Abstract
Orthodontically induced inflammatory root resorption (OIIRR) is an undesirable complication of orthodontic treatment (OT) with an ambiguous aetiologic mechanism. This study aimed to identify OIIRR-associated biomarkers in the gingival crevicular fluid (GCF) using proteomic analysis. In this randomized clinical trial, the upper first premolars (UFP) were exposed either to light or heavy force. The GCF was collected at 1 h, 1 day, 7 days, 14 days, 21 days, and 28 days following force application. After extraction of UFP, roots were imaged and resorption premolar, was used to deliver either light forcecraters were measured. Proteomic analysis of GCF was performed using 2D gel electrophoresis with MALDI-TOF/TOF MS/MS. Results were further analyzed by bioinformatics analyses showing the biological functions and predicted pathways. The predicted canonical pathways showed that the expression of immunoglobulin kappa (IGKC), neutrophil gelatinase-associated lipocalin (NGAL), neurolysin mitochondrial (NEUL), keratin, type II cytoskeletal 1 (K2C1), S100-A9, and the extracellular calcium-sensing receptor (CASR) were significantly associated with a range of biological and inflammatory processes. In conclusion, up-regulation of S100A9, CASR, and K2C1 suggested a response to force-related inflammation, chemotactic activities, osteoclastogenesis, and epithelial cell breakdown. Meanwhile, the up-regulation of IGKC, NGAL, and K2C1 indicated a response to the inflammatory process, innate immunity activation, and epithelial cell breakdown.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Orthodontically induced inflammatory root resorption (OIIRR) is an undesirable and unpredictable complication of orthodontic treatment (OT) (Nieto-Nieto et al. 2017). Despite many attempts in the last decades to understand the nature of OIIRR, however, its aetiologic mechanism is still ambiguous. Nevertheless, it is agreed that the etiology for the development and progression of this condition is multi-factorial involving patient- and treatment-related factors. Force magnitudes and duration were the most evident-associated factors (Currell et al. 2019).
The OIIRR is typically symptomless, but when the root structure loss is significant, clinical symptoms emerge with tooth mobility as the worst one (Dudic et al. 2009; Brezniak and Wasserstein 2019). Therefore, the clinical significance of root resorption is closely correlated to its early detection, using various methods to evaluate both the orthodontic and biological issues that may lead to root resorption (Dindaroğlu and Doğan 2016). The most common methods for detecting OIIRR clinically are two-dimensional (2D) and three-dimensional radiographs, i.e., cone-beam computed tomography. However, both methods cannot assess whether resorption is stopped or still active (Samandara et al. 2019).
Gingival crevicular fluid (GCF) is one of the biological fluids that can reflect the ongoing biological process in situ through the analysis of its content and composition as well as flow rate (Barros et al. 2016). The use of GCF biomarkers for the prediction, diagnosis, and monitoring of different dental and periodontal conditions has shown a positive potential as an effective, non-invasive, and safe tool. In addition, recent data suggested a therapeutic measure from the validated GCF biomarkers to overcome OIIRR (Buduneli 2019).
The discovery and identification of novel biomarkers are highly aided by advanced molecular biology tools, including proteomic strategies and mass spectrometry (MS) (Khurshid et al. 2016; Ghallab 2018). In orthodontics, comparative proteomics MS can be used to monitor and compare GCF proteome profiles and identify potential OIIRR biomarkers (Rody et al. 2014). Most of the previous studies for early detection of OIIRR have been performed to test individual biomarkers that are representative of a single biological pathway. To the best of our knowledge, limited studies have employed GCF proteomics analysis to detect possible diagnostic biomarkers for OIIRR. Therefore, this clinical trial aimed to identify candidate biomarkers, with different proteome profiling in GCF, associated with early OIIRR which could help orthodontists precisely predict, diagnose, and monitor OT.
Materials and methods
Study design and settings
This two-arm split-mouth, randomized-controlled clinical trial was approved by the Ethics Committee of the College of Dentistry, University of Baghdad (ID #023,418, January 9, 2019) with clinical trial registration #NCT04291443 on 02/03/2020. The study commenced from November 2018 to June 2020. All participants were provided with comprehensive methodological information during enrollment and written informed consent was gathered before the study was started.
Study population and outcomes
Adult patients attended to College of Dentistry, the University of Baghdad seeking OT and analogous positive controls of periodontitis patients were recruited.
-
I.
Inclusion criteria
Adult (18–32 years), with no history of any medical disease/condition, who needed bilateral extraction of upper first premolar (UFP) as a part of OT. Patients had impacted third molars exhibiting good oral hygiene and a healthy periodontium (Chapple et al. 2018), and no radiographic signs of pre-existing apical root resorption of the teeth were included in the study. Both UFPs should have two roots, with nearly similar shapes and nearly equal lengths and are normally positioned within the arch.
-
II.
Exclusion criteria
Patients with tooth impaction other than third molars. Smoker individuals and patients on anti-inflammatory drugs within the last 3 months and during the study. Pregnant/lactating female or on contraceptives, history of bruxism and clenching, previous OT, traumatized, carious and dentally restored UFP, any oral pathology, and those who are not willing to consent.
The primary outcomes were the degree of root resorption and the associated GCF biomarkers following the application of orthodontic force. The secondary outcome was measuring protein levels in the GCF.
Randomization and masking
A split-mouth study design was followed with an allocation ratio of 1:1 (right and left sides). A simple randomization method was used in which the two forces were consecutively applied on each side among participants by the main investigator (H.S.MS.). GCF and extracted teeth were coded by the main investigator and blindly analyzed by another investigator and an independent bio-statistician.
Interventions and measurements
The root length (mm) of UFP and pre-existing apical root resorption of the extracted teeth were assessed radiographically. Additionally, demographic data, study model, and photographs were collected. All participants received full-mouth ultrasonic scaling and polishing with oral hygiene instructions 2 weeks before OT. Baseline data were obtained before the placement of elastic separators and 2 weeks following ultrasonic scaling and polishing. GCF samples were conducted at 1 h, 1 day, 7 days, 14 days, 21 days, and 28 days following force application. At the endpoint of the trial, extractions of the UFP were carried out.
Bilateral sectional fixed orthodontic appliances were bonded on the molars and UFP, which consisted of TPA fabricated from 1-inch hard stainless-steel wire. This was soldered with the 0.022-inch slot Roth bands cemented on the 1st molars and a self-ligating bracket with a 0.022″ × 0.028″ slot size was bonded on UFP using light-cured bond cement. A cantilever spring, 0.016″ and 0.019″ × 0.025″ TMA wires, extended from the 1st molar to the UFP, bypassing the 2nd premolar, was used to deliver either light force (LF) (25 g) or heavy force (HF) (225 g), respectively (Fig. 1 I). Each spring was prepared on a study model before the day of bonding, and a coil with an internal diameter of 3 mm was integrated next to the ipsilateral 1st molar. The spring was activated and adjusted to be loaded to the decided magnitude. The measurement was done via a strain gauge that measures to the nearest gram. To inhibit extra forces on the UFP, light-curing band cement material was added onto the mandibular 1st molars’ occlusal surfaces to open the bite lightly. During 28 days, there was no reactivation to the spring nor orthodontic appliance was bonded on the mandibular arch.
