweet Avalanche and Wangxifeng varieties played important regulatory roles under salt and drought stress. The analysis of protein structure and active sites indicate that the potential different roles of RcHSP90-1–1, RcHSP90-5–1, and RcHSP90-6–1 in salt and drought stresses may come from the differences of corresponding protein structures and activation sites. These data will provide information for the breeding of rose varieties with high stress resistance.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Rosa chinensis belongs to the Rosaceae family. It is one of the most important ornamental plants globally, with significant economic, and cultural value (Omidi et al. 2022). The growth of roses, including R. chinensis, is closely related to changes in the external environment. Drought and salt stress limit their growth and outdoor landscape applications, leading to a significant decline in yield and quality (Jia et al. 2021; Su et al. 2023; Geng et al. 2023). Plants are highly sensitive to environmental changes. Changes in environmental conditions such as drought, heat, excess water, and salinity usually affect plant growth and development in a combined manner. Plants adapt to stress by regulating their signaling pathways and changing the expression of related genes (Shang et al. 2013). Therefore, exploring abiotic stress response genes in roses will help to improve their stress tolerance.
Heat shock proteins (HSPs) are chaperones that are present in many prokaryotes and eukaryotes. HSPs are typical stress response proteins. Their gene activity and production can be induced by stimulating environmental parameters such as high temperature, high salt levels, drought, and other stresses, with important biological significance in the adaptation of organisms to stress (Zhang et al. 2013). HSPs are divided into five families based on their molecular weight: sHSP (small molecule heat shock protein), HSP60, HSP70, HSP90, and HSP100 (Wang et al. 2004; Sajad et al. 2022). HSP90 is a heat shock protein family with an approximate molecular weight of 90 kD. It is a homologous dimer chaperone regulated by adenine riboside triphosphate (ATP). HSP90 comprises an N-terminal domain that binds and hydrolyzes ATP, an intermediate domain that recognizes substrate proteins and companion molecules, and a C-terminal domain that participates in dimer formation (Kitaoku et al. 2022).
Known members of the Hsp90 gene family in model plants including seven in Arabidopsis (Krishna & Gloor 2001), 21 in tobacco (Song et al. 2019), seven in tomato (Zai et al. 2015), 10 in poplar (Zhang et al. 2013), six in cucumber (Zhang et al. 2021), 12 in heading cabbage (Sajad et al. 2022), 35 in Brassica napus (Wang et al. 2022b), and 10 in pumpkin (Hu et al. 2023b). The Hsp90 genes of different plants are generally divided into three to five subgroups that play different roles in plant growth. Hsp90 has important roles in plant seed germination, seedling growth, and the folding, processing, transportation, and degradation of proteins (Reddy et al. 1998; Wang et al. 2004, 2011). Arabidopsis AtHsp90-1 is strongly expressed in mature seed embryos (Prasinos 2005). AtHsp90.6 is an essential protein for embryogenesis (Luo et al. 2019). Inhibited production of HSP90s in Arabidopsis affects fruiting rate, flowering time, and other characteristics (Sangster & Queitsch 2005).
Hsp90 is also important in the response of plants to abiotic stress. HSP90 can control plant growth and development under high-temperature stress by activating the SGT1b TIR1 protein complex (Wang et al. 2016). HSP90 plays an important role in the YODA cascade that mediates the heat stress response and regulation of stomatal formation to adapt to high-temperature environments (Samakovli et al. 2020a, b; Samakovli et al. 2020a, b). Arabidopsis HSP90.1 can activate the substrate protein ZTL, thereby reducing insoluble protein aggregation, refolding erroneous proteins, and improving plant heat tolerance (Gil et al. 2017); HSP90.1 interacts with ROF1 and NBR1 to enhance heat tolerance through heat stress memory (Meiri & Breiman 2009; Thirumalaikumar et al. 2021). Heat stress results in the significantly upregulated expression of CmoHSP90-3 and CmoHSP90-6 in pumpkin, while the expression levels of CmoHSP90-6 and CmoHSP90-10 are significantly upregulated by cold stress (Hu et al. 2023b). The overexpression of five GmHSP90s genes in Arabidopsis can increase proline content and enhance the resistance of the plants to abiotic stresses, such as high temperature and drought (Xu et al. 2013). Overexpression of AtHsp90.2, AtHsp90.5, and AtHsp90.7 can enhance drought resistance in Arabidopsis (Song et al. 2009). In tobacco, NtHSP90-4, NtHSP90-5, and NtHSP90-9 are upregulated in response to induction by abscisic acid (ABA), and drought, salt, cold, and heat stress, while NtHSP90-6 and NtHSP90-7 are not induced by these conditions (Song et al. 2019). MeHSP90.9 can recruit MeWRKY20 and MeCatalase1 proteins to regulate the drought resistance of cassava (Wei et al. 2020). In soybeans, plants transformed with 12 GmHSP90s are strongly induced by high-temperature treatment and exhibit stronger heat and drought resistance (Xu et al. 2013). Finally, in terms of plant immunity, wheat plants overexpressing TaHsp90.2 and TaHsp90.3 exhibit significant resistance to stripe rust (Wang et al. 2011). In rice, a companion complex composed of cytoplasmic Hsp90 and its copartner Hop/Sti1 is involved in chitin response and antifungal immunity (Chen et al. 2010).
In-depth research on the function of plant HSP90 genes under stress is important to understanding plant stress signal transduction and multiple stress response mechanisms, and for improving plant stress resistance. The abiotic stress response mechanisms and expression patterns of the HSP90 gene family in ornamental plants are still unclear, with few studies performed on the HSP90 gene in rose plants. In our study, bioinformatics methods were used to identify and analyze the HSP90 family members of rose. The findings provide a foundation for further studies on the functions of the HSP90 gene and a theoretical reference for studies seeking to breed more stress-resistant rose varieties.
Materials and methods
Plant materials and treatments
'Wangxifeng' and 'Sweet Avalanche' hybrid potted rose varieties were purchased from AIBIDA Horticultural Technology Co., Ltd. (Yunnan, China). These two rose hybrids are the main varieties sold in the market, and the leaves of Wangxifeng are leathery, and the leaves of Sweet Avalanche are paper.
