Abstract
In the present study, Streptomyces spp. were isolated, characterized, and the efficacy was tested against Groundnut bud necrosis orthotospovirus (GBNV) in tomato. Among the three inoculation methods viz., pre-, post-, and simultaneous inoculation, tested for antiviral efficacy, pre-inoculation spray of the three Streptomyces spp. viz., Streptomyces mutabilis, Streptomyces rochei, and Streptomyces chrestomyceticus (SAT1, SAT4, and STR2) recorded the least disease severity index (DSI) of GBNV in tomato. In the pot culture, seed treatment of liquid consortium of three Streptomyces spp. @ 2 ml/g of seeds along with seedling dip at 10 ml/lit followed by soil drenching at 10 ml/lit on 7 days after transplanting (DAT) and foliar application at 0.5% on 15 DAT, 30 DAT, and 45 DAT recorded the least GBNV infection of 15% DSI and 16.67% DSI in trial I and II respectively. Besides, under field conditions, the disease incidence was reduced to 14.44% recording a higher yield of 76.67 t/ha in the treated plants against 63.99 t/ha in control. Upregulation of defense genes viz., PR1, PR2, PR6, WRKY, MAPKK, and NPR1 during tripartite interaction between tomato, Streptomyces, and GBNV was analyzed by qRTPCR, indicating that the consortia could decrease the virus severity through induced systemic resistance pathways. Thus, it is concluded that Streptomyces spp. can be used for the management of GBNV in tomato.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Tomato (Solanum lycopersicum L., Family: Solanaceae) is one of the most important and popular vegetable crops grown throughout the world. In India, tomato is being cultivated in an area of 851.69 thousand ha with a production of 21,002.81 thousand tons and productivity of 24.66 MT/ha (Indiastat, 2021) Biotic and abiotic stresses are the major limiting factors in tomato cultivation (AbuQamar et al. 2009), among which diseases, such as damping off, Fusarium wilt, late blight, and bacterial wilt destroy the crop throughout the world (Brahimi et al. 2017). Besides, viral diseases viz., Tomato mosaic virus (TMV), Tomato leaf curl virus (TLCV), Tomato spotted wilt virus (TSWV), and Groundnut bud necrosis orthotospovirus (GBNV) are considered as major constraints causing huge economic losses in commercial cultivation (Scholthof et al. 2011). More than 29 Tospoviruses have been reported globally (Zhu et al. 2019), among which Groundnut bud necrosis orthotospovirus (GBNV) infecting tomato cause severe destruction to the crop (Mandal et al. 2012). GBNV incidence was first reported by Reddy et al. (1968) in India from Groundnut. Severe destruction of crop upto 80% is reported in States like Tamil Nadu, Karnataka, Andhra Pradesh, etc., in India due to GBNV leading to huge economical loss (Dasgupta et al. 2003). Among the different crop growth stages, flowering and fruit formation are most susceptible to GBNV infection in tomato (Umamaheswaran et al. 2003).
Various management strategies viz., installing yellow sticky traps, raising border crops, spraying systemic insecticides targeting the insect vectors have been followed to curtail the disease to a certain extent. However, repeated spraying of insecticides will develop resistance to the molecule besides polluting the environment. Alternatively, inducing immune response in the host through application of bioagents is a promising defensive and ecofriendly strategy (Van wees et al. 2008; Harish et al. 2009). Plant growth-promoting rhizobacteria (PGPR) and endophytic bacteria trigger the induced systemic resistance (ISR) pathways in plants and in turn provide resistance against virus. Besides, beneficial bacteria like Streptomyces, Bacillus, and Pseudomonas possess growth-promoting genes for auxin and cytokinin which induces the growth of the plant. Approximately, two-thirds of the thousands of naturally occurring antibiotics have been isolated from actinobacteria, and about 75% are produced by members of the genus Streptomyces (Basilio et al. 2003). Our previous studies revealed that PR proteins such as chitinase, β-1,3-glucanase, phenylalanine ammonia lyase, polyphenol oxidase, peroxidase and phenolic compounds were triggered by Bacillus spp. and provided resistance against Banana bunchy top virus infection (Harish et al. 2008, 2009). Recently, our research group reported that application of nano-encapsulated Bacillus sp. consortia suppressed GBNV infection in tomato (Kishorkumar et al. 2024). Besides, consortia of Bacillus spp. triggered the defense gene expression and reduced the incidence of CMV in ridge gourd (Karthikeyan et al. 2024). Pseudomonas fluorescens strains (Pf1 and CHAO) reduced leaf crinkle infection in black gram by activating defense enzymes such as peroxidase, phenylalanine ammonia lyase, and polyphenol oxidase (Karthikeyan et al. 2009). Culture filtrate of Streptomyces ovatisporus LC597360 exhibited 93.9% biocontrol efficacy against Tomato mosaic virus (ToMV) through increased activity of defense-related enzymes such as ascorbate oxidase, catalase, peroxidase, and polyphenol oxidase and plant growth promotion (Taha et al. 2021). Pre-inoculation of soil with Streptomyces pactum Act12 agent enhanced activity of both peroxidase and chitinase and induced expression of plant systemic resistance genes in tomato against Tomato yellow leaf curl virus (Li et al. 2019). Accordingly, in this study, Streptomyces spp. was isolated from tomato ecosystem, characterized through molecular techniques and exploited for the management of Groundnut bud necrosis virus in tomato.
Materials and methods
Virus inoculum
The virus isolate was collected from Coimbatore, Tamil Nadu, India and maintained in cowpea var. CO7. Pathogenicity was proved by sap inoculation on tomato var. PKM 1 and was further characterized through scanning electron microscope and molecular methods as described in Rahul et al. (2022). The purified virus isolates (Accession No. ON529555; ON529556) were used for further studies.
Isolation and characterization of Streptomyces spp.
Actinobacteria was isolated from the rhizosphere soil by serial dilution plate technique (Johnson and Curl 1972). Along with the isolated cultures, Streptomyces isolates STR2 (Streptomyces chrestomyceticus), ACT18 (Streptomyces rochei), and ACT30 (Streptomyces griseoviridis) were obtained from the culture collection Centre, Department of Plant Pathology, Tamil Nadu Agricultural University, Coimbatore, Tamil Nadu, India. A section of 10-day-old Streptomyces colony grown on starch casein agar (SCA) plates was mounted on a stub and sputter coated with gold, and the images were taken using SEM (FEI Quanta, 250) available at the Department of Nano technology, TNAU, Coimbatore (Abidin et al. 2016).
Biochemical characterization of Streptomyces spp.
Gram staining
To determine the Gram reaction of the isolate, a thin layer of Streptomyces culture was applied on the glass slide and heat fixed. After floating a drop of crystal violet on the bacteria for 30 s, the slide was rinsed with flowing water. Then it was treated with Gram’s iodine solution for 30 s, rinsed with water, and decolorized with 95 percent ethyl alcohol. The smears were immersed in an oil solution containing safranin for 30 s rinsed, blotted dry, and viewed using a 100 X oil immersion microscope (Hucker and Conn 1923).