A Heavy force (225 g) side using 0.019″ × 0.025″ TMA wire, B occlusal view of the appliance with a trans-palatal arch in situ, and C light force (25 g) side using 0.016″ TMA wire. D Fiji (Image-J, Inc.) software was used to analyze a straight-on 2D root image to measure the surface area of resorption craters at each determined site. The pink delineated area indicates the total area of the analyzed site (i.e., cervical), and the blue delineated areas indicate the resorption craters at each site (scale bar 1 mm)
GCF collection for proteomics analysis
GCF samples were collected by infraclavicular method during the day between 9:00 am and 2:30 pm using PerioPaper™ for healthy periodontium and PerioColpaper™ strips for periodontitis patients (OraFlow Inc. New York, USA) from four sides (mesiobuccally, distobuccally, mesiopalataly, and distopalataly) per tooth (Saloom et al. 2017). GCF elution, processing, and storage followed the procedure described in a previous study (Saloom et al. 2017; Mohammed-Salih and Saloom 2022).
Teeth extraction and imaging method for root resorption quantification
On day 28, extractions of the UFP were performed only with forceps avoiding any contact to the cervical root cementum. Processing, disinfection, storage, and imaging of the teeth were performed as previously described (Malek et al. 2003; Chutimanutskul et al. 2006). The resorption crater's surface area was measured by absolute straight-on images using Stereozoom Microscope (Nikon SMZ 745 T, Model C-LEDS, China) with 10 × . Buccal, palatal, mesial, and distal root surfaces were photographed using a digital camera (Q Imaging, Surrey, Canada). Three images per surface were taken in the cervico-apical direction which was divided into cervical, middle, and apical thirds. Buccal, palatal, distal, and mesial surfaces for each third were determined, namely buccal cervical (BC), buccal middle (BM), buccal apical (BA), palatal cervical (PC), palatal middle (PM), palatal apical (PA), mesial cervical (MC), mesial middle (MM), mesial apical (MA), distal cervical (DC), distal middle (DM), and distal apical (DA). Each resorption crater area was encircled and measured together with the whole surface area using software (Fiji, Image-J, Inc.) (Fig. 1 II). The teeth were allocated into high resorption (HR) and low resorption (LR) groups according to the total percentage of resorption surface area per tooth. Subsequently, GCF samples for the HR group (HR0 to HR6) and LR group (LR0 to LR6), in addition, to the periodontitis group, were prepared for proteomics analysis.
GCF proteomics analysis
Determination of protein concentration using Bradford assay
All steps for protein quantification by Bradford assay were performed according to a previous work of Mohammed-Salih and Saloom (Mohammed-Salih and Saloom 2022). Experiments were run in triplicate.
Determination of GCF proteins purity by SDS-PAGE
To assess the purity and integrity of the GCF proteins, an SDS-PAGE was run as described by Mansor and coauthors (Mansor et al. 2020). First, 2 µg of each GCF sample was mixed with SDS-reducing buffer (1:1) and denatured at 95 °C for 5 min. Samples were then electrophoresed in a 5% stacking gel followed by 12% resolving gel at 100 V for 50 min using Mini-Protean III System (Bio-Rad Laboratories, Hercules, CA, USA). In the first lane of each gel, Precision Plus Protein™ Dual Color Standards (Bio-rad Laboratories, Hercules, CA, USA) were loaded to estimate the molecular weight (MW) of resolved proteins. Silver staining was performed as previously described (Yan et al. 2000).
Two-dimensional gel electrophoresis with MALDI-TOF/TOF MS/MS
GCF samples of the orthodontic group at T0, T4, and T6 time-points and the periodontitis group were further analyzed by 2D gel electrophoresis coupled with MALDI-TOF/TOF MS/MS. The samples were subjected to 2DE for their 2D protein separation and extraction. The 2DE was run in quadruplicate for each group.
A. First-dimensional separation/isoelectric focusing
Protein separation was done according to their isoelectric point (pI). Ettan IPGphor II Isoelectric Focusing (IEF) System and 7 cm immobile pH gradient strips (IPG) (GE Healthcare, Chicago, IL, USA), pH 4–7, were used. Each sample was thawed and gently vortexed for 30 s. Then, 80 μg of protein was mixed with 45µL thiourea rehydration solution. The sample was collectively employed for at least 12 h on the strips using gradual pipe-turning onto the rehydration tray. Then, strips were transported on the ceramic collector before the Ettan IPGphor II device was electrically oriented using the following program: 500 V/1 h, 1000 V/1 h, 5000 V/1 h, 8000 V/1 h, and lastly at 29,000 V/1 h at 20 °C. IEF was done at 50Amp.
B. Second-dimensional separation
Before loading on 12% PAGE, the IPG strips were incubated in 10 mL equilibration buffer. The SDS running buffer was applied to the strips for rinsing followed by loading into the second-dimension gel. The electrophoresis of the gel was carried out using the Ruby SE 600 electrophoresis system (GE Healthcare, Chicago, IL, USA) at 20 °C. The overlay polyacrylamide gel was removed and replaced with the SDS running buffer until it was assembled. A 0.5% warm agarose replaced the running SDS buffer which covered the gel to seal the strip. The parameters for electrophoresis were rendered at 12% SDS-PAGE for 15 min at 10 mA/gel and 3.5 h at 20 mA/gel using Precision Plus Protein™ Dual Color Standards Ladder (10-250 kDa). The gel was fixed overnight with 40% methanol and 10% acetic acid in H2O. Staining was done with a silver staining solution as previously described (Al-Obaidi et al. 2017).
Gel image acquisition
The stained gels, SDS-PAGE and 2DE, were digitally visualized by Calibrated Densitometer GS-800 Mode gel scanner (Biorad Laboratories, Hercules, CA, USA) supplied with software Quantity one-4.6.9 (Basic). The 2DE gels images were automatically analyzed using the Progenesis SameSpot 3.1v (Nonlinear Dynamic Ltd., Durham, NC, USA) software. After evaluating the existence of every spot in all gels, the number of spots was counted for each condition.
MALDI-TOF/TOF MS/MS and database query
-
1.