After removing the soil from the selected potted roses that exhibit normal and consistent growth were individually placed in a vase filled with deionized water (Table 1 and Figure S1), and cultivated at 22 ± 1 °C and 60% relative humidity using cycles of 16 h light/8 h dark. Refer to Geng (Geng et al. 2022) and Su's (Su et al. 2023) methods, the experiment adopts a single-factor randomized block design, salt and drought stresses were simulated with 200 mM NaCl and 10% PEG-6000, respectively, for 72 h. Deionized water was used as a control for the same length of time. Leaves were collected taken at 0, 3, 6, 12, 24, 48, and 72 h. The third leaf at the top of each treatment rose was quick-frozen in liquid nitrogen and stored at -80℃, and RNA was extracted for further experiments. Each treatment had three biological replicates.
Quantitative real-time reverse transcription polymerase chain reaction (qRT-PCR)
Total RNA was extracted with an RNA Easy Plant Tissue Kit (DP452, Tiangen, Beijing, China). Genomic DNA contaminants were removed by digestion with RNase-free DNase I as described RNA Easy Plant Tissue Kit. RNA concentration and integrity were measured using Nanodrop 2000(Thermo Fisher Scientific Inc, USA). Reverse transcription of RNAs was performed using the HiScript®II 1st Strand cDNA Synthesis Kit (+ gDNA wiper) (Vazyme Biochemical Technology Co., Ltd., Nanjing, China). The cDNA was stored at -20℃. The relative expression level of the target gene was analyzed using qRT-PCR. The SYBR Green detection kit and AceQ qPCR SYBR Green Master Mix (Vazyme, Nanjing, China) were used in the experiments with the QuantStudio6 system (ABI, Waltham, MA, USA) using the rose gene RcTCTP (Remay et al. 2009) as the internal control. The program comprised 5 min at 95 °C, then 40 cycles at 95 °C for 10 s and 60 °C for 20 s. The comparative 2–ΔΔCT method was used to calculate relative gene expression (Hu et al. 2023a). All the primers used in the present study (Table S1) were synthesized by Gene Create (Wuhan, China).
Identification of HSP90 family members
Genome sequences of R. chinensis, Arabidopsis thaliana, and Oryza sativa were downloaded from Ensemble Plants (http://plants.ensembl.org/index.html). The whole genome sequence of Ru. idaeus was downloaded from the Genome Database for Rosaceae (https://www.rosaceae.org/). The typical HSP90 domain (PF00183) was downloaded from InterPro (https://www.ebi.ac.uk/interpro/) as the search model. As previously described (Xu et al. 2021), a new hidden Markov model was developed to ensure accurate search results, using hmmbuild (E-value < 1 × 10−5) in the HMMERv3 suite to search for the HSP90 gene in the whole genomes of rose, Arabidopsis, rice, and raspberry. The Simple Molecular Structure Research Tool (SMART) (http://smart.embl-heidelberg.de/) (Letunic et al. 2021) and National Center for Biotechnology Information-Conserved Domains Database (NCBI-CDD) (https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi) (Lu et al. 2020) were used to examine candidate sequences, reserving sequences containing the complete HSP90 domain for further analysis (Table S2).
Analysis of physical and chemical properties, phylogeny, gene structure, and conserved motifs
RcHSP90s were submitted to Expasy (https://web.expasy.org/compute_pi/) for the physical and chemical properties prediction protein sequences, and to Plant-mPLoc (http://www.csbio.sjtu.edu.cn/bioinf/plant-multi/) for subcellular localization prediction. A phylogenetic tree (1000 bootstraps) was constructed using the maximum likelihood method of MEGA X software (Kumar et al. 2018) and visualized with EvolView (He et al. 2016). The GFF3 file of roses was used for exon–intron structure analysis. The MEME program (http://memesuite.org/tools/meme) was used for motif analysis. The maximum number of motifs was 20 and the optimal width of each motif was 6–50 residues (Xu et al. 2021). TBtools was used for visualization analysis (Chen et al. 2020).
Macrocollinearity, gene duplication and cis-acting element analyses
The Multicollinearity Scanning Toolkit (MCScanX) was used to identify tandem genes and segmental duplications of the HSP90 gene in roses, as well as macrocollinearity between roses and other species (Wang et al. 2012; Xie et al. 2018). The upstream promoter 2000 bp sequence of HSP90 gene family members was extracted from the rose genome using TBtools. Their cis-regulatory elements were predicted by PlantCARE (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) and visualized by TBtools software (Chen et al. 2020).
Heatmap of RcHSP90s expression based on RNA-seq data
Using the method described by Liu et al. (https://biotec.njau.edu.cn/plantExp/) (Tian et al. 2018; Liu et al. 2023a), the corresponding expression data of RcHSP90s at different stages in flowers, leaves, and roots under ABA, drought, and salt stress treatments were extracted, and used to construct a heatmap using TBtools (Chen et al. 2020).
HSP90 protein structure, active sites, and functional diversification
Secondary structure analysis of proteins was performed using SPOMA (https://npsa-prabi.ibcp.fr/cgi-bin/npsa_automat.pl?page=npsa_sopma.html). SPPIDER (http://sppider.cchmc.org/) was used to perform protein active site prediction. As described by Xu et al. (2021), the functional diversification among the subgroups was analyzed using DIVERGE v3.0 software based on the selected protein sequences (Gu et al. 2013).
Results
Identification, phylogenetic and physicochemical analyses of HSP90 family members
Six, eight, seven, and seven HSP90 genes were identified from the genomes of rose, raspberry, Arabidopsis, and rice, respectively. Members of the HSP90 family were named according to their gene locations (Table S2). The comparative results of phylogenetic evolutionary trees are shown in Fig. 1. The 28 members were divided into two groups and four subgroups (Classes 1a, 1b, 2a, and 2b). RiHSP90-5–1 was not included in any subgroup. Among them, Class 1b and 2b have the most and fewest gene members (nine and four, respectively). Among the four subgroups, except for Class 1a, which does not contain rice HSP90 genes, all other subgroups contain HSP90 genes from four species. In each subgroup, the HSP90 members of rose and raspberry, which both belong to the Rosaceae family, cluster together, indicating a closer evolutionary relationship of HSP90 genes between the two species.