IAA, siderophore, and phosphate solubilization
The actinobacterial isolates were tested for the production of IAA, siderophore and ability to solubilize phosphorus. A 20 ml of Kenknight broth that had been modified with 0.1 percent tryptophan was used to assess the IAA production. Actinobacterial cultures were inoculated to the broth, and incubated for 5 days at 28 °C. Cell-free culture was mixed with 2 ml of the Salkowski reagent (0.5 M FeCl2 in 50 ml of 35% perchloric acid), and the mixture was left in the dark for 30 min. The development of pink color suggests the production of IAA. The amount of IAA generated was measured using a spectrophotometer at 530 nm and represented as μg/ml using a standard curve (Patten and Glick 1996). Siderophore synthesis by actinobacteria was tested on Chrome Azurol S blue agar as per the methodology of Schwyn and Neilands (1987). Actinobacterial isolates were streaked on CAS agar medium and incubated for 5 days at 28 °C in dark condition. The development of a color zone ranging from yellow to light orange around the colonies indicates the production of siderophores. The actinobacteria isolates were tested for the solubilization of phosphate as per the protocol of Nautiyal (1999). The actinobacterial isolates were streaked on a sterile Petri plate containing Pikovskaya’s agar medium, and incubated at 30 °C for 4 days. The presence of phosphorus solubilizing capacity was assessed by the clear zone formation in the medium.
Molecular characterization of Streptomyces spp.
The genomic DNA of actinobacteria was isolated following the cetyl trimethyl ammonium bromide (CTAB) method (Clark 2013). Pure culture of actinobacteria was grown on starch casein broth for 7 days. The bacterial suspension was centrifuged for 10 min at 8000 rpm. The supernatant was discarded and the pellet was resuspended with 500 μl CTAB buffer, mixed thoroughly and incubated at 55 °C for 60 min in a water bath. Then 5 M NaCl (100 μl) was added and incubated at 55 °C for 10 min, followed by addition of 500 μl chloroform and shaken intermittently for 30 min and centrifuged at 12,000 rpm for 15 min. To the supernatant, 0.1 vol. sodium acetate and 3 vol. ice cold isopropanol were added and incubated at –20 °C for 4 h. The DNA was pelleted by centrifugation at 12,000 rpm for 15 min at 4 °C following incubation. The pellet was washed with 70% ethanol, air dried, and dissolved in 50 μl of milli-Q water.
Sequencing and phylogenetic analysis
Polymerase chain reaction (PCR) of the genomic DNA of actinobacteria was carried out using genus-specific primers Sm6F (5′-GGTGGCGA6AGGCGGA-3′) and Sm5R (5′- GAACTGAGACCGGCTTTTTGA-3′) as followed by Santos-Cervantes et al. (2017). The reactions were carried out in 10 μl reaction mixture containing 5 μl of master mix, 1 μl of forward primer (1 µM), 1 μl of reverse primer (1 µM), 1 μl of template DNA (100 ng), and 2 μl of milli-Q water. DNA samples were amplified on a DNA thermal cycler (C1000 BioRad Ltd.) using the PCR conditions: initial denaturation at 94 °C for 10 min; followed by 32 cycles of denaturation at 94 °C for 1 min, 57 °C for 1 min and 72 °C for 1 min with a final extension of 72 °C for 10 min. The PCR products were separated by electrophoresis on 1.2% agarose gel and sequenced at Biokart India Pvt. Limited, Bangalore, India. The sequence was deposited in the NCBI and the accession number was obtained. Nucleotide sequences were aligned using CLUSTAL W 1.81 and phylogenetic analysis of the sequence was carried out by comparing the reference sequences retrieved from the Genbank database with 1,000 bootstrap replicates using MEGA 11 software (Tamura et al. 2021).
Preparation of Streptomyces suspension
Each unique actinobacterial strain was individually inoculated into starch casein broth to ensure uniform development and incubated in an orbital shaker at 150 rpm for 5 days at 37 °C (Rahila et al. 2023). Later to the culture broth, Tween 20 (1%), glycerol (1%), trehalose (1%), and polyvinylpyrrolidone (1%) were added. This mixture was incubated in an orbital shaker for 5 min at 150 rpm (Vinodkumar et al. 2018) and used for further experiments.
In vitro screening of Streptomyces spp. against GBNV
To assess the antiviral efficacy of Streptomyces spp. against GBNV, three methodologies viz., pre-inoculation, post-inoculation spray, and simultaneous application were carried out in tomato cv. PKM 1 (Sangeetha et al. 2020). The experiment was carried out in completely randomized design (CRD) with eight treatments replicated thrice. In the pre-inoculation experiment, 0.5 percent (5 ml/L of water) culture broth of the individual isolate was sprayed 24 h prior to the virus inoculation and GBNV was sap inoculated. In the post-inoculation experiment, 0.5 percent culture broth was sprayed 24 h after the inoculation of GBNV through sap; and in the simultaneous inoculation, the virus and the actinobacteria were inoculated at the same time. The treated plants were incubated in the glasshouse at 28 °C for symptoms expression. The disease severity index (DSI) in tomato was calculated based on the symptom severity grade chart (Sain and Chadha 2012) (Fig. 1). The experiment was conducted twice to ensure the reproducibility of the experiment. The DSI was calculated based on the grading using the formula:
Plant growth promotion by roll towel method
The roll towel technique was used to analyze the plant growth-promoting potential of eight Streptomyces spp. Twenty-five tomato seeds of cv. PKM 1 were surface sterilized with one percent sodium hypochlorite solution, rinsed three times with sterile water, and dried. Seeds were soaked in the Streptomyces suspension @ 107 cfu/ml containing 100 mg of carboxymethylcellulose for 12 h, and kept on a germination paper. The seeds immersed in sterile water were used as control. The experiment was kept in a growth chamber at 30°C. The germination percentage and the morphological characters of the seedlings viz., shoot length and root length were measured (Meena et al. 2022), and the vigor index was calculated using the formula:
Development and testing of liquid formulation of Streptomyces spp. consortium against GBNV
Based on their antiviral nature, the compatibility of the best isolates was tested to form a consortium using the cross-streak technique (Fuentes et al.2016). The compatibility of the isolates was assessed by the growth of the isolate. After testing the compatibility, the individual bacterial strains viz., SAT1, SAT4, and STR2 were co-inoculated into starch casein broth and cultured for 5 days at 37°C and 150 rpm in an orbital shaker. The formulation was prepared as described earlier and the concentration was assessed via serial dilution. The experiment was conducted in a CRD with five treatments and four replications with two plants per replication in the glass house. Foliar spray of the Streptomyces consortium @ 0.5% was carried out on 30 days after transplantation followed by inoculation of GBNV after 24 h. The plants inoculated with GBNV and buffer was used as positive and negative control, respectively. As a chemical inducer, foliar spray of salicylic acid at 200 ppm was used. The extra sap was removed using sterile distilled water after the sap inoculation of GBNV. The plant samples were taken in a polythene bag at various time intervals, including 0, 24, 48, 72, and 96 h following inoculation and stored in –20 °C for analyzing the defense gene expression. The treatment details were as follows: T1–uninoculated control, T2–foliar application of Streptomyces consortium @ 0.5% alone, T3–inoculated control (GBNV alone), T4–foliar application of Streptomyces consortium @ 0.5% + GBNV, and T5–foliar application of salicylic acid @ 200 ppm + GBNV.