In-gel digestion
The chosen spots were excised manually in a clean laminar airflow safety cabinet from the 2DE gels. Then, they were washed [50% of Acetonitrile (ACN) in 100 mM of (NH4) HCO3] and reduced to a 60 °C water bath containing a reduction solution [100 mM of DTT in 100 mM of (NH4) HCO3] for 30 min. The reduced samples were placed in a solution of alkylates [55 mM of IAA in 100 mM of (NH4) HCO3] and incubated in darkness for 20 min, allowed to dry for 1 h with the ScanSpeed MiniVac Evaporator (Saur, Reutlingen, Germany). This was followed by washing and incubating in 100%ACN for 15 min. Samples were digested by adding 7 μg/μL trypsin solution and incubated at 30 °C for 24 h. The digested peptides were mixed and spun with 100%ACN; ScanSpeed MiniVac Evaporator (Saur, Reutlingen, Germany) was used for drying the collected supernatants for 1 h. The resultant dried peptides were stored at − 80 °C for further MS analysis.
-
2.
Protein desalting and identification
The dried peptides were substituted in the desalting solution of TA30 (0.1%TFA in 30%ACN), using Zip-Tip Cµ18 (Millipore, Bedford, MA, USA). To determine the identity of the peptides, the purified desalted peptides were analyzed by Ultraflextreme MALDI-TOF/TOF MS/MS (Bruker, Bremen, Germany), a positive-ion mode reflector. All spectra were baseline processed using Bruker Flex-Analysis software (version 3.3) and then analyzed using MASCOT search engine version 3.5 (Matrix Science: http://www.matrixscience.com, Boston, MA, USA) using taxonomy Homo sapiens entries against the SwissProt database to identify the abundant proteins of interest. The parameters used for database searches were as follows: trypsin was set as the enzyme with one missed cleavage allowed, with a 1.0 Da fragment mass tolerance, 200 ppm a peptide mass tolerance, and peptide charge (+ 1, + 2, and + 3), the variable modification was methionine oxidation, the fixed modification was carbamidomethylation, and monoisotopic as mass values.
Bioinformatics analyses
Hierarchical clustering/heat map
All significant proteins were clustered and presented in a heat map generated by Genesis software (Sturn et al. 2002) to visualize the abundance pattern of the significantly altered proteins. Distinct patterns were seen in the altered abundance GCF proteins following orthodontically induced inflammatory root resorption for both heavy and light forces (HF and LF). The red boxes represented upregulated proteins, while down-regulated proteins were shown in green. The higher intensity of the color is proportionate to the higher protein abundance.
Gene ontology (GO) enrichment
For functional classification, Gene Ontology (GO) enrichment analysis was performed by the Protein Analysis Through Evolutionary Relationships (PANTHER) classification system. PANTHER includes all annotations provided by the Gene Ontology Consortium (http://geneontology.org) to enable the identification of genes or proteins datasets related to their specific molecular functions, roles in biological processes, and roles as cellular components (Mi et al. 2012).
Ingenuity Pathway Analysis (IPA) software
Ingenuity Pathways Analysis (IPA) (Ingenuity® Systems, Redwood City, CA, USA) software (www.ingenuity.com) was used to analyze the predicted canonical pathways of altered abundance GCF proteins following orthodontically induced inflammatory root resorption for both heavy and light forces (HF and LF). Results were imported into the IPA software. The “Core Analysis of IPA” was used to interpret the data in the context of biological processes, pathways and networks (Mahmood et al. 2021). Both up-and-down-regulated proteins were defined as value parameters for the analysis. Each protein was mapped and overlaid onto a global molecular network developed from information contained in the Ingenuity Knowledge Base. The network of proteins was then algorithmically generated based on their connectivity. Right-tailed Fisher’s exact test was used to calculate a p value to indicate the probability of each biological function assigned to the network was due to chance.
Statistical analysis
For descriptive statistics, means, median, standard deviations, standard errors, frequencies, and percentages were used. Normality distribution was checked by Shapiro–Wilk’s test. Accordingly, a two-tailed unpaired t test to test the root length difference between the buccal and palatal roots of one side compared to that on the other side. While ANOVA was used for multiple comparisons of significant differences within groups. The Mann–Whitney U test was used to test the significance of the differences in total resorption percentage between force groups. Freidman test and Games–Howell post hoc test were used to compare the significance of the differences in resorption percentage within each group. A p value < 0.05 was assigned as the significant level at a 95% confidence interval. Statistical Package for the Social Science (SPSS, version 22, IBM, New York, USA) was used for data analysis.
Results
Demographic characteristics
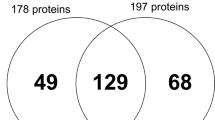
A total of 102 patients were assessed for eligibility, and 38 participants were recruited and divided into OT (n = 20) and periodontitis (n = 18) groups (Fig. 2). Demographic data for all patients at baseline and a comparison of root length and any pre-existing apical root resorption of UFP are illustrated in Table 1.
Quantification of relative root resorption surface area
For the HF group, the highest percentage of surface resorption was observed at the BC, BA, PC, and PA surfaces. In addition, the BC showed severe resorption (29%) of the total site surface area. Further, 37% of the PA surfaces were resorbed (Table 2). Whereas in the LF group, less than 2% of the BC sites were resorbed and the BA sites of the root LF exhibited minor resorption (< 1.5%) of the total site surface area (Table 2). For HF intra-group comparisons, there was a significant increase in the percentage of resorption at different sites of BM and PM surfaces (Table 2). Furthermore, the buccal sites showed a significant resorption percentage as compared to other sites, whereas no significant difference between the BM and BA sites. On the palatal sites, the PA showed a significant resorption percentage than the remaining sites with no significant difference between the PC and PM sites (Table 2). A comparison of the percentage of resorption between the force groups at different sites showed significant differences between the groups at all sites except the MC surface (Table 3).
Determination of protein concentration in GCF
The highest protein concentrations were detected at 14 days for HR (0.95 ± 0.08 µg/µL) and LR group after 21 days (0.94 ± 0.06 µg/µL). The lowest average value showed after 1 day and 28 days in HR and LR groups (0.55 ± 0.06 and 0.52 ± 0.02 µg/µL, respectively). The protein concentration of the periodontitis group was 0.49 ± 0.04 µg/µL. For both resorption groups, the GCF-protein concentration was decreased following immediate force application with a sharp increase after 7 days and 14 days in LR and HR groups, respectively. Protein concentration continued to gradually decline in the LR group till the 28 days, whereas in the HR group, a sharp decrease in protein concentration occurred at 21 days followed by a slight increase after 28 days (Supplementary Fig. 1).
Proteomics analysis
The results indicated that all GCF gels exhibited protein spots with the highest intensity closely clustered together in the pI region (pH 5–6) and two distinct regions of spots with MW around 10–15 kDa and 25–100 kDa were resolved. The HR group showed the highest total number of protein spots after 14 days and 28 days after force application followed by the periodontitis group. The least number of total spots was associated with the LR group 14 days following force application. After computerized analysis, the matching of each spot in all gels with a significant difference (P < 0.05) had more than 1.5-fold changes, and the total number of protein spots was equal to 39 spots (Fig. 3). Twenty nine were clinically resolved and excised from the gels for analysis. Spot profiles were electronically expressed and clinically resolved on each gel with their log normalized volume together with experimental pI and MW (Table 4).