Phylogenetic relationship analysis of 28 HSP90 proteins from Oryza sativa, Arabidopsis thaliana, Rosa chinensis, and Rubus idaeus. The diverse classes of HSP90 proteins are marked with different colors. The HSP90 proteins of Oryza sativa (Os), Arabidopsis thaliana (At), Rosa chinensis (Rc), and Rubus idaeus (Ri) were represented by green circles, blue checkmarks, red stars, and green checkmarks, respectively. Gene IDs of all the analyzed genes are presented in Table S2
The predicted physicochemical properties of rose HSP90 family member proteins are shown in Table 2. The protein encoded by family members was found to range in length from 447 to 839 amino acids (aa), with a relative molecular weight of 51,719.03 to 94,796.23 D and an isoelectric point of 4.93 to 5.52. RcHSP90-4–1, RiHSP90-2–1, RiHSP90-4–1, and RiHSP90-7–1 are unstable proteins (instability coefficient > 40); all belong to the Class 1a and Class 2a subgroups. The other proteins are stable proteins.
Chromosomal distribution and collinearity analysis of HSP90 family members
The distribution of HSP90 on chromosomes of rose and raspberry is shown in Fig. 2. Six members of Rosa chinensis are distributed on five chromosomes and eight members of Raspberry are distributed on six chromosomes. One HSP90 gene was located on chromosome 1, 2, 4, and 5 of roses, and two HSP90 genes were on chromosome 6. Raspberry has one HSP90 gene on chromosome 3, 4, 5, and 6, and two HSP90 genes on chromosome 2 and 7. No tandem genes or segmental duplication genes were presented in the HSP90 gene of roses. RiHSP90-5–1 and RiHSP90-7–1 in raspberry were segmental duplication genes, and RiHSP90-7–1 and RiHSP90-7–2 were tandem duplication genes.
Location, homologous gene pairs, and tandem duplication of HSP90 genes in Rosa chinensis and Rubus idaeus. All HSP90 genes were mapped to their respective loci in Rosa chinensis (A) and Rubus idaeus (B). MCScanX software was used to analyze tandem duplication and homologous gene pairs. The genes in blue font indicate genes are generated through tandem repeats, while the genes at both ends of the red line are homologous gene pairs
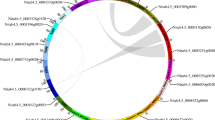
To further explore the relationship between HSP90 family members among different plants, collinearity maps of roses and rice, roses and Arabidopsis, and roses and raspberries were constructed (Fig. 3). The collinearity numbers of HSP90 family members were zero between roses and rice, three between roses and Arabidopsis, and six between roses and raspberries. Compared to rice, roses have more HSP90 collinearity genes than raspberries and with Arabidopsis.
Macro-collinearity analysis of HSP90 genes between Oryza sativa, Arabidopsis thaliana, Rosa chinensis, and Rubus idaeus. Collinearity relationships are shown between two adjacent species. Gray lines in the background indicate the collinear blocks within different genomes and the red lines highlight the syntenic HSP90 gene pairs
Analysis of motif sequence and gene structure of HSP90 family members
Motifs of HSP90 genes in rose, raspberry, Arabidopsis, and rice were analyzed using MEME software. Results of the analyses of structural characteristics of the family members are shown in Fig. 4. Motifs 1, 2, 3, 4, 8, 9, 10, 12, 15, and 18 were presented in HSP90 genes in all four plants. Motifs 17 and 19 were presented in Class 1, while Motif 17 (except AtHSP90-4–1) and 19 were absent in Class 2. All HSP90 members except RiHSP90-2–1 contain Motifs 5, 6, 7, 11, 13, and 16. Unlike Class 2b, Class 2a contained Motif 14. The motif composition of members of the HSP90 gene family in the four plants is similar, with slightly different motif compositions among members of different subgroups. The similarity in motif composition among HSP90 members of the same subgroup is greater.
Conserved motifs and gene structure of HSP90 genes in different species. A Phylogenetic tree constructed using the neighbor-joining method with 1000 bootstrap replicates by MEGA X software. B Motif composition of HSP90 proteins. (C) Exon–intron structure of HSP90s. Yellow boxes indicate exons and black lines indicate introns
A significant difference was evident in the number of exons between Classes 1 and Class 2 (Fig. 4C). The number of exons ranges from one to four in Class 1 and from 15 to 20 in Class 2. In Class 1a (except for RiHSP90-2–1, which contained one exon) all contained four exons. In Class 1b (except for OsHSP90-8–2, which has two exons) all contained three exons. Class 2a contained 19–20 exons and Class 2b contained 15 exons and 14 introns. The similar composition of exons and introns in the same subgroup of the HSP90 gene in rose samples confirms the conservatism of the HSP90 gene.
Analysis of cis elements in rose HSP90 family members
The upstream sequences of HSP90 family members contained many non-biological stress elements, photolight-response elements, and hormone-responsive elements(Fig. 5). In the Class 1a subgroup, RcHSP90-1–1 and RcHSP90-6–1 possess abiotic stress response elements involved in ABA and gibberellin reactions, as well as defense and low-temperature reactions. In the Class 1b subgroup, RcHSP90-5–1 has abiotic stress response elements involved in low temperature. In the Class 1b subgroup, RcHSP90-5–1 has abiotic stress response elements involved in low temperature. In the Class 2a subgroup, RcHSP90-4–1 has cis-acting elements involved in defense and stress response. Finally, in the Class 2b subgroup, RcHSP90-6–2 has cis-acting elements involved in low-temperature induction.
Functional diversification analysis
The functional differentiation analysis findings are shown in Table 3. Among them, the Type-I θ value was 0.1999–0.7810, with a corresponding standard error (SE) of 0.1129. Except for 1b/1a, the p-values of all other subgroups were less than 0.01, indicating a significant difference in their evolutionary rates. Type II analysis revealed θ between 0.0079 and 0.7556, with an SE of 0.2548. Except for 1b/1a, the p-values of all other subgroups were less than 0.01, indicating both a significant level of difference in type II functional differentiation and the presence of functional differentiation caused by amino acid changes in the aforementioned subgroups. The above results indicated that the functional differences between Class 1 and Class 2 genes may due to the changes in evolutionary rate and from changes in certain key amino acid sites. This also applies to the Class 2a and 2b subgroups.