Evaluation of Streptomyces consortium against GBNV in tomato under glass house conditions
The efficacy of Streptomyces consortium against GBNV was evaluated in two pot culture experiments in tomato cv. PKM1. The experiment was carried out in a completely randomized design (CRD) with eight treatments replicated thrice and five plants per replication. Surface-sterilized tomato seeds cv. PKM 1 were soaked in the liquid formulation of Streptomyces spp. consortium (2 ml/g) for 12 h and dried in shade for 30 min. Twenty-five days after sowing, tomato seedling was dipped in the bioformulation (10 ml/L) for 2 h and planted. For soil drenching, 10 ml of the bioformulation (10 ml/L) was dispensed into each pot 7 days after planting, and 25 ml of bioformulation/pot (0.5%) was sprayed onto the leaves 15 days after transplanting. Foliar application of salicylic acid (200 ppm), Benzothiadiazole, BTH (1 mM), thiamethoxam 25 WG (0.5 g/L), and water were also maintained. Virus was inoculated in the plants using sap transmission method after 24 h of application of bioformulation. The effectiveness of the bioformulation on the incidence of GBNV was recorded 6–21 days after inoculation, which is graded on a 0–4 scale. Disease severity index (DSI) was calculated based on the grading.
Evaluation of Streptomyces consortium against GBNV under field conditions
A field trial was conducted in the farmer’s field at Poochanari (10.6363° N, 76.9045° E) village of Coimbatore district, Tamil Nadu, India during April 2022 to July 2022 to test the plant growth promotion and efficacy of Streptomyces spp. consortium against GBNV in tomato. The trial was conducted in the tomato cv. Heemsohna. The experiment was laid out in a randomized block design (RBD) with eight treatments with a plot size of 5 m × 3 m. Three replications were maintained for each treatment. All the agronomic practices were carried out as per the standard recommendations. Observations were made on per cent disease incidence and growth parameters.
Defense genes expression by quantitative real-time PCR
Tomato leaf samples of all the five treatments from varying time intervals (0, 24, 48, 72, and 96 h) were collected. The total RNA from all these samples was extracted using Trizol method following the protocol of Chomczynski and Sacchi (1987). Further, the Revert Aid first strand cDNA synthesis kit (Thermo scientific) was used to synthesize cDNA. The defense genes’ (PR1, PR2, PR6, WRKY, MAPKK, and NPR1) expression during tritrophic interaction was analyzed using Roche 96 well Light Cycler. The reaction was performed using 5 μl of SYBR green (KAPA SYBR FAST LC480, cat. PGK025-A, Sigma Aldrich), 1.5 μl forward primer, 1.5 μl reverse primer and 2 μl of cDNA. For each gene, qPCR was conducted under pre-incubation of 95 °C for 10 min, three steps amplification of 95 °C for 30 s, 60 °C for 30 s, and 72 °C for 30 s followed by standard melting temperature analysis. Actin was used as a housekeeping reference gene to normalize the defense gene expression. The uninoculated control was fixed at scale 1.0 and utilized as a calibrator. Comparative 2 –ΔΔCT method was used to analyze the relative fold change of the target gene, according to Livak and Schmittgen (2001).
Statistical analysis
The data were statistically analyzed using IBM SPSS Statistics software version 28.0.0.0. The percentage values of the disease incidence are arcsine-transformed. Data were subjected to analysis of variance (ANOVA) at two significant levels (P < 0.05 and P < 0.01), and means were compared by least significant difference (LSD).
Results
In the current study, 18 actinobacteria were isolated from rhizosphere soil collected from various places in Tamil Nadu. Out of 18, 5 Streptomyces spp. viz. SAT1, SAT4, SAT5, SAT11, and SAT18 were selected based on various in vitro antagonistic assays. Besides, three Streptomyces spp. STR2, ACT18, and ACT30 obtained from the Department of Plant Pathology, TNAU, Coimbatore were used for the current research.
Cultural and morphological identification of Streptomyces spp.
All the isolated actinobacteria had shown good growth rate in starch casein agar plates. The substrate and aerial mycelium color varied from shades of white, grey, yellow and to maroon (Fig. 2a). Streptomyces isolates viz., SAT1, SAT4, SAT5, SAT11, SAT18, STR2, ACT30, and ACT18 were further observed with good aerobic growth rate, powdery texture, and earthy odor (Supplementary File 1). The spore produced by the Streptomyces spp. was observed through a phase contrast microscope (40X magnification) which were found to be spherical, hyaline, and arranged in chains (Fig. 2b).
Morphological characterization of Streptomyces isolate. The Streptomyces isolate was grown in starch casein agar medium and the morphological characters were observed. The substrate and aerial mycelium colour varied from shades of white, grey, yellow and to maroon. The spore produced by the Streptomyces spp. was observed through a phase contrast microscope (40X magnification). a. Growth of Streptomyces colony on starch casein agar medium, b. microscopic observation at 40X magnification
Scanning electron microscopy (SEM) of Streptomyces spp.
Morphological confirmation for effective Streptomyces spp. viz. SAT1, SAT4, and STR2 was done through SEM imaging. The average diameter of spores was 617.5 nm in SAT1, 460.8 nm in SAT4, and 704.8 nm in STR2. The SEM images have shown fully grown conidial chains for the isolate SAT1 with a magnification of 25000X (Fig. 3).
Biochemical characterization of Streptomyces spp.
Gram staining
Gram staining was done in the Streptomyces isolates which resulted in retention of crystal violet color indicating all the isolates are Gram positive in nature.