A representative 2DE electrophoretic gel after silver staining showing the resulting image of all GCF samples [high resorption (HR) and low resorption (LR) (in 3 time-points, 0, baseline; 4, 14 days; 6, 28 days); and periodontitis], analyzed by Progenesis SameSpot software. Black circles indicate the identified spots. Protein spots are differentially expressed in all replicas of all groups with significant differences [ANOVA, P < 0.05] and fold changes > 1.5
Identification of proteins using MALDI-TOF/TOF MS/MS analysis
Out of 19 spots, it was able to detect eight identified proteins. The identified proteins were as follows immunoglobulin kappa (IGKC) and its isoform (spots 648 and 1068, respectively) with a base peak at 1797.919 m/z ratio for both spots, neutrophil gelatinase-associated lipocalin (NGAL; spot 1115, peak at 1432.002 m/z ratio), neurolysin mitochondrial (NEUL; spot 649, peak at 724.117 m/z ratio), keratin, type II cytoskeletal 1 (K2C1) and its isoforms (spot 1315 and 1328; base peaks at 725.736 and 1143.569 m/z ratio, respectively), and S100-A9 (spot 1320; base peak value at 1863.948 m/z ratio). The last identified protein was the extracellular calcium-sensing receptor (CASR; spot 1356, base peak at 1884.829 m/z ratio). Table 4 shows the summary for these identified proteins that were cited from UniProt Knowledgebase/SwissProt/Proteomes at the molecular level.
Differential abundance of identified proteins in 2D gel electrophoresis
The IGKC and its isoform were considered abundant proteins that were resolved in different spots and exhibited different profiles between groups. The spot with ID:648 showed down-regulated protein profile (fold change = 7.1, P = 0.003, Supplementary Table 1) in the resorption groups after force application in comparison to the baseline. Whereas its level in the periodontitis group was higher than HR and LR at T0, T4 and T6 (Fig. 4A). Additionally, the other spot (1068) exhibited a different profile among all groups (fold change = 6.3, P = 0.001, Supplementary Table 1) (Fig. 4B). NGAL also showed changes in its profile (fold change = 5.7, P = 0.007, Supplementary Table 1); it was upregulated in HR than LR group. However, its level in both resorption groups was higher than in the periodontitis group at all time-points. The only identified upregulated protein in the LR group after 14 days of force application was the NEUL (fold change = 9.7, P = 0.011, Supplementary Table 1). It was down-regulated in the HR group throughout the trial, while in the periodontitis group, its level was higher than in other groups (Fig. 4A). K2C1 and its isoform (fold change = 4.4; 5.9, P = 0.013; 0.00001, respectively), S-100 and extracellular CASR (fold change = 5.8; 5.0, P = 0.0008; 0.013, respectively) (Supplementary Table 1) were the three upregulated proteins in HR group after force application. In the LR group, these proteins were returned to their normal level after 28 days. These proteins revealed a low level in the periodontitis group in comparison to the other groups at different time-points (Fig. 4A).
A Summary of up-and-downregulation of identified proteins at different time-points, with the color-coded comparison of their abundance between the resorption groups (HR, high resorption; LR low resorption) and periodontitis (P). Color-coded: red indicates the highest protein expression; yellow indicates less than the highest and blue indicates the least expression. B Heat maps showing a distinct pattern in the altered abundance GCF proteins following orthodontically induced inflammatory root resorption for both heavy and light forces (HF and LF). All significant proteins were clustered in a heat map generated by Genesis software. The protein abundances are presented in red (upregulated) and green (down-regulated). C Illustration of the molecular correlations between the proteins with a different abundance that linked them to the top biological functions in response to the expression of proteins in the highly resorbed teeth compared to the respective baseline. Nodes in red represent upregulated proteins, while green nodes represent down-regulated proteins. Nodes with various shapes represent the functional class of the proteins. Edges with various labels describe the nature of the relationship between the nodes
Bioinformatics analyses
Clustering of the altered abundance proteins
The heat map was then generated using Genesis software to show the abundance pattern of proteins and their changes in LR, HR, and CP at T0, T4, and T6. Distinct abundance patterns are seen in all groups ranging from LR and HR compared to CP (Fig. 4B).
Top biological functions of the significantly identified proteins
Detailed information on the significantly altered abundance proteins and their respective Gene Ontology functions are outlined in Table 5. Briefly, the significantly altered proteins were involved in major biological processes, molecular functions, and cellular components derived from the GO database namely extracellular space, defense response, cell recognition, innate immune response, calcium ion transport, peptide metabolic process, and epidermal cell differentiation.
Figure 4C shows a graphical representation of the predicted molecular relationships between the identified proteins which linked them to the top four biological functions. Overall, the up-regulation of IGKC, S100-A9 and CASR downregulation of NGAL, NEUL and keratin, K2C1 were significantly associated with inflammatory response, plaque psoriasis, degranulation of neutrophils, oscillation of Ca2+, and signal transduction.
Discussion
The OT-associated OIIRR is a common pathological problem with undesirable consequences; therefore, its early detection is crucial for achieving satisfactory outcomes. This study aimed to identify OIIRR-associated GCF biomarkers exhibiting specific differential protein profiling following different OF applications. Eight spots (IGKC, NGAL, NEUL-mitochondrial, K2C1, S100-A9 and CASR) were identified, using MALDI-TOF/TOF MS/MS, with differential protein profiling in each time point as compared to the baseline. Most of these proteins were related to the inflammatory process, immune triggering factors, and mechanical stress. Although, these proteins were previously detected in GCF of patients with gingivitis and periodontitis. However, their expressions of OIIRR have received limited attention.
Results indicated downregulation of IGKC and its isoform in resorption groups after 14 days of force application but upregulated in the periodontitis group. The IGKC functions as an adaptive immune response enhancer that influences oral microbiota by interfering with bacterial adherence and metabolism (Zouali 2017; Figueredo et al. 2019). This is consistent with this study in which the protein downregulation in resorption groups after force application was detected, while in periodontitis, it was highly expressed. The NGAL is another protein identified which possesses the ability to form a complex with bacterial MMP-9, thus mediating innate immunity response (Nasioudis and Witkin 2015; Westerlund et al. 1996). In addition, IL-1β was found to upregulate NGAL in human epithelial cells and is involved in the pathogenesis and severity of periodontal disease (Nakajima et al. 2019; Morelli et al. 2014; Tan et al. 2020). In the current study, the level of NGAL was down-regulated after 14 days of force application in the resorption groups, followed by up-regulation after 28 days. Whereas its level was down-regulated in the periodontitis group compared to healthy subjects. Upregulation of NGAL at 28 days could be an indicator of an ongoing immunological response to the bone remodeling process. A recent study showed that the incidence and severity of the disease are strongly associated with increased levels of NGAL and clinical parameters, as well as serum and salivary IL-1β and IL-10 (Tan et al. 2020).