Expression analysis of RcHSP90s
To elucidate the potential function of RcHSP90s, the expression patterns of family members in different organs at different stages and under drought salt stress were assessed. The findings are shown in Fig. 6. The expression level of RcHSP90s showed an overall trend of and initial increase followed by a decrease from the bud stage and flowering stage to aging stage (Fig. 6A). Compared with the expression levels in the bud stage without treatment, the expression levels of RcHSP90-5–1 increased by 237.91% and 161.62% in the flowering and senescence stages, respectively. Upon ABA treatment, the expression levels increased by 212.58% and 120.19%, respectively. Compared with the expression levels during the bud stage without treatment, the expression levels of RcHSP90-6–2 decreased by 71.37% and 60.95% during the flowering and senescence stages, respectively. Upon ABA treatment, the expression levels decreased by 73.54% and 71.61%, respectively. The expression of RcHSP90s in leaves and roots under salt stress is shown in Figs. 6C and D. In salt-tolerant varieties, with increasing salt stress time, the expression level of RcHSP90-5–1 in leaves first increased and then decreased. In roots the reverse trend was evident. Compared with 0 days, the expression level of RcHSP90-5–1 in leaves increased by 127.23% at 7 days, while the expression level of RcHSP90-5–1 in roots decreased by 61.26% at 4 days. Upon imposition of salt stress, expression of RcHSP90-6–2 first decreased and then increased in both leaves and roots. In salt-sensitive varieties, with increasing salt stress time, the expression levels of RcHSP90-5–1, RcHSP90-6–2, and RcHSP90-4–1 in leaves first decreased and then increased. Compared with 0 days, the expression of three genes respectively decreased by 71.31%, 75.27%, and 77.75% at 2 days. With increasing salt stress time, the expression levels of RcHSP90-5–1, RcHSP90-6–2, and RcHSP90-4–1 in the roots all decreased. In drought treatment, compared with the control, the expression level of RcHSP90s first decreased, then increased, and then decreased with increased of stress time. Compared with the control, the expression levels of RcHSP90-1–1, RcHSP90-6–1, and RcHSP90-5–1 in leaves increased by 2636.80%, 1092.68%, and 273.82%, respectively. The collective findings indicate that some members of the HSP90 family in roses may have positive roles in regulating drought and salt stress.
Heatmap of expression profiling of rose HSP90 genes under different stress. The color scale bar represents the expression values (in log2-based tags per million values) of the genes. The values in square frames represent the tags per million values. T, Salt tolerant varieties; F, Salt sensitive varieties
To further investigate the expression changes of RcHSP90s under salt and drought stresses, Two different varieties Sweet Avalanche of paper leaves and Wangxifeng of leathery leaves were used for qRT-PCR. The expression status of HSP90 gene family members under salt and drought stress conditions is shown in Fig. 7. Compared with 0 h, the expression levels of RcHSP90-1–1 and RcHSP90-6–1 in the Class 1a subgroup of the two varieties showed a decreasing trend with increasing drought stress time; the difference became significant. Under salt stress, compared with 0 h, the expression levels of RcHSP90-1–1 in the two varieties also showed a decreasing trend; the difference became significant. However, the expression of RcHSP90-6–1 differed in the two varieties, with a decreasing trend in Sweet Avalanche that was significantly different. In Wangxifeng, RcHSP90-6–1 expression first decreased and then increased, with a significant difference. Both RcHSP90-1–1 and RcHSP90-6–1 were strongly induced by drought and salt stresses in both Wangxifeng and Sweet Avalanche, indicating that these two genes may actively participate in the adaptive regulation of drought and salt stresses. In the Class 1b subgroup, compared to 0 h, the expression levels of RcHSP90-5–1 in the two varieties showed an overall decreasing trend under drought and salt stresses, with the difference reaching a significant level. In the Class 2a subgroup, compared to 0 h, RcHSP90-2–1 expression of the two varieties showed significant differences under salt and drought stresses. The expression of RcHSP90-6–2 in Class 2b subgroup of Sweet Avalanche was 8.37 times that of 0 h under salt stress, while the expression of RcHSP90-6–2 in Wangxifeng was 23.80 times that of 0 h under salt stress at 48 h. The expression of RcHSP90-6–2 in Sweet Avalanche and Wangxifeng increased significantly under drought stress, and the expression of RcHSP90-6–2 in Wangxifeng at 24 h was 12.23 times that at 0 h under drought stress. This indicates that both Wangxifeng and RcHSP90-6–2 in Sweet Avalanche were strongly induced under salt stress and participated in the adaptive regulation of salt stress, but RcHSP90-6–2 in Wangxifeng was more strongly induced. In salt stress, the expression levels of RcHSP90-1–1, RcHSP90-5–1, and RcHSP90-6–1 in Sweet Avalanche were significantly down-regulated, while those of Wangxifeng first decreased and then increased. This indicateed that these three genes play different roles in the adaptive regulation of salt stress in the two varieties.
Expression analysis of RcHSP90s by qRT-PCR. Error bars indicate the standard error. Asterisks indicate significant differences (assessed using Duncan’s honestly significant difference test), **P < 0.01. All the raw data for qRT-PCR are listed in Table S3
Analysis of HSP90s protein structure and active sites
The secondary structure of HSP90 protein in rose and raspberry samples is shown in Table 4. In the secondary structure, the alpha helix comprises the largest proportion, ranging from 46.48% to 55.19%, followed by a random coil at approximately 30%. Beta turn has the lowest proportion, ranging from 3.80 to 5.59%. The average proportion of random coils in the first category (Class 1a and 1b) is 28.93%, while the average proportion of proteins in the second category is relatively high, reaching 30.96%. Correspondingly, the alpha helix in the first category accounts for 52.59%, higher than the 50.49% in the second category (Class 2a and 2b). The number of beta turns for Class 1a, 1b, and 2b were all 19, while Class 2a has 21 (except for RcHSP90-2–1). Except for RiHSP90-2–1, there were 25 alpha helices in Class 1a, 24 in Class 1b, 24–29 in Class 2a, and 32–35 in Class 2b. Compared with the first type of HSP90 protein in roses and raspberries, the quantity and proportion of the second type of alpha helix were higher.
The active sites of HSP90 protein are shown in Fig. 8, S2–S4. Including the proteins of roses and raspberries, the structure of HSP90s in the same subgroup is similar, while the protein structure between different subgroups is slightly different. These findings indicated that the structural differences of homologous genes in HSP90 during evolution were smaller than those between HSP90 proteins in the same family species (roses and raspberries). The active sites of HSP90s were mainly concentrated in four parts (Fig. 8A–D), three were concentrated in the alpha helix and its surroundings (Figs. 8A, B, D), and one was concentrated on the loop (Fig. 8C). The active sites of the second type of HSP90 (Class 2a and 2b, Figures S3-S4) were concentrated in the longer N-terminal extension chain and alpha helix, while the N-terminal extension chain of the first type (Class 1a and 1b, Figs. 8 and S2) is shorter and does not contain alpha helix. The N-terminal of Class 2b protein contains two alpha helices with approximately 15 amino acids; the second one does not feature the distribution of an active site, while the alpha helices of Class 2a were shorter, which is the main difference in the structure and active sites of these two sub histones. The collective findings reveal differences in protein structure and active sites among the subgroups of roses and raspberries to a certain extent, although the similarity among members within the same subgroup is relatively high.