IAA production analysis
The Streptomyces isolates were tested for indole-3-acetic acid production. All the study isolates turned light pink to dark pink color indicating positive result for IAA production. The control has shown yellow colour reaction indicating the experimental set up was precise. The IAA production varied from 70.56 µg/ml in SAT1 followed by 57.47 µg/ml in SAT4, 49.32 µg/ml in STR2 and the least production of 6.83 µg/ml in ACT 18 (Fig. 4a).
a IAA production analysis by different Streptomyces isolates. A Kenknight broth modified with 0.1 percent tryptophan was inoculated with Streptomyces isolate and the IAA production was assessed. All the isolates turned light pink to dark pink colour. The amount of IAA generated was measured using a spectrophotometer at 530 nm and represented as μg/ml using a standard curve: IAA Standard curve. Comparison of IAA production by different isolates. b Siderophore production assay of Streptomyces isolates. c Phosphate solubilization assay of Streptomyces isolates. Actinobacterial isolates were streaked on CAS agar medium and incubated for 5 days at 28°C in dark, and the development of a colour zone ranging from yellow to light orange around the colonies indicates the production of siderophores. The actinobacterial isolates were streaked on a sterile Petri plate containing Pikovskaya’s agar medium and incubated at 30°C for 4 days. The presence of phosphorus solubilizing capacity was assessed by the clear zone formation in the medium
Siderophore production assay
The Streptomyces isolates were tested for siderophore production using CAS medium. The isolates viz., SAT1, SAT4, and STR2 have shown positive reaction to siderophore production as they formed yellow–orange coloured halo around the colony in blue-colored CAS medium. In the control plate, no halo zone was observed (Fig. 4b).
Phosphate solubilization assay
The Streptomyces isolates were tested for phosphate solubilization in Pikovskaya’s medium. The isolates, SAT1 and STR2, formed clear halo zone around the colony indicating positive reaction to phosphate solubilization (Fig. 4c).
Molecular characterization, sequencing, and phylogenetic analysis of Streptomyces spp.
A PCR reaction was performed using Streptomyces genus-specific primers to amplify the 16S rRNA gene of the isolates viz. SAT1, SAT4, SAT5, SAT11, and SAT18. STR2 (Streptomyces chrestomyceticus Accession No-MW50590) was used as a positive control. All the isolates produced an amplicon of 620 bp. PCR products of two effective isolates SAT1 and SAT4 were sequenced and analyzed. The BLAST analysis revealed the isolate SAT1 is having 99.37% sequence similarity with Streptomyces mutabilis (LM644089) and SAT4 is having 99.64% identity with Streptomyces rochei (MN631088). The sequences were deposited in GenBank and is available under the accession numbers OP115789 (SAT1) and OP115790 (SAT4). The phylogenetic analysis revealed the characterized isolates SAT1 and SAT4 were clustered under a single group, while the isolate STR2 formed a separate group (Fig. 5).
Phylogenetic tree of effective Streptomyces isolates. Polymerase chain reaction (PCR) of the genomic DNA of actinobacteria was carried out using genus specific primers. The phylogenetic analysis was performed using MEGA 11.0 software using neighbor joining method. Tree branch support was determined using 1000 replications. Boot strap values are displayed at the nodes. Bacillus subtilis was used as an out-group
Compatibility test for formulating Streptomyces spp. consortium liquid formulation
The ability of the effective Streptomyces spp. on plant growth promotion by standard roll towel method, revealed high vigour index in STR2 (1078.17), followed by SAT4 (1038.72) and SAT1 (1025.80) which were on par with each other. The compatibility of three best isolates viz., SAT1, SAT4, and STR2 was checked but no inhibition zone is found between the tested isolates. Therefore, these three isolates having plant growth promotion and antiviral activity are compatible with each other and could be co-inoculated to form a consortium.
Efficacy of Streptomyces spp. consortium against GBNV in glasshouse conditions
Disease severity index
The disease severity index (DSI) was calculated based on the observations made after 21 days of virus inoculation. In trail I, sequential application of seed treatment and seedling dip, soil drenching and foliar application of Streptomyces consortium (T4) recorded the lowest DSI of 15.00 with 82.69% virus inhibition over control. The inoculated control recorded a DSI of 86.67, whereas salicylic acid recorded 50.00 DSI with 42.31% virus inhibition (Table 1). In trial II, the sequential application of Streptomyces consortium recorded the lowest DSI of 16.67% with 81.48% virus inhibition. All the treatments were significantly different with respect to the virus inoculated control.
Profiling of defense genes response in the tripartite interaction
The priming effect of Streptomyces sp. against GBNV was assessed through q-RT PCR. Six defense-related genes viz., PR1, PR2, PR6, WRKY, MAPKK1, and NPR1 genes of tomato were used as targets with the house keeping gene actin as internal control.
-
i.
PR1
In the present study, the consortium-treated plants challenged with GBNV shown upregulation of PR1 gene by 2.41-fold at 24 h, 2.70-fold at 48 h, peak expression of 6.75-fold at 72 h, and 5.25-fold at 96 h after virus inoculation. In the present study, a significant difference with respect to uninoculated control was observed in the relative expression pattern of PR1 gene, which confirms that Streptomyces consortium has consistently induced the PR1 defense gene expression in virus challenged tomato plants (Fig. 6a).
Expression analysis of defense genes in tomato upon application of Streptomyces consortium and challenged with GBNV through real-time PCR. The defense genes (PR1, PR2, PR6, WRKY, MAPKK, and NPR1) expression during tritrophic interaction was analyzed through real-time PCR using specific primers. Actin was used as a housekeeping reference gene. Comparative 2 –ΔΔCT method was used to analyze the relative fold change of the target gene
-
ii.
PR2
PR2 gene was found to produce a proteinaceous pathogenic inhibitor β-1,3-endoglucanases that play a significant role in defense mechanism. PR2 gene expression pattern altered in all the treated plants compared to inoculated and uninoculated control plants. The consortium-treated plants challenged with GBNV shown upregulation like a sigmoidal curve by 2.22-fold at 24 h, 2.45-fold at 48 h, and 0.36-fold at 72 h, whereas the gene transcripts in inoculated control plants were downregulated over time. Salicylic acid-treated plants were initially downregulated and later reached its peak at 72 h by 2.63-fold and then decreased (Fig. 6b).
-
iii.
PR6
PR6 gene is reported to be a salicylic acid marker gene. The expression profile of PR6 gene was significantly different on all days. The consortium-treated plants challenged with GBNV initially did not show any altered gene expression, but later the transcripts showed upregulation by 2.11-fold at 24 h, 2.77-fold at 48 h, 3.71-fold at 72 h, and then downregulated by 3.36-fold at 96 h. Salicylic acid treatment showed the highest defense gene activation at 72 h with a 4.00-fold increase (Fig. 6c).
-
iv.
WRKY
In the present study, expression pattern of WRKY gene was altered in all the treated plants compared to inoculated and uninoculated control plants. WRKY gene was initially unaltered and then downregulated at 24 h and 48 h when compared to the uninoculated control plants. However, at 72 h and 96 h, the consortium-treated plants challenged with GBNV shown 2.94-fold and 1.00-fold increase in gene expression, respectively. Salicylic acid treatment showed highest defense gene activation at 72 h with 2.72-fold increase. The fold increase in WRKY gene expression of consortium-treated plants was higher than the salicylic-acid-treated plants (Fig. 6d).