Although the physiological role of NEUL is known (Rioli et al. 1998); however, its pathological role is still elusive despite experimental models indicating its correlation with angiogenesis and tumor growth (Paschoalin et al. 2007), sepsis (Piliponsky et al. 2008), stroke (Rashid et al. 2014), and metabolic disorders (Cavalcanti et al. 2014). Neuronal regulation for bone formation observed in distant bones of the skeleton suggests that neuropeptides, similar to the mechanical loading effect, have a crucial role through RANKL/OPG pathway in suppressing bone resorption (Sample et al. 2008; Yoo et al. 2014). Interestingly, in this study, NEUL was found to be upregulated in LR group after 14 days of force application and in the periodontitis group as well, while the opposite was observed in HR group throughout the trial. This could be due to that the resorption activity, in HR group, may be enhanced through RANKL/OPG pathway associated with continuous downregulation of NEUL enzyme. In previous investigations comparing protein content between periodontitis and healthy subjects, K2C1 was highly expressed in the GCF of periodontitis patients which potentially reflects periodontal tissue destruction (McLaughlin et al. 1996; Silva-Boghossian et al. 2013). In the current study, the heavy buccally directed force in HR group may accelerate the junctional epithelium destruction; thereby, expression of K2C1 was upregulated.
S100A9 is the protein of interest in several proteomics studies correlating its level in periodontal diseases compared to healthy periodontium (Wei et al. 2019). Proteomics and transcriptomic studies showed controversies about the level of salivary S100A9 in periodontitis patients (Haigh et al. 2010; Salazar et al. 2013; Karna et al. 2019). However, most of the proteomics studies indicated up-regulation of S100A9 expression which is strongly related to the severity of periodontitis (Choi et al. 2011; Kido et al. 2012; Silva-Boghossian et al. 2013; Carneiro et al. 2012). However, other studies indicated that S100A9 was highly upregulated in patients with healthy periodontium (Preianò et al. 2018; Yaprak et al. 2018). This could be due to the regulatory role of S100A9 in protecting the healthy periodontium as a tissue response to microbial invasion. The results of this study suggested that S100A9 enhanced severe osteoclastogenesis and osteoclast/odontoclast differentiation in response to HF application. While, in LR group, S100A9 is mostly related to bone remodeling rather than inflammation in response to mechanical loading. CASR was another highly expressed protein in HR group. CASR was found to be expressed in human PDL cells (Koori et al. 2014) and dental pulp cells (Mizumachi et al. 2017; Chen et al. 2019). Koori et al. (2014) indicated that up-regulation of CASR is contributing to the proliferation and osteo/odontogenic differentiation of human PDL stem cells. In addition, the CASR triggers several signaling downstream, thereby triggering different intracellular cascades (Brown and MacLeod 2001). For instance, the CASR guides myeloid osteoclast precursors' migration to bones allowing bone remodeling to initiate (Boudot et al. 2010). CASR has been hypothesized to regulate immune function by regulating the activation of the inflammasome pathway (Lee et al. 2012). Canton et al. (2016) concluded that CASR can act at injury and inflammation sites in the migration to and acquisition of antigens, thereby helping the immune surveillance function of antigen-presenting cells and cell death by pyroptosis (Canton et al. 2016). These findings could explain the increased expression of CASR in this study, particularly in HR resorption group after 14 days of fore application followed by a sharp decrease in its expression at the end of the study.
Bioinformatics analyses revealed that inflammatory response was activated by the up-and downregulation of IGKC, S100-A9, CASR, NGAL, and K2C1 which eventually affected the oscillation of Ca2+, degranulation of neutrophils, plaque psoriasis, and signal transduction. As reported in other previous studies, root resorption from orthodontic complications induced oral inflammatory responses (Lee and Lee 2016; Umashetty et al. 2015). During periodontitis, studies found increased calcium released from shallow regions of the alveolar bone and eventually flowing into the GCF, reflecting the progression of inflammation (Kuraji et al. 2021). Moreover, neutrophil infiltration is the established hallmark of acute inflammation as well as immune system modulation during periodontitis. Neutrophils are also acknowledged as important players in chronic inflammatory disorders, including periodontitis and psoriasis (Hajishengallis et al. 2016). To date, no studies have reported on the OIIRR effects on psoriasis, but our analyses showed that changes in the expression of S100-A9, NGAL, and K2C1 were associated with plaque psoriasis. The oral psoriasis symptoms include small red dots in the mouth, bleeding, oral lesions, and tooth decay. Most cases of oral psoriatic lesions are often underdiagnosed, because they are asymptomatic, but the increased oral mucosal lesions during periodontitis confirmed the diagnosis of oral psoriasis (Ganzetti et al. 2014). Hence, there is an increased association between periodontitis and oral psoriasis, which may suggest common underlying pathogenic risk factors. In addition, changes in the expression of CASR and NEUL were associated with signal transduction. It remains largely unknown how NEUL is involved in signal transduction during OIIRR. However, the signal transduction between pro-inflammatory cytokines and mediators is one of the mechanisms regulating the increased susceptibility to periodontitis (Souza et al. 2012). Interestingly, the changes in the expression of this protein during OIIRR-induced oral inflammatory events are highly associated with the pathogenesis of periodontitis.
The small sample size is one of the limitations that affect the generalizability of the results. The tiny volume of GCF and evaporation issues along with rapid protein denaturation are common problems associated with this biofluid. Further, OIIRR is a multi-factorial condition; therefore, without a standardized study design that considered all these confounders, false-positive biomarkers may be identified. However, the results of this study were promising and supportive of using the biomarkers available in GCF as a safer alternative to radiographs. Identification of root resorption non-invasively with biomarkers is highly important as it can enable orthodontists to achieve optimum OT with minimum adverse effects.
Overall, S100A9, CASR, and K2C1 up-regulation suggested a response to force-related inflammation, chemotactic activities, and osteoclastogenesis enhancement along with epithelial cell breakdown during the early phase of OT. Meanwhile, the up-regulation of IGKC, NGAL, and K2C1 indicated a response to the inflammatory process, innate immunity activation, and epithelial cell breakdown. Clinically, these proteins could be used as predictive/diagnostic biomarkers for OIIRR to avoid undesirable consequences of this condition. Further validations for the identified proteins will be recommended for future research to overcome this study's limitation and to ensure the diagnostic significance of the identified potential protein biomarker.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author upon request.