Discussion
Different replication modes of RcHSP90s and RiHSP90s may be the reason for the difference in their quantity
As an important economic ornamental flower globally, the rose accounts for more than 30% of the total global flower trade. The plant is subject to a variety of stresses during production and transportation to the market, which affect its yield and quality; salt and drought are important threats (Liu et al. 2023b; Zhao et al. 2023). The important functions of Hsp90 in plant growth and development regulation, stress response, and other life activities have been studied in many plants, but there are few studies in rose and raspberry. Whole genome identification has identified six Hsp90 family members in rose and eight in raspberry, similar to the findings of seven in Arabidopsis (Krishna & Gloor 2001) and seven in tomato (Zai et al. 2015); less than the 35 in Brassica napus (Wang et al. 2022b), 21 in tobacco (Song et al. 2019), 12 in cabbage (Sajad et al. 2022), and 10 in pumpkin (Hu et al. 2023b); and more than the four in Dendrobium officinale (Wang et al. 2022a). RcHsp90s and RiHsp90s were divided into two major categories and four subgroups (Fig. 1). The amino acid length, isoelectric point, and instability index of Hsp90 members in different subgroups of roses and raspberries were different. The unstable proteins all belong to Class 1a and 2a subgroups (Table 2), indicating that the physicochemical properties of members in each subgroup are also different.
Macro-collinearity analysis showed that the number of Hsp90 collinearity genes between roses and raspberries, roses and Arabidopsis, and roses and rice decreased sequentially, with 6, 3, and 0 genes, respectively (Fig. 3). Both roses and raspberries belong to the Rosaceae family, while they belong to the dicotyledonous family, which is consistent with their close relationship. There are six RcHSP90s, with collinearity with HSP90 in raspberries of also six. There are eight RiHSP90s, among them, RcHSPs do not have tandem duplication genes or segmental duplication genes, while RiHSP90-7–1 and RiHSP90-7–2 are tandem duplication genes, and RiHSP90-5–1 and RiHSP90-7–1 are segmental duplication genes (Fig. 2). In other plants, Csa1G569270/Csa1G569290 in cucumber HSP90 are tandem duplication genes, and no segmental duplication genes (Zhang et al. 2021). B. napus HSP90 replication includes, proximal, dispersed, and whole genome duplication (Wang et al. 2022b). Pumpkin HSP90 contains four pairs of segmental duplication, and no tandem duplication (Hu et al. 2023b). PtHsp90-5a/PtHsp90-5b is the only pair of segmental duplication genes in Populus (Zhang et al. 2013). During the evolution of plant genome and genetic systems, gene duplications have been one of the leading causes of the expansion of gene families (Cannon et al. 2004; Hu et al. 2022, 2023a). The amplification of the HSP90 gene family in each species was performed in a specific way (Zhang et al. 2021), which may be an important reason for the difference in the number of HSP90s between roses and raspberries.
Different protein structures, active sites, and specific expression of HSPs lead to diverse biological functions
Salt stress is one of the most severe environmental factors that restrict crop production globally (Yokoi et al. 2002), and drought has a significant impact on the growth and quality of roses (Su et al. 2023; Zhao et al. 2023; Geng et al. 2023). RcHSP90s contains abundant abiotic stress, hormone, and photolight-response acting elements (Fig. 5). The qRT-PCR results of Sweet Avalanche and Wangxifeng (Fig. 7) showed a decreasing trend of the expression levels of HSP90 genes in Class 1a and 1b subgroups showed under drought and salt stress; the difference became significant, while the expression levels of RcHSP90-6–2 in Class 2b increased significantly. In summary, RcHSP90-1–1, RcHSP90-5–1, and RcHSP90-6–1 in the first category may play important roles in the response of rose to abiotic stresses, such as salt and drought. At the same time, the changing trends of the three genes in the two varieties of Sweet Avalanche and Wangxifeng were not similar, this indicated that the three genes play different roles in the adaptive regulation of drought and salt stress in the two varieties. Previous studies have shown that stiff, leathery leaves are widespread in species adapted to drought occurring in various environments throughout the world, and that waxy coriaceous leaves may help minimize water loss during high water-stress conditions (De Micco and Aronne 2012; Medeiros et al. 2017; Wright et al. 2021), this may be one of the reasons for the differential gene expression between two different leaf type varieties, we will also conduct further research on their differences. HSP90.2 (AT5G56010) and HSP90.3 in Class 1b play important roles in the response to low temperature stress (Bao et al. 2014). The expressions of PtHsp90-1a, PtHsp90-1b, and PtHsp90-3 in the first class of poplar Hsp90 were upregulated under almost all drought stresses, and PtHsp90-1a and PtHsp90-1b were immediately and highly induced under high temperature conditions (Zhang et al. 2013). In general, different sequences encode different proteins and ultimately perform different functions, while homologous genes have similar gene structures and functions, and sequences with high similarity often play similar roles in different species (Chen et al. 2007). Homologous genes may change in the course of evolution due to changes in environment or selection pressure, and intron gain and/or intron loss, and intron densities were proposed to play an important role in the evolution of large eukaryote genomes (Jeffares et al. 2006). Motif composition and gene structure of Hsp90 genes in the same subgroup of Dendrobium officinale, B. napus, and cucumber are highly similar, with differences between different subgroups (Zhang et al. 2021; Wang et al. 2022a, 2022b). In the present study, the motif composition, gene structure, and protein structure of HSP90 within each subgroup of roses were highly similar, with differences between subgroups (Figs. 2, 8, S2–S4). However, compared to the first type, the second type of protein has a longer random coil in the N segment, and these irregular curls were distributed with a large number of active sites. Compared with Class 2a, the HSP90 protein of Class 2b also features two alpha helices with a length of approximately 15 amino acids, although no active sites were distributed on the second helix. Based on this, the basic structure and active sites of HSP90 protein ensure the consistency of protein basic functions, such as abiotic stress. The difference in N-terminal structure and active sites may be one of the reasons for the differentiation of protein functions.