-
xxii.
MAPKK
Initially the expression is unaltered but upregulated at 24 h by 2.46-fold and thereafter there is a decline in gene expression on consortium treatment challenged with GBNV. When compared to the salicylic acid control, the Streptomyces consortium-treated plants expressed MAPKK genes at the earliest on 24 h, whereas the salicylic acid control attained its peak on 48 h and then downregulated. There was an alternate upregulation and downregulation of MAPKK transcription factor from 24 to 96 h. Overall, the treatments differed significantly with respect to the uninoculated control indicating the MAPKK gene expression was induced by Streptomyces consortium treatment (Fig. 6e).
-
vi.
NPR1
The expression pattern of NPR1 gene in uninoculated control and in the consortium-treated plants challenged with GBNV was significantly different. It was observed that NPR1 gene expression in the consortium-treated plants challenged with GBNV was 2.11-, 2.81-, 0.81-, and 0.46-fold on 24, 48, 72 and 96 h, respectively, which followed a sigmoidal pattern. When compared to the salicylic acid control, the Streptomyces consortium-treated plants showed a highest defense gene activation at 48 h with 2.81-fold increase, whereas the salicylic acid control attained its peak on 48 h with 1.78-fold increase and then downregulated. Then gene expression was steadily declining over time and the treatments differed significantly compared to the uninoculated control (Fig. 6f).
A heat map of the defense gene expression during the tripartite interaction was constructed (Fig. 7). The data revealed that the defense gene expression varied at different time intervals. PR1 and PR6 were found to be high during 24 h and 96 h, whereas the expression of MAPKK and WRKY was high in 48 h and NPR1 and PR2 was higher in 72 h. This shows the differential expression of defense/resistance genes at varied time intervals.
Efficacy of Streptomyces spp. consortium against GBNV in tomato cv. Heemsohna under field conditions
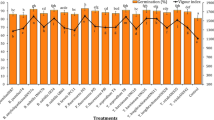
The Streptomyces consortium was applied through different methods, and the percent incidence was calculated at 80 days after transplanting in the field. The percent incidence was 14.44% in sequential application of seed treatment and seedling dip, soil drenching and foliar application of Streptomyces consortium (T4). The control plot recorded a PI of 37.78% and salicylic acid treatment recorded a PI of 27.78% with 26.47% virus inhibition (Table 2). The yield attributes viz., number of fruits per plant per picking, average weight of fruit, fruit length, fruit girth and yield were calculated. The highest average plant height of 150.67 cm was observed in sequential application of seed treatment and seedling dip, soil drenching and foliar application of Streptomyces consortium (T4). The control (T8) had the shortest plant height of 137.47 cm and salicylic acid treatment recorded 144.1 cm. The fruits per plant per picking (6.33 fruits/picking), average weight of fruit (101.13 g), fruit length (8.97 cm), and fruit girth (19.77 cm) were the highest in Streptomyces consortium. Yield of sequential application (T4) was 76.67 t/ha, while the untreated control recorded an yield of 63.99 t/ha. Salicylic acid treatment (T5) recorded a yield of 70.52 t/ha (Table 2).
Discussion
Groundnut bud necrosis orthotospovirus (GBNV) is a well-known viral pathogen that affects a wide range of hosts including groundnut, peanut, tomato, potato, onion, and pulses (Mandal et al. 2012). GBNV has been reported to cause up to 100% yield loss in chilli crops, posing a significant threat to vegetable production in India (Kunkalikar et al. 2011), and similarly huge yield loss was observed in tomato. Current management strategies for GBNV focus mainly on controlling the vector, primarily thrips, which leads to substantial environmental hazards due to the extensive use of chemical pesticides. Efforts to develop and deploy resistant cultivars have been largely unsuccessful, highlighting the urgent need for ecologically safe management strategies for GBNV. Thereby, biological control using Streptomyces spp. presents a promising strategy for managing GBNV in tomato. Streptomyces spp. are renowned for producing a vast array of bioactive compounds, including antibiotics, antifungals, and plant growth-promoting substances. Evidence for the effectiveness of Streptomyces in managing plant diseases was provided by Nasr-Eldin et al. (2019), who demonstrated that crude filtrates from Streptomyces netropsis, S. ambofaciens, and S. actuosus significantly reduced disease severity and virus concentration of Potato Virus Y (PVY) in potato. These microorganisms play a dual role by directly inhibiting pathogen growth and by inducing systemic resistance in plants, thereby enhancing their overall health and resilience against infections.
In the present study, the confirmation of 18 Streptomyces spp. isolates as Gram-positive aligns with previous observations regarding the Gram-positive nature of antimicrobial Streptomyces spp. (Arasu et al. 2008). This consistency supports the reliability of our findings and adds evidence on the fundamental characteristics of Streptomyces species. The production of IAA among the isolates varied, with SAT1 yielding the highest amount (70.56 µg/ml) and ACT18 the lowest (6.83 µg/ml), consistent with the findings of Gopalakrishnan et al. (2011) on the plant growth-promoting properties of Streptomyces spp. In addition, siderophore production was observed in SAT1, SAT4, and STR2, as indicated by the formation of an orange zone in the CAS medium, supporting Gopalakrishnan et al. (2011) observations. These characteristics highlight the potential of Streptomyces spp. to promote plant growth and act as biocontrol agents by improving nutrient uptake and suppressing phytopathogens through iron sequestration. Furthermore, phosphate solubilization was noted in SAT1 and STR2, forming clear halo zones in Pikovskaya’s agar, consistent with Gangwar et al. (2012) findings on Streptomyces lavendul Streptomyces lavendulae R22. This process enhances soil fertility and promotes plant growth by making phosphorus more available to plants, crucial for crop productivity.
SAT1 was identified as Streptomyces mutabilis and SAT4 as Streptomyces rochei through molecular characterization. The cross-streak method showed no inhibition zone between SAT1 and SAT4, which line-up with the results of Ankati et al. (2021) who developed Streptomyces consortia based on compatibility studies. In our study, the disease severity index (DSI) for SAT1, SAT4, and STR2 treatments during pre-inoculation spray was 20%, 30%, and 33.33%, respectively, with corresponding virus inhibition rates of 77%, 66%, and 63%. These results are consistent with Vinodkumar et al. (2018), who observed that simultaneous inoculation of bacterial suspensions with Tobacco streak virus (TSV) significantly reduced lesions in cowpea. Furthermore, the present study indicates that Streptomyces spp. can produce a variety of secondary metabolites that enhance plant immunity and inhibit pathogen growth, supporting their potential as biocontrol agents in sustainable agriculture.