Code availability
Fiji (Image-J, Inc.) was used to analyze a straight-on 2D root image to measure the surface area of root resorption. Progenesis SameSpot software was used to analyze 2DE gel images and Ingenuity Pathways Analysis (IPA) (Ingenuity® Systems, Redwood City, CA, USA) software (www.ingenuity.com) was used to predict the biological functions associated with the differentially expressed proteins, details described in detail in the Materials and methods section.
References
Al-Obaidi JR, Hussin SNIS, Saidi NB, Rahmad N, Idris AS (2017) Comparative proteomic analysis of Ganoderma species during in vitro interaction with oil palm root. Physiol Mol Plant Pathol 99:16–24. https://doi.org/10.1016/j.pmpp.2017.02.001
Barros SP, Williams R, Offenbacher S, Morelli T (2016) Gingival crevicular fluid as a source of biomarkers for periodontitis. Periodontology 70(1):53–64. https://doi.org/10.1111/prd.12107
Boudot C, Saidak Z, Boulanouar AK, Petit L, Gouilleux F, Massy Z, Brazier M, Mentaverri R, Kamel S (2010) Implication of the calcium sensing receptor and the Phosphoinositide 3-kinase/Akt pathway in the extracellular calcium-mediated migration of RAW 264.7 osteoclast precursor cells. Bone 46(5):1416–1423. https://doi.org/10.1016/j.bone.2010.01.383
Brezniak N, Wasserstein A (2019) Apical root shortening versus root resorption-Is there a difference? Am J Orthod Dentofac Orthop 156(2):164–165. https://doi.org/10.1016/j.ajodo.2019.04.028
Brown EM, MacLeod RJ (2001) Extracellular calcium sensing and extracellular calcium signaling. Physiol Rev 81(1):239–297. https://doi.org/10.1152/physrev.2001.81.1.239
Buduneli N (2019) Biomarkers in periodontal health and disease. Rationale, benefits, and future directions. https://doi.org/10.1007/978-3-030-37317-7
Canton J, Schlam D, Breuer C, Gütschow M, Glogauer M, Grinstein S (2016) Calcium-sensing receptors signal constitutive macropinocytosis and facilitate the uptake of NOD2 ligands in macrophages. Nat Commun 7:11284. https://doi.org/10.1038/ncomms11284
Carneiro LG, Venuleo C, Oppenheim FG, Salih E (2012) Proteome data set of human gingival crevicular fluid from healthy periodontium sites by multidimensional protein separation and mass spectrometry. J Periodontal Res 47(2):248–262. https://doi.org/10.1111/j.1600-0765.2011.01429.x
Cavalcanti DM, Castro LM, Rosa Neto JC, Seelaender M, Neves RX, Oliveira V, Forti FL, Iwai LK, Gozzo FC, Todiras M, Schadock I, Barros CC, Bader M, Ferro ES (2014) Neurolysin knockout mice generation and initial phenotype characterization. J Biol Chem 289(22):15426–15440. https://doi.org/10.1074/jbc.M113.539148
Chapple ILC, Mealey BL, Van Dyke TE, Bartold PM, Dommisch H, Eickholz P, Geisinger ML, Genco RJ, Glogauer M, Goldstein M, Griffin TJ, Holmstrup P, Johnson GK, Kapila Y, Lang NP, Meyle J, Murakami S, Plemons J, Romito GA, Shapira L, Tatakis DN, Teughels W, Trombelli L, Walter C, Wimmer G, Xenoudi P, Yoshie H (2018) Periodontal health and gingival diseases and conditions on an intact and a reduced periodontium: consensus report of workgroup 1 of the 2017 world workshop on the classification of periodontal and peri-implant diseases and conditions. J Periodontol 89(Suppl 1):S74-s84. https://doi.org/10.1002/jper.17-0719
Chen Y, Gao Y, Tao Y, Lin D, An S (2019) Identification of a calcium-sensing receptor in human dental pulp cells that regulates mineral trioxide aggregate-induced mineralization. J Endod 45(7):907–916. https://doi.org/10.1016/j.joen.2019.03.019
Choi YJ, Heo SH, Lee JM, Cho JY (2011) Identification of azurocidin as a potential periodontitis biomarker by a proteomic analysis of gingival crevicular fluid. Proteome Sci 9:42. https://doi.org/10.1186/1477-5956-9-42
Chutimanutskul W, Ali Darendeliler M, Shen G, Petocz P, Swain MV (2006) Changes in the physical properties of human premolar cementum after application of 4 weeks of controlled orthodontic forces. Eur J Orthod 28(4):313–318. https://doi.org/10.1093/ejo/cji115
Currell SD, Liaw A, Blackmore Grant PD, Esterman A, Nimmo A (2019) Orthodontic mechanotherapies and their influence on external root resorption: a systematic review. Am J Orthod Dentofac Orthop 155(3):313–329. https://doi.org/10.1016/j.ajodo.2018.10.015
Dindaroğlu F, Doğan S (2016) Root resorption in orthodontics. Turk J Orthod 29(4):103–108. https://doi.org/10.5152/TurkJOrthod.2016.16021
Dudic A, Giannopoulou C, Leuzinger M, Kiliaridis S (2009) Detection of apical root resorption after orthodontic treatment by using panoramic radiography and cone-beam computed tomography of super-high resolution. Am J Orthod Dentofa Orthop 135(4):434–437. https://doi.org/10.1016/j.ajodo.2008.10.014
Figueredo CM, Lira-Junior R, Love RM (2019) T and B cells in periodontal disease: new functions in A complex scenario. Int J Mol Sci 20(16):3949. https://doi.org/10.3390/ijms20163949
Ganzetti G, Campanati A, Santarelli A, Pozzi V, Molinelli E, Minnetti I, Brisigotti V, Procaccini M, Emanuelli M, Offidani A (2014) Periodontal disease: an oral manifestation of psoriasis or an occasional finding? Drug Dev Res 75(S1):S46–S49. https://doi.org/10.1002/ddr.21194
Ghallab NA (2018) Diagnostic potential and future directions of biomarkers in gingival crevicular fluid and saliva of periodontal diseases: review of the current evidence. Arch Oral Biol 87:115–124. https://doi.org/10.1016/j.archoralbio.2017.12.022
Haigh BJ, Stewart KW, Whelan JR, Barnett MP, Smolenski GA, Wheeler TT (2010) Alterations in the salivary proteome associated with periodontitis. J Clin Periodontol 37(3):241–247. https://doi.org/10.1111/j.1600-051X.2009.01525.x
Hajishengallis G, Moutsopoulos NM, Hajishengallis E, Chavakis T (2016) Immune and regulatory functions of neutrophils in inflammatory bone loss. Semin Immunol 28(2):146–158. https://doi.org/10.1016/j.smim.2016.02.002