In summary, the HSP90 members of roses and raspberries, both belonging to the Rosaceae family, have greater similarities. Different replication modes are the reason for the quantity difference between the two proteins. The same subgroup features a similar structure. Differences in motif composition, protein structure, and active sites among different subgroups may be the causes of functional differentiation. RcHSP90-1–1, RcHSP90-5–1, and RcHSP90-6–1 may play important roles in the salinity and drought stress of rose These results provide theoretical support for studies of the evolution and function of the HSP90 family of genes and in breeding rose varieties with strong stress resistance.
Data availability
All relevant data are within the manuscript and its Additional files.
References
Bao F, Huang X, Zhu C, Zhang X, Li X, Yang S (2014) Arabidopsis HSP90 protein modulates RPP4-mediated temperature-dependent cell death and defense responses. New Phytol 202:1320–1334. https://doi.org/10.1111/nph.12760
Cannon SB, Mitra A, Baumgarten A, Young ND, May G (2004) The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol 4:10. https://doi.org/10.1186/1471-2229-4-10
Chen F, Mackey AJ, Vermunt JK, Roos DS (2007) Assessing performance of orthology detection strategies applied to eukaryotic genomes. PLoS ONE 2:e383. https://doi.org/10.1371/journal.pone.0000383
Chen L, Hamada S, Fujiwara M, Zhu T, Thao NP, Wong HL, Krishna P, Ueda T, Kaku H, Shibuya N, Kawasaki T, Shimamoto K (2010) The Hop/Sti1-Hsp90 chaperone complex facilitates the maturation and transport of a PAMP receptor in rice innate immunity. Cell Host Microbe 7:185–196. https://doi.org/10.1016/j.chom.2010.02.008
Chen C, Chen H, Zhang Y, Thomas HR, Frank MH, He Y, Xia R (2020) TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol Plant 13:1194–1202. https://doi.org/10.1016/j.molp.2020.06.009
De Micco V, Aronne G (2012) Morpho-anatomical traits for plant adaptation to drought. Plant responses to drought stress. Springer Berlin Heidelberg, Berlin, Heidelberg, pp 37–61
Geng L, Su L, Wang Y, Geng Z, Lin S, Zhang Y, Yu S, Fu L, Liu Q, Cheng C, Jiang X (2022) Role of RcTINY2 in the regulation of drought and salt stress response in arabidopsis and rose. Horticulturae 8(8):747. https://doi.org/10.3390/horticulturae8080747
Geng L, Yu S, Zhang Y, Su L, Lu W, Zhu H, Jiang X (2023) Transcription factor RcNAC091 enhances rose drought tolerance through the abscisic acid–dependent pathway. Plant Physiol 193:1695–1712. https://doi.org/10.1093/plphys/kiad366
Gil K-E, Kim W-Y, Lee H-J, Faisal M, Saquib Q, Alatar AA, Park C-M (2017) ZEITLUPE contributes to a thermoresponsive protein quality control system in Arabidopsis. Plant Cell 29:2882–2894. https://doi.org/10.1105/tpc.17.00612
Gu X, Zou Y, Su Z, Huang W, Zhou Z, Arendsee Z, Zeng Y (2013) An update of DIVERGE software for functional divergence analysis of protein family. Mol Biol Evol 30:1713–1719. https://doi.org/10.1093/molbev/mst069
He Z, Zhang H, Gao S, Lercher MJ, Chen W-H, Hu S (2016) Evolview v2: an online visualization and management tool for customized and annotated phylogenetic trees. Nucleic Acids Res 44:W236–W241. https://doi.org/10.1093/nar/gkw370
Hu P, Ren Y, Xu J, Wei Q, Song P, Guan Y, Gao H, Zhang Y, Hu H, Li C (2022) Identification of ankyrin-transmembrane-type subfamily genes in Triticeae species reveals TaANKTM2A-5 regulates powdery mildew resistance in wheat. Front Plant Sci 13:943217. https://doi.org/10.3389/fpls.2022.943217
Hu P, Ren Y, Xu J, Luo W, Wang M, Song P, Guan Y, Hu H, Li C (2023a) Identification of acyl-CoA-binding protein gene in Triticeae species reveals that TaACBP4A-1 and TaACBP4A-2 positively regulate powdery mildew resistance in wheat. Int J Biol Macromol 246:125526. https://doi.org/10.1016/j.ijbiomac.2023.125526
Hu Y, Zhang T, Wang P, Li Y, Wang M, Zhu B, Liao D, Yun T, Huang W, Chen Y, Zhang W, Zhou Y (2023b) Genome-wide characterization of HSP90 Gene family in chinese pumpkin (Cucurbita moschata Duch.) and their expression patterns in response to heat and cold stresses. Agronomy 13:430. https://doi.org/10.3390/agronomy13020430
Jeffares DC, Mourier T, Penny D (2006) The biology of intron gain and loss. Trends Genet 22:16–22. https://doi.org/10.1016/j.tig.2005.10.006
Jia X, Feng H, Bu Y, Ji N, Lyu Y, Zhao S (2021) Comparative transcriptome and weighted gene co-expression network analysis identify key transcription factors of rosa chinensis ‘Old blush’ after exposure to a gradual drought stress followed by recovery. Front Genet 12:690264. https://doi.org/10.3389/fgene.2021.690264
Kitaoku Y, Taira T, Numata T, Ohnuma T, Fukamizo T (2022) Structure, mechanism, and phylogeny of LysM-chitinase conjugates specifically found in fern plants. Plant Sci 321:111310. https://doi.org/10.1016/j.plantsci.2022.111310
Krishna P, Gloor G (2001) The Hsp90 family of proteins in Arabidopsis thaliana. Cell Stress Chaper 6(3):238. https://doi.org/10.1379/1466-1268(2001)006%3c0238:thfopi%3e2.0.co;2
Kumar S, Stecher G, Li M, Knyaz C, Tamura K (2018) MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol 35:1547–1549. https://doi.org/10.1093/molbev/msy096
Letunic I, Khedkar S, Bork P (2021) SMART: recent updates, new developments and status in 2020. Nucleic Acids Res 49:D458–D460. https://doi.org/10.1093/nar/gkaa937