The present study explored the tripartite interaction of tomato, GBNV, and Streptomyces spp. consortium, revealing defense-related gene expression patterns. Six defense-related genes (PR1, PR2, PR6, WRKY, MAPKK1, and NPR1) in tomatoes were examined, with the actin gene serving as an internal control (Mascia et al. 2010). The present study shows that a significant upregulation of the PR1 gene in tomato plants challenged with GBNV and treated with Streptomyces consortium was evident, suggesting a potential role in preventing virus systemic movement through hypersensitive cell death. Pathogenesis-related protein-1 gene (PR1) expression greatly impacts defense by inducing hypersensitive cell death. PR-1 protein was associated with the plant cell wall appositions, suggesting that it may play a role in preventing the spread of pathogens by strengthening the host cell walls. Similar findings were reported by Vanthana et al. (2019) in Bacillus-treated tomato plants with GBNV inoculation. Furthermore, the PR6 gene exhibited increased expression upon inoculation with Pectobacterium carotovorum subsp. carotovorum, supporting its involvement in plant defense (Djami‐Tchatchou et al. 2019). The present study also observed altered PR6 gene expression, with a 2.11- to 3.36-fold increase from 24 to 96 h post-inoculation. PR-2 and PR-3 protein levels increased in tomato plants exposed to Pseudomonas fluorescens strain CHAO (Kandan et al. 2005). In our study, there was an upregulation of these genes at various time points, suggesting potential restriction of viral cell-to-cell movement due to callose accumulation at host plasmodesmata. WRKY transcription factors displayed a complex expression pattern in response to GBNV, with an initial downregulation followed by upregulation at 96 h. This pattern is consistent with the findings in Bacillus-treated tomato plants, indicating their involvement in defense mechanisms (Vanthana et al. 2019). MAPKK gene upregulation at 24 h, followed by a decline, suggests the production of transducing signals inducing resistance during the tripartite interaction of tomato, GBNV, and Streptomyces spp. consortium. Similar findings were reported in tomato under biotic and abiotic stress conditions by Wu et al. (2014). NPR1, associated with the ISR pathway, exhibited an initial upregulation, followed by a decline. This pattern is in line with the reported resistance against GBNV induced by Bacillus amyloliquefaciens in chilli, supporting NPR1 role in plant defense mechanisms (Rajamanickam and Nakkeeran 2020; Pieterse et al. 1998).
Streptomyces spp. have demonstrated significant efficacy as elicitors of plant systemic resistance against various phytopathogens (Kaari et al. 2022). Sequential application of a Streptomyces consortium through seed treatment, seedling dip, soil drenching, and foliar application effectively managed bud necrosis disease (GBNV) in both glasshouse and field trials. The disease severity index (DSI) was notably low at 15.00 and 16.67 in trial I and trial II, respectively. This aligns with previous studies of Harish et al. (2009) showing the superior biocontrol efficacy of Streptomyces mixtures compared to single isolates against different plant viruses. In addition, observations on Cucumber mosaic virus inhibition and reduced infection rates of Tomato yellow leaf curl disease further support the potential of Streptomyces in disease management (Galal 2006). In the field trial, the sequential application of Streptomyces consortium resulted in a significantly lower GBNV incidence of 14.44% (61.76% virus inhibition). These findings are consistent with other studies reporting varied levels of disease reduction in response to biocontrol agents against GBNV, Banana bunchy top virus (BBTV), Cucumber mosaic virus (CMV), and Poty virus Y (Vanthana et al. (2022).
Biocontrol of pathogens primarily involves stimulating plant growth and suppressing harmful microbes in the rhizosphere. Microbes, producing auxins, gibberellins, cytokinins, ethylene, volatile chemicals, and enzymes, enhance plant defense and improve yield attributes (Olanrewaju et al 2017). In field conditions, soil application and foliar spray of B. amyloliquefaciens (VB7) decreased TSV incidence by 52% (Vinodkumar et al. 2018). Bacillus consortia applied to tomato plants under field conditions reduced GBNV incidence by 10.5%, increasing plant height, fruits per plant per picking, individual fruit weight, and overall tomato yield (Vanthana et al. 2022). Likewise, in our study, the sequential application of Streptomyces consortium resulted in the highest average plant height (150.67 cm), fruits per plant per picking (6.33 fruits/picking), average fruit weight (101.13 g), fruit length (8.97 cm), and fruit girth (19.77 cm), along with an increased yield (76.67 tons/ha). The sequential application of the Streptomyces consortium in our study not only demonstrated substantial improvements in plant height, fruit characteristics, and overall yield, paralleling the positive effects observed with Bacillus consortia. These findings highlight the promising role of Streptomyces spp. as effective biocontrol agents and growth enhancers, paving way for their broader use in sustainable agricultural practices.
Conclusion
In the present study, the potential of Streptomyces spp. against Groundnut bud necrosis orthotospovirus was revealed in glasshouse and field experiments. We reported the natural occurrence of GBNV in tomato and confirmed its identity through molecular methods. Three Streptomyces spp. viz., Streptomyces mutabilis, Streptomyces rochei, and Streptomyces chrestomyceticus were found to possess enhanced PGPR activity in tomato as evident by growth promotion and antiviral activity. Our current investigation presented a clear idea about the effect of induced systemic resistance by Streptomyces sp. consortium against GBNV infection in tomato. As evidenced by qPCR, application of Streptomyces spp. consortium had triggered defense gene expression viz., PR1, PR2, PR6, WRKY, MAPKK, and NPR1 in tomato which in turn effectively reduced the severity of GBNV in tomato. In addition, plant growth metrics and yield attributes of tomato were also increased when the consortium was applied in the field. The current research findings indicated that Streptomyces spp. can be effectively utilized for managing bud necrosis disease and increasing the yield in tomato.
Data availability
The data will be made available on request.