Karna S, Shin YJ, Kim S, Kim HD (2019) Salivary S100 proteins screen periodontitis among Korean adults. J Clin Periodontol 46(2):181–188. https://doi.org/10.1111/jcpe.13059
Khurshid Z, Zohaib S, Najeeb S, Zafar MS, Rehman R, Rehman IU (2016) Advances of proteomic sciences in dentistry. Int J Mol Sci 17(5):728. https://doi.org/10.3390/ijms17050728
Kido J, Bando M, Hiroshima Y, Iwasaka H, Yamada K, Ohgami N, Nambu T, Kataoka M, Yamamoto T, Shinohara Y, Sagawa I, Nagata T (2012) Analysis of proteins in human gingival crevicular fluid by mass spectrometry. J Periodontal Res 47(4):488–499. https://doi.org/10.1111/j.1600-0765.2011.01458.x
Koori K, Maeda H, Fujii S, Tomokiyo A, Kawachi G, Hasegawa D, Hamano S, Sugii H, Wada N, Akamine A (2014) The roles of calcium-sensing receptor and calcium channel in osteogenic differentiation of undifferentiated periodontal ligament cells. Cell Tissue Res 357(3):707–718. https://doi.org/10.1007/s00441-014-1918-5
Kuraji R, Wu YH, Hashimoto S, Miyashita Y, Mishiro S, Ito H, Kamarajan P, Kapila Y, Numabe Y (2021) Periodontal inflammation triggers a site-specific and wide radius of calcium metabolic effects on alveolar bone. J Periodonatal Res 56(2):314–329. https://doi.org/10.1111/jre.12824
Lee YJ, Lee TY (2016) External root resorption during orthodontic treatment in root-filled teeth and contralateral teeth with vital pulp: a clinical study of contributing factors. Am J Orthod Dentofac Orthop 149(1):84–91. https://doi.org/10.1016/j.ajodo.2015.06.027
Lee GS, Subramanian N, Kim AI, Aksentijevich I, Goldbach-Mansky R, Sacks DB, Germain RN, Kastner DL, Chae JJ (2012) The calcium-sensing receptor regulates the NLRP3 inflammasome through Ca2+ and cAMP. Nature 492(7427):123–127. https://doi.org/10.1038/nature11588
Mahmood RI, Abbass AK, Razali N, Al-Saffar AZ, Al-Obaidi JR (2021) Protein profile of MCF-7 breast cancer cell line treated with lectin delivered by CaCO3NPs revealed changes in molecular chaperones, cytoskeleton, and membrane-associated proteins. Int J Biol Macromol 184:636–647
Malek S, Darendeliler MA, Rex T, Kharbanda OP, Srivicharnkul P, Swain MV, Petocz P (2003) Physical properties of root cementum: part 2. effect of different storage methods. Am J Orthod Dentofacial Orthop 124(5):561–570. https://doi.org/10.1016/s0889-5406(03)00398-6
Mansor M, Al-Obaidi JR, Jaafar NN, Ismail IH, Zakaria AF, Abidin MAZ, Selamat J, Radu S, Jambari NN (2020) Optimization of protein extraction method for 2DE proteomics of goat’s milk. Molecules 25(11):2625
McLaughlin WS, Kirkham J, Kowolik MJ, Robinson C (1996) Human gingival crevicular fluid keratin at healthy, chronic gingivitis and chronic adult periodontitis sites. J Clin Periodontol 23(4):331–335. https://doi.org/10.1111/j.1600-051x.1996.tb00554.x
Mi H, Muruganujan A, Thomas PD (2012) Panther in 2013: modeling the evolution of gene function, and other gene attributes, in the context of phylogenetic trees. Nucl Acids Res 41(D1):D377–D386
Mizumachi H, Yoshida S, Tomokiyo A, Hasegawa D, Hamano S, Yuda A, Sugii H, Serita S, Mitarai H, Koori K, Wada N, Maeda H (2017) Calcium-sensing receptor-ERK signaling promotes odontoblastic differentiation of human dental pulp cells. Bone 101:191–201. https://doi.org/10.1016/j.bone.2017.05.012
Mohammed-Salih HS, Saloom HF (2022) Collection, storage and protein extraction method of gingival crevicular fluid for proteomic analysis. Baghdad Sci J 19(2):0368. https://doi.org/10.21123/bsj.2022.19.2.0368
Morelli T, Stella M, Barros SP, Marchesan JT, Moss KL, Kim SJ, Yu N, Aspiras MB, Ward M, Offenbacher S (2014) Salivary biomarkers in a biofilm overgrowth model. J Periodontol 85(12):1770–1778. https://doi.org/10.1902/jop.2014.140180
Nakajima M, Hosojima M, Tabeta K, Miyauchi S, Yamada-Hara M, Takahashi N, Miyazawa H, Matsuda-Matsukawa Y, Sato K, Sugita N, Komatsu Y, Ishikawa T, Akiishi K, Yamazaki K, Kato K, Saito A, Yoshie H (2019) β (2)-Microglobulin and neutrophil gelatinase-associated lipocalin, potential novel urine biomarkers in periodontitis: A Cross-Sectional Study in Japanese. Int J Dent 2019:1394678. https://doi.org/10.1155/2019/1394678
Nasioudis D, Witkin SS (2015) Neutrophil gelatinase-associated lipocalin and innate immune responses to bacterial infections. Med Microbiol Immunol 204(4):471–479. https://doi.org/10.1007/s00430-015-0394-1
Nieto-Nieto N, Solano JE, Yañez-Vico R (2017) External apical root resorption concurrent with orthodontic forces: the genetic influence. Acta Odontol Scand 75(4):280–287. https://doi.org/10.1080/00016357.2017.1294260
Paschoalin T, Carmona AK, Rodrigues EG, Oliveira V, Monteiro HP, Juliano MA, Juliano L, Travassos LR (2007) Characterization of thimet oligopeptidase and neurolysin activities in B16F10-Nex2 tumor cells and their involvement in angiogenesis and tumor growth. Mol Cancer 6:44. https://doi.org/10.1186/1476-4598-6-44
Piliponsky AM, Chen CC, Nishimura T, Metz M, Rios EJ, Dobner PR, Wada E, Wada K, Zacharias S, Mohanasundaram UM, Faix JD, Abrink M, Pejler G, Pearl RG, Tsai M, Galli SJ (2008) Neurotensin increases mortality and mast cells reduce neurotensin levels in a mouse model of sepsis. Nat Med 14(4):392–398. https://doi.org/10.1038/nm1738
Preianò M, Maggisano G, Murfuni MS, Villella C, Pelaia C, Montalcini T, Lombardo N, Pelaia G, Savino R, Terracciano R (2018) An analytical method for assessing optimal storage conditions of gingival crevicular fluid and disclosing a peptide biomarker signature of gingivitis by MALDI-TOF MS. Proteom Clin Appl 12(5):e1800005. https://doi.org/10.1002/prca.201800005
Rashid M, Wangler NJ, Yang L, Shah K, Arumugam TV, Abbruscato TJ, Karamyan VT (2014) Functional up-regulation of endopeptidase neurolysin during post-acute and early recovery phases of experimental stroke in mouse brain. J Neurochem 129(1):179–189. https://doi.org/10.1111/jnc.12513