Liu J, Zhang Y, Zheng Y, Zhu Y, Shi Y, Guan Z, Lang K, Shen D, Huang W, Dou D (2023a) PlantExp: a platform for exploration of gene expression and alternative splicing based on public plant RNA-seq samples. Nucleic Acids Res 51:D1483–D1491. https://doi.org/10.1093/nar/gkac917
Liu X, Zhou X, Li D, Hong B, Gao J, Zhang Z (2023b) Rose WRKY13 promotes disease protection to Botrytis by enhancing cytokinin content and reducing abscisic acid signaling. Plant Physiol 191:679–693. https://doi.org/10.1093/plphys/kiac495
Lu S, Wang J, Chitsaz F, Derbyshire MK, Geer RC, Gonzales NR, Gwadz M, Hurwitz DI, Marchler GH, Song JS, Thanki N, Yamashita RA, Yang M, Zhang D, Zheng C, Lanczycki CJ, Marchler-Bauer A (2020) CDD/SPARCLE: the conserved domain database in 2020. Nucleic Acids Res 48:D265–D268. https://doi.org/10.1093/nar/gkz991
Luo A, Li X, Zhang X, Zhan H, Du H, Zhang Y, Peng X (2019) Identification of AtHsp90 6 involved in early embryogenesis and its structure prediction by molecular dynamics simulations. R Soc Open Sci 6(5):190219. https://doi.org/10.1098/rsos.190219
Medeiros CD, Falcão HM, Almeida-Cortez J, Santos DYAC, Oliveira AFM, Santos MG (2017) Leaf epicuticular wax content changes under different rainfall regimes, and its removal affects the leaf chlorophyll content and gas exchanges of Aspidosperma pyrifolium in a seasonally dry tropical forest. S Afr J Bot 111:267–274. https://doi.org/10.1016/j.sajb.2017.03.033
Meiri D, Breiman A (2009) Arabidopsis ROF1 (FKBP62) modulates thermotolerance by interacting with HSP90.1 and affecting the accumulation of HsfA2-regulated sHSPs. Plant J 59:387–399. https://doi.org/10.1111/j.1365-313X.2009.03878.x
Mooers BHM (2016) Simplifying and enhancing the use of PyMOL with horizontal scripts. Protein Sci 25:1873–1882. https://doi.org/10.1002/pro.2996
Omidi M, Khandan-Mirkohi A, Kafi M, Zamani Z, Ajdanian L, Babaei M (2022) Biochemical and molecular responses of Rosa damascena mill. cv. Kashan to salicylic acid under salinity stress. BMC Plant Biol 22:373. https://doi.org/10.1186/s12870-022-03754-y
Prasinos C (2005) Tight regulation of expression of two Arabidopsis cytosolic Hsp90 genes during embryo development. J Exp Bot 56:633–644. https://doi.org/10.1093/jxb/eri035
Reddy RK, Chaudhary S, Patil P, Krishna P (1998) The 90 kDa heat shock protein (hsp90) is expressed throughout Brassica napus seed development and germination. Plant Sci 131:131–137. https://doi.org/10.1016/S0168-9452(97)00254-9
Remay A, Lalanne D, Thouroude T, Le Couviour F, Hibrand-Saint Oyant L, Foucher F (2009) A survey of flowering genes reveals the role of gibberellins in floral control in rose. Theor Appl Genet 119:767–781. https://doi.org/10.1007/s00122-009-1087-1
Sajad S, Jiang S, Anwar M, Dai Q, Luo Y, Hassan MA, Tetteh C, Song J (2022) Genome-wide study of Hsp90 gene family in cabbage (Brassica oleracea var capitata L.) and their imperative roles in response to cold stress. Front Plant Sci 13:908511. https://doi.org/10.3389/fpls.2022.908511
Samakovli D, Tichá T, Šamaj J (2020a) HSP90 chaperones regulate stomatal differentiation under normal and heat stress conditions. Plant Signal Behav 15:1789817. https://doi.org/10.1080/15592324.2020.1789817
Samakovli D, Tichá T, Vavrdová T, Ovečka M, Luptovčiak I, Zapletalová V, Kuchařová A, Křenek P, Krasylenko Y, Margaritopoulou T, Roka L, Milioni D, Komis G, Hatzopoulos P, Šamaj J (2020b) YODA-HSP90 module regulates phosphorylation-dependent inactivation of SPEECHLESS to control stomatal development under acute heat stress in Arabidopsis. Mol Plant 13:612–633. https://doi.org/10.1016/j.molp.2020.01.001
Sangster TA, Queitsch C (2005) The HSP90 chaperone complex, an emerging force in plant development and phenotypic plasticity. Curr Opin Plant Biol 8:86–92. https://doi.org/10.1016/j.pbi.2004.11.012
Shang H, Li W, Zou C, Yuan Y (2013) Analyses of the NAC transcription factor gene Family in Gossypium raimondii Ulbr.: chromosomal location, structure, phylogeny, and expression patterns. J Integr Plant Biol 55:663–676. https://doi.org/10.1111/jipb.12085
Song H, Zhao R, Fan P, Wang X, Chen X, Li Y (2009) Overexpression of AtHsp90.2, AtHsp90.5 and AtHsp90.7 in Arabidopsis thaliana enhances plant sensitivity to salt and drought stresses. Planta 229:955–964. https://doi.org/10.1007/s00425-008-0886-y
Song Z, Pan F, Yang C, Jia H, Jiang H, He F, Li N, Lu X, Zhang H (2019) Genome-wide identification and expression analysis of HSP90 gene family in Nicotiana tabacum. BMC Genet 20:35. https://doi.org/10.1186/s12863-019-0738-8
Su L, Zhang Y, Yu S, Geng L, Lin S, Ouyang L, Jiang X (2023) RcbHLH59-RcPRs module enhances salinity stress tolerance by balancing Na+/K+ through callose deposition in rose (Rosa chinensis). Hortic Res 10:uhac291. https://doi.org/10.1093/hr/uhac291
Thirumalaikumar VP, Gorka M, Schulz K, Masclaux-Daubresse C, Sampathkumar A, Skirycz A, Vierstra RD, Balazadeh S (2021) Selective autophagy regulates heat stress memory in Arabidopsis by NBR1-mediated targeting of HSP90.1 and ROF1. Autophagy 17:2184–2199. https://doi.org/10.1080/15548627.2020.1820778
Tian X, Wang Z, Zhang Q, Ci H, Wang P, Yu L, Jia G (2018) Genome-wide transcriptome analysis of the salt stress tolerance mechanism in Rosa chinensis. PLoS ONE 13(7):e0200938. https://doi.org/10.1371/journal.pone.0200938
Wang W, Vinocur B, Shoseyov O, Altman A (2004) Role of plant heat-shock proteins and molecular chaperones in the abiotic stress response. Trends Plant Sci 9:244–252. https://doi.org/10.1016/j.tplants.2004.03.006