References
Abidin ZAZ, Ahmad A, Latip J, Usup G (2016) Marine Streptomyces sp. UKMCC PT15 producing undecylprodigiosin with algicidal activity. J Technol 78:55–60. https://doi.org/10.11113/jt.v78.9944
AbuQamar SH, Luo K, Laluk MV, Mickelbart MT (2009) Crosstalk between biotic and abiotic stress responses in tomato is mediated by the AIM1 transcription factor. Plant J 58:347–360. https://doi.org/10.1111/j.1365-313X.2008.03783.x
Ankati S, Srinivas V, Pratyusha S, Gopalakrishnan S (2021) Streptomyces consortia-mediated plant defense against Fusarium wilt and plant growth-promotion in chickpea. Microb Pathog 157:104961. https://doi.org/10.1016/j.micpath.2021.104961
Arasu MV, Duraipandiyan V, Agastian P, Ignacimuthu S (2008) Antimicrobial activity of Streptomyces spp. ERI-26 recovered from Western Ghats of Tamil Nadu. Med Mycol J 18:147–153. https://doi.org/10.1016/j.mycmed.2008.07.004
Basilio A, Gonzalez I, Vicente M, Gorrochategui J, Cabello A, Gonzalez A, Genilloud O (2003) Patterns of antimicrobial activities from soil actinomycetes isolated under different conditions of pH and salinity. J Appl Microbiol 95:814–823. https://doi.org/10.1046/j.1365-2672.2003.02049.x
Brahimi M, Boukhalfa K, Moussaoui A (2017) Deep learning for tomato diseases: classification and symptoms visualization. Appl Artif Intell 31:299–315. https://doi.org/10.1080/08839514.2017.1315516
Chomczynski P, Sacchi N (1987) Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem 162:156–159. https://doi.org/10.1016/0003-2697(87)90021-2
Clark MS (2013) Plant molecular biology-a laboratory manual. Springer Science & Business Media
Dasgupta I, Malathi V, Mukherjee S (2003) Genetic engineering for virus resistance. Curr Sci 84: 341–354. https://www.jstor.org/stable/24107418
Djami-Tchatchou AT, Matsaunyane LB, Ntushelo K (2019) Gene expression responses of tomato inoculated with Pectobacterium carotovorum sub sp carotovorum. Microbiol Open 8:e911. https://doi.org/10.1002/mbo3.911
Fuentes MS, Colin VL, Amoroso MJ, Benimeli CS (2016) Selection of an actinobacteria mixed culture for chlordane remediation. Pesticide effects on microbial morphology and bioemulsifier production. J Basic Microbiol 56:127–137. https://doi.org/10.1002/jobm.201500514
Gangwar M, Rani S, Sharma N (2012) Investigating endophytic actinomycetes diversity from rice for plant growth promoting and antifungal activity. Int J Adv Lif Sci 1:10–21
Gopalakrishnan S, Pande S, Sharma M, Humayun P, Kiran BK, Sandeep D, Vidya MS, Deepthi K, Rupela O (2011) Evaluation of actinomycete isolates obtained from herbal vermicompost for the biological control of Fusarium wilt of chickpea. Crop Prot 30:1070–1078. https://doi.org/10.1016/j.cropro.2011.03.006
Harish S, Kavino M, Kumar N, Saravanakumar D, Soorianathasundaram K, Samiyappan R (2008) Biohardening with plant growth promoting rhizosphere and endophytic bacteria induces systemic resistance against banana bunchy top virus. Appl Soil Ecol 39(2):187–200. https://doi.org/10.1016/j.apsoil.2007.12.006
Harish S, Kavino M, Kumar N, Balasubramanian P, Samiyappan R (2009) Induction of defense-related proteins by mixtures of plant growth promoting endophytic bacteria against Banana bunchy top virus. Biol Control 51:16–25. https://doi.org/10.1016/j.biocontrol.2009.06.002
Hucker G, Conn H (1923) Methods of gram staining. Tech Bulletin. New York Agricultural Experiment Station (93)
Indiastat (2021) https://www.indiastat.com/
Johnson LF, Curl EA (1972) Methods for research on the ecology of soil-borne plant pathogens. Methods for research on the Ecology of Soil-Borne Plant Pathogens. Burgess Publishing Company, Minneapolis
Kaari M, Joseph J, Manikkam R, Sreenivasan A, Venugopal G (2022) Biological control of Streptomyces sp. UT4A49 to suppress tomato bacterial wilt disease and its metabolite profiling. J King Saud Univ Sci 34:101688. https://doi.org/10.1016/j.jksus.2021.101688
Kandan A, Ramiah M, Vasanthi V, Radjacommare R, Nandakumar R, Ramanathan A, Samiyappan R (2005) Use of Pseudomonas fluorescens-based formulations for management of Tomato spotted wilt virus (TSWV) and enhanced yield in tomato. Biocontrol Sci Techn 15:553–569. https://doi.org/10.1080/09583150500088546
Karthikeyan G, Doraisamy S, Rabindran R (2009) Pseudomonas fluorescens mediated systemic resistance against Urdbean leaf crinkle virus in blackgram (Vigna mungo). Arch Phytopathol Pflanzenschutz 42:201–212. https://doi.org/10.1080/03235400600982519
Karthikeyan G, Barkavi G, Harish S, Varanavasiappan S (2024) Expression of defense responsive genes in tripartite interaction of cucumber mosaic virus and plant growth promoting rhizobacteria in ridge gourd (Luffa acutangula (L.) Roxb). Physiol Mol Plant Pathol 129:102176. https://doi.org/10.1016/j.pmpp.2023.102176
Kishorkumar C, Harish S, Karthikeyan G, Sharmila DJS, Nivedha M (2024) Harnessing nanoencapsulated Bacillus spp. consortia to combat Groundnut bud necrosis orthotospovirus in Tomato. ACS Appl Mater Interfaces 16(9):11185–11193. https://doi.org/10.1021/acsami.3c16145
Kunkalikar SR, Poojari S, Arun BM, Rajagopalan PA, Chen TC, Yeh SD, Ravi KS (2011) Importance and genetic diversity of vegetable-infecting tospoviruses in India. Phytopathology 101:367–376. https://doi.org/10.1094/phyto-02-10-0046
Li Y, Guo Q, Li Y, Sun Y, Xue Q, Lai H (2019) Streptomyces pactum Act12 controls tomato yellow leaf curl virus disease and alters rhizosphere microbial communities. Biol Fertil Soils 55:149–169. https://doi.org/10.1007/s00374-019-01339-w
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2− ΔΔCT method. Methods 25:402–408. https://doi.org/10.1006/meth.2001.1262
Mandal B, Jain R, Krishnareddy M, Krishna Kumar N, Ravi K, Pappu H (2012) Emerging problems of tospoviruses (Bunyaviridae) and their management in the Indian subcontinent. Plant Dis 96:468–479. https://doi.org/10.1094/PDIS-06-11-0520
Mascia T, Santovito E, Gallitelli D, Cillo F (2010) Evaluation of reference genes for quantitative reverse-transcription polymerase chain reaction normalization in infected tomato plants. Mol Plant Pathol 11:805–816. https://doi.org/10.1111/j.1364-3703.2010.00646.x
Meena LI, Rajeswari E, Ahiladevi P, Kamalakannan A, Kalaiselvi T (2022) Antifungal potential of Streptomyces rameus GgS 48 against mungbean root rot [Rhizoctonia bataticola (Taub.) Butler]. J Biosci 47:1–17. https://doi.org/10.1007/s12038-021-00244-5