Rioli V, Kato A, Portaro FC, Cury GK, Te Kaat K, Vincent B, Checler F, Camargo AC, Glucksman MJ, Roberts JL (1998) Neuropeptide specificity and inhibition of recombinant isoforms of the endopeptidase 3.4. 24.16 family: comparison with the related recombinant endopeptidase 3.4. 24.15. Biochem Biophys Res Commun 250(1):5–11
Rody WJ Jr, Holliday LS, McHugh KP, Wallet SM, Spicer V, Krokhin O (2014) Mass spectrometry analysis of gingival crevicular fluid in the presence of external root resorption. Am J Orthod Dentofac Orthop 145(6):787–798. https://doi.org/10.1016/j.ajodo.2014.03.013
Salazar MG, Jehmlich N, Murr A, Dhople VM, Holtfreter B, Hammer E, Völker U, Kocher T (2013) Identification of periodontitis associated changes in the proteome of whole human saliva by mass spectrometric analysis. J Clin Periodontol 40(9):825–832. https://doi.org/10.1111/jcpe.12130
Saloom HF, Papageorgiou SN, Carpenter GH, Cobourne MT (2017) Impact of obesity on orthodontic tooth movement in adolescents: a prospective clinical cohort study. J Dent Res 96(5):547–554
Samandara A, Papageorgiou SN, Ioannidou-Marathiotou I, Kavvadia-Tsatala S, Papadopoulos MA (2019) Evaluation of orthodontically induced external root resorption following orthodontic treatment using cone beam computed tomography (CBCT): a systematic review and meta-analysis. Eur J Orthod 41(1):67–79. https://doi.org/10.1093/ejo/cjy027
Sample SJ, Behan M, Smith L, Oldenhoff WE, Markel MD, Kalscheur VL, Hao Z, Miletic V, Muir P (2008) Functional adaptation to loading of a single bone is neuronally regulated and involves multiple bones. J Bone Miner Res 23(9):1372–1381. https://doi.org/10.1359/jbmr.080407
Silva-Boghossian CM, Colombo AP, Tanaka M, Rayo C, Xiao Y, Siqueira WL (2013) Quantitative proteomic analysis of gingival crevicular fluid in different periodontal conditions. PLoS ONE 8(10):e75898. https://doi.org/10.1371/journal.pone.0075898
Souza JACd, Rossa C Jr, Garlet GP, Nogueira AVB, Cirelli JA (2012) Modulation of host cell signaling pathways as a therapeutic approach in periodontal disease. J Appl Oral Sci 20(2):128–138. https://doi.org/10.1590/s1678-77572012000200002
Sturn A, Quackenbush J, Trajanoski Z (2002) Genesis: cluster analysis of microarray data. Bioinformatics 18(1):207–208
Tan A, Gürbüz N, Özbalci F, Koşkan Ö, Yetkin Ay Z (2020) Increase in serum and salivary neutrophil gelatinase-associated lipocalin levels with increased periodontal inflammation. J Appl Oral Sci 28:e20200276. https://doi.org/10.1590/1678-7757-2020-0276
Umashetty G, Hoshing U, Patil S, Ajgaonkar N (2015) Management of inflammatory internal root resorption with biodentine and thermoplasticised Gutta-Percha. Case Rep Dent 2015:452609. https://doi.org/10.1155/2015/452609
Wei L, Liu M, Xiong H (2019) Role of calprotectin as a biomarker in periodontal disease. Mediators Inflamm 2019:3515026. https://doi.org/10.1155/2019/3515026
Westerlund U, Ingman T, Lukinmaa PL, Salo T, Kjeldsen L, Borregaard N, Tjäderhane L, Konttinen YT, Sorsa T (1996) Human neutrophil gelatinase and associated lipocalin in adult and localized juvenile periodontitis. J Dent Res 75(8):1553–1563. https://doi.org/10.1177/00220345960750080601
Yan JX, Wait R, Berkelman T, Harry RA, Westbrook JA, Wheeler CH, Dunn MJ (2000) A modified silver staining protocol for visualization of proteins compatible with matrix-assisted laser desorption/ionization and electrospray ionization-mass spectrometry. Electrophoresis 21(17):3666–3672. https://doi.org/10.1002/1522-2683(200011)21:17%3c3666::Aid-elps3666%3e3.0.Co;2-6
Yaprak E, Kasap M, Akpınar G, Kayaaltı-Yüksek S, Sinanoğlu A, Guzel N, Demirturk Kocasarac H (2018) The prominent proteins expressed in healthy gingiva: a pilot exploratory tissue proteomics study. Odontology 106(1):19–28. https://doi.org/10.1007/s10266-017-0302-9
Yoo YM, Kwag JH, Kim KH, Kim CH (2014) Effects of neuropeptides and mechanical loading on bone cell resorption in vitro. Int J Mol Sci 15(4):5874–5883. https://doi.org/10.3390/ijms15045874
Zouali M (2017) The emerging roles of B cells as partners and targets in periodontitis. Autoimmunity 50(1):61–70. https://doi.org/10.1080/08916934.2016.1261841
Acknowledgements
The authors would like to express their appreciation to Hassanien Ahmed Hadi, a maxillo-facial surgeon, for his unceasing help and efforts in the clinical aspect of this project. Thanks to Proteomic and Histology lab members at Agro-Biotechnology Institute (ABI/NIBM, Malaysia) for their scientific and laboratory assistance in biochemical and proteomics analysis.
Funding
Not applicable.
Author information
Authors and Affiliations
Contributions
Study conception and design: HSMS and HFS; data collection: HSMS and HAA.; analysis and interpretation of results: HSMS, JRA., and NR.; draft manuscript preparation: HSMS and AAA; proofreading and critical review: AAA; validation: HAA; supervision: HFS. All authors reviewed the results and approved the final version of the manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
All authors have no relevant interests to declare.
Ethical approval
Ethical approval for this study was obtained following the Declaration of Helsinki, by the Ethics Committee of College of Dentistry, University of Baghdad (ID #023418, January 9, 2019) with clinical trial registration #NCT04291443 on 02/03/2020.
Consent to participate
Control subjects and involved individuals provided written informed consent to the study protocol of the project before starting this research.
Consent for publication
All authors agree to publish this work.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Mohammed-Salih, H.S., Al-lami, H.A., Saloom, H.F. et al. Detection of orthodontically induced inflammatory root resorption-associated biomarkers from the gingival crevicular fluid by proteomics analysis: a randomized-controlled clinical trial. 3 Biotech 13, 157 (2023). https://doi.org/10.1007/s13205-023-03572-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s13205-023-03572-5