Wang G, Wei X, Fan R, Zhou H, Wang X, Yu C, Dong L, Dong Z, Wang X, Kang Z, Ling H, Shen Q, Wang D, Zhang X (2011) Molecular analysis of common wheat genes encoding three types of cytosolic heat shock protein 90 (Hsp90): functional involvement of cytosolic Hsp90s in the control of wheat seedling growth and disease resistance. New Phytol 191:418–431. https://doi.org/10.1111/j.1469-8137.2011.03715.x
Wang Y, Tang H, DeBarry JD, Tan X, Li J, Wang X, Lee T-H, Jin H, Marler B, Guo H, Kissinger JC, Paterson AH (2012) MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res 40:e49–e49. https://doi.org/10.1093/nar/gkr1293
Wang R, Zhang Y, Kieffer M, Yu H, Kepinski S, Estelle M (2016) HSP90 regulates temperature-dependent seedling growth in Arabidopsis by stabilizing the auxin co-receptor F-box protein TIR1. Nat Commun 7:10269. https://doi.org/10.1038/ncomms10269
Wang H, Dong Z, Chen J, Wang M, Ding Y, Xue Q, Liu W, Niu Z, Ding X (2022a) Genome-wide identification and expression analysis of the Hsp20, Hsp70 and Hsp90 gene family in Dendrobium officinale. Front Plant Sci 13:979801. https://doi.org/10.3389/fpls.2022.979801
Wang L, Liu F, Ju L, Xue B, Wang Y, Wang D, Hou D (2022b) Genome structures and evolution analysis of Hsp90 gene family in brassica napus reveal the possible roles of members in response to salt stress and the infection of Sclerotinia sclerotiorum. Front Plant Sci 13:854034. https://doi.org/10.3389/fpls.2022.854034
Wei Y, Liu W, Hu W, Yan Y, Shi H (2020) The chaperone MeHSP90 recruits MeWRKY20 and MeCatalase1 to regulate drought stress resistance in cassava. New Phytol 226:476–491. https://doi.org/10.1111/nph.16346
Wright CL, De Lima ALA, De Souza ES, West JB, Wilcox BP (2021) Plant functional types broadly describe water use strategies in the Caatinga, a seasonally dry tropical forest in northeast Brazil. Ecol Evol 11:11808–11825. https://doi.org/10.1002/ece3.7949
Xie T, Chen C, Li C, Liu J, Liu C, He Y (2018) Genome-wide investigation of WRKY gene family in pineapple: evolution and expression profiles during development and stress. BMC Genom 19:490. https://doi.org/10.1186/s12864-018-4880-x
Xu J, Xue C, Xue D, Zhao J, Gai J, Guo N, Xing H (2013) Overexpression of GmHsp90s, a heat shock protein 90 (Hsp90) gene family cloning from soybean, decrease damage of abiotic stresses in Arabidopsis thaliana. PLoS ONE 8:e69810. https://doi.org/10.1371/journal.pone.0069810
Xu J, Hu P, Tao Y, Song P, Gao H, Guan Y (2021) Genome-wide identification and characterization of the Lateral Organ Boundaries Domain ( LBD ) gene family in polyploid wheat and related species. PeerJ 9:e11811. https://doi.org/10.7717/peerj.11811
Yokoi S, Bressan RA, Hasegawa PM (2002) Salt Stress tolerance of plants. JIRCAS Work Rep 23(1):25–33
Zai WS, Miao LX, Xiong ZL, Zhang HL, Ma YR, Li YL, Chen YB, Ye SG (2015) Comprehensive identification and expression analysis of Hsp90s gene family in Solanum lycopersicum. Genet Mol Res 14:7811–7820. https://doi.org/10.4238/2015.July.14.7
Zhang J, Li J, Liu B, Zhang L, Chen J, Lu M (2013) Genome-wide analysis of the populus Hsp90 gene family reveals differential expression patterns, localization, and heat stress responses. BMC Genom 14:532. https://doi.org/10.1186/1471-2164-14-532
Zhang K, He S, Sui Y, Gao Q, Jia S, Lu X, Jia L (2021) Genome-wide characterization of HSP90 gene family in cucumber and their potential roles in response to abiotic and biotic stresses. Front Genet 12:584886. https://doi.org/10.3389/fgene.2021.584886
Zhao X, Lin S, Yu S, Zhang Y, Su L, Geng L, Cheng C, Jiang X (2023) Exogenous calcium enhances the physiological status and photosynthetic capacity of rose (Rosa hybrida L.) under drought stress. Hortic Plant J. https://doi.org/10.1016/j.hpj.2023.01.010
Acknowledgements
This study was funded by the National Natural Science Foundation of China (No. 31901538), the Basic Research Special Project of Key Research Projects in Higher Education Institutions of Henan Province (No. 24ZX013), and the Henan Science and Technology Research Program (No. 212102110477).
Author information
Authors and Affiliations
Contributions
Jun Xu, Ping Hu, and Shuangwei Liu completed the experimental design, experimental process, data analysis, initial draft of the paper, and writing revisions for this study. Yueming Ren completed the experimental process, experimental data, and paper writing revisions. Yang You, Zhifang Wang, Yongqiang Zhang, and Xinjie Zhu participated in experimental design and paper writing revision. All authors have read and agreed with the final text.
Corresponding author
Supplementary Information
Below is the link to the electronic supplementary material.
13205_2024_4052_MOESM2_ESM.pdf
Supplementary file2 Figure S1 Potted rose and experimental treatments. Left: potted rose, 4-6 cuttings per pot; Medium: distilled water was the solution of the control group; Right treatment group: 10% PEG6000 or 200mM NaCl solution (PDF 1589 KB)
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Xu, J., Liu, S., Ren, Y. et al. Genome-wide identification of HSP90 gene family in Rosa chinensis and its response to salt and drought stresses. 3 Biotech 14, 204 (2024). https://doi.org/10.1007/s13205-024-04052-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s13205-024-04052-0