Nasr-Eldin M, Messiha N, Othman B (2019) Induction of potato systemic resistance against the potato virus Y (PVYNTN), using crude filtrates of Streptomyces spp. under greenhouse conditions. Egypt J Biol Pest Control 29:62. https://doi.org/10.1186/s41938-019-0165-1
Nautiyal CS (1999) An efficient microbiological growth medium for screening phosphate solubilizing microo66rganisms. FEMS Microbiol Lett 170:265–270. https://doi.org/10.1111/j.1574-6968.1999.tb13383.x
Olanrewaju OS, Glick BR, Babalola OO (2017) Mechanisms of action of plant growth promoting bacteria. World J Microbiol Biotechnol 33:1–16. https://doi.org/10.1007/s11274-017-2364-9
Patten CL, Glick BR (1996) Bacterial biosynthesis of indole-3-acetic acid. Can J Microbiol 42:207–220. https://doi.org/10.1139/m96-032
Pieterse CM, Van Wees SC, Van Pelt JA, Knoester M, Laan R, Gerrits H, Weisbeek PJ, Van Loon LC (1998) A novel signaling pathway controlling induced systemic resistance in Arabidopsis. Plant Cell 10:1571–1580. https://doi.org/10.1105/tpc.10.9.1571
Rahila R, Harish S, Kalpana K, Anand G, Arulsamy M, Kalaivanan R (2023) Antifungal metabolites of Streptomyces chrestomyceticus STR-2 inhibits Magnaporthe oryzae, the incitant of rice blast. Curr Microbiol 80:107. https://doi.org/10.1007/s00284-023-03205-3
Rahul Dev AS, Harish S, Karthikeyan G, Varanavasiappan S, Nivedha M (2022) Molecular characterization of Groundnut bud necrosis virus in Tamil Nadu. Pest Mgmt Hortl Ecosyst 28:161–167. https://doi.org/10.5958/0974-4541.2022.00030.3
Rajamanickam S, Nakkeeran S (2020) Flagellin of Bacillus amyloliquefaciens works as a resistance inducer against Groundnut bud necrosis virus in chilli (Capsicum annuum L.). Arch Virol 165:1585–1597. https://doi.org/10.1007/s00705-020-04645-z
Reddy M, Reddy D, Apparao A (1968) A new record of virus disease on Groundnut. Plant Dis Rep 52:494–495
Sain S, Chadha M (2012) Evaluation of improved lines of tomato for yield performance and disease resistance under open field conditions. Indian J Hortic 69:185–194
Sangeetha B, Krishnamoorthy AS, Renukadevi P, Malathi VD, Sharmila DJS, Amirtham D (2020) Antiviral activity of basidiomycetous fungi against Groundnut bud necrosis virus in tomato. Pestic Biochem Physiol 166:104570. https://doi.org/10.1016/j.pestbp.2020.104570
Santos-Cervantes ME, Felix-Gastelum R, Herrera-Rodríguez G, Espinoza-Mancillas MG, Mora-Romero AG, Leyva-López NE (2017) Characterization, pathogenicity and chemical control of Streptomyces acidiscabies associated to potato common scab. Am Potato J 94:14–25. https://doi.org/10.1007/s12230-016-9541-5
Scholthof KBG, Adkins S, Czosnek H, Palukaitis P, Jacquot E, Hohn T, Hohn B, Saunders K, Candresse T, Ahlquist P (2011) Top 10 plant viruses in molecular plant pathology. Mol Plant Pathol 12:938–954. https://doi.org/10.1111/j.1364-3703.2011.00752.x
Schwyn B, Neilands J (1987) Universal chemical assay for the detection and determination of siderophores. Anal Biochem 160:47–56. https://doi.org/10.1016/0003-2697(87)90612-9
Taha M, Ghaly M, Atwa H, Askoura M (2021) Evaluation of the effectiveness of soil Streptomyces isolates for induction of plant resistance against Tomato mosaic virus (ToMV). Curr Microbiol 78:3032–3043. https://doi.org/10.1007/s00284-021-02567-w
Tamura K, Stecher G, Kumar S (2021) MEGA11: molecular evolutionary genetics analysis version 11. Mol Biol Evol 38:3022–3027. https://doi.org/10.1093/molbev/msab120
Umamaheswaran K, Jain R, Bhat A, Ahlawat Y (2003) Biological and molecular characterization of a Tospovirus isolate from tomato and its relationship with other Tospoviruses. Indian Phytopathol 56:168–173. https://epubs.icar.org.in/index.php/IPPJ/article/view/18288
Van Wees SC, Van der Ent S, Pieterse CM (2008) Plant immune responses triggered by beneficial microbes. Curr Opin Plant Biol 11:443–448. https://doi.org/10.1016/j.pbi.2008.05.005
Vanthana M, Nakkeeran S, Malathi V, Renukadevi P, Vinodkumar S (2019) Induction of in planta resistance by flagellin (Flg) and elongation factor-TU (EF-Tu) of Bacillus amyloliquefaciens (VB7) against Groundnut bud necrosis virus in tomato. Microb Pathog 137:103757. https://doi.org/10.1016/j.micpath.2019.103757
Vanthana M, Nakkeeran S, Malathi V, Renukadevi P, Vinodkumar S, Sivakumar U, Suganthi A (2022) Flagellin and elongation factor of Bacillus velezensis (VB7) reprogramme the immune response in tomato towards the management of GBNV infection. J Virol Methods 30:1114438. https://doi.org/10.1016/j.jviromet.2021.114438
Vinodkumar S, Nakkeeran S, Renukadevi P, Mohankumar S (2018) Diversity and antiviral potential of rhizospheric and endophytic Bacillus species and phyto-antiviral principles against tobacco streak virus in cotton. Agric Ecosyst Environ 267:42–51. https://doi.org/10.1016/j.agee.2018.08.008
Wu J, Wang J, Pan C, Guan X, Wang Y, Liu S, He Y, Chen J, Chen L, Lu G (2014) Genome-wide identification of MAPKK and MAPKKK gene families in tomato and transcriptional profiling analysis during development and stress response. PLoS One 9:e103032. https://doi.org/10.1371/journal.pone.0103032
Zhu M, Van Grinsven IL, Kormelink R, Tao X (2019) Paving the way to tospovirus infection: multilined interplays with plant innate immunity. Annu Rev Phytopathol 57:41–62
Acknowledgements
The authors are grateful to the Department of Plant Pathology, TNAU, Coimbatore and DST-SERB No: CRG/2020/004589 for providing infrastructure facilities for conducting the experiments.
Funding
The authors declare that there is no financial or non-financial interests that are directly or indirectly related to the work submitted for publication.
Author information
Authors and Affiliations
Contributions
ASR: experimentation, methodology, and writing––original draft; SH: conceptualization, supervision, review and editing; GK: supervision, editing; MN and CS: performed data analysis and data curation.
Corresponding author
Ethics declarations
Conflict of interest
There are no conflict of interest. The manuscript was reviewed by all the authors before submission.
Ethical approval
This article does not contain any studies with human participants or animals performed by the authors.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Dev, A.S.R., Harish, S., Karthikeyan, G. et al. Consortia of Streptomyces spp. triggers defense/PAMP genes during the interaction of Groundnut bud necrosis orthotospovirus in tomato. 3 Biotech 14, 196 (2024). https://doi.org/10.1007/s13205-024-04030-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s13205-024-04030-6