Abstract
Crop improvement is a continuous process in agriculture which ensures ample supply of food, fodder and fiber to burgeoning world population. Despite tremendous success in plant breeding and transgenesis to improve the yield-related traits, there have been several limitations primarily with the specificity in genetic modifications and incompatibility of host species. Because of this, new breeding techniques (NBTs) are gaining worldwide attention for crop improvement programs. Among the NBTs, genome editing (GE) using site-directed nucleases (SDNs) is an important and potential technique that overcomes limitations associated with classical breeding and transgenesis. These SDNs specifically target a compatible region in the gene/genome. The meganucleases (MgN), zinc finger nucleases (ZFN), transcription activator-like effectors nucleases (TALENs), and clustered regularly interspaced short palindromic repeats (CRISPR) and CRISPR-associated endonuclease (Cas) are being successfully employed for GE. These can be used for desired or targeted modifications of the native endogenous gene(s) or targeted insertion of cis/trans elements in the genomes of recipient organisms. Applications of these techniques appear to be endless ever since their discovery and several modifications in original technologies have further brought precision and accuracy in these methods. In this review, we present an overview of GE using SDNs with an emphasis on CRISPR/Cas system, their advantages, limitations and also practical considerations while designing experiments have been discussed. The review also emphasizes on the possible applications of CRISPR for improving economic traits in crop plants.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Plant breeding for crop improvement involves selection and hybridization, which are largely dependent on homologous recombination between chromosomes to generate the genetic diversity which has been in practice since the ancient time. To increase the range of natural variation for traits, plant breeders also have been exploring chemicals or irradiation methods. Although, classical and mutational breeding have resulted in significant improvements in various agronomic traits, viz., yield, quality, nutrition, biotic and abiotic stress resistance, there are certain limitations in terms of labor, time and precise knowledge of selection. Recent advances in plant genetics and breeding approaches led to the development of marker-assisted selection or marker-aided breeding where traits are linked with specific DNA markers on the genome which facilitate in rapid and accurate selection for those traits. Further advancements in biotechnological methodologies enabled the researchers to add desirable traits in crops by inserting genes-of-interest from other organisms into the plant genome which is popularly known as transgenesis. The use of transgene (or foreign gene) and its non-specific integration in the host genome, the use of bacterial-origin selection markers and possibilities of somaclonal variations are the issues which raise concern for biosafety. A potential alternative to transgenic is cisgenics wherein variation is created at predetermined sites in the native gene(s) of an organism (creation of novel alleles).
Conversely, new breeding techniques (NBTs) offer additional options to replace conventional breeding and transgenic technology. NBTs which include a range of methods, viz., site-directed nucleases (SDNs), cisgenesis, RNA-dependent DNA methylation, grafting (non-genetically modified (GM) scion on GM rootstock), reverse breeding, agro-infiltration, etc. (Lusser et al. 2012), established for introduction of targeted changes in plant genome to modify the economic traits of crops. Among those, use of SDNs such as ZFNs (zinc finger nucleases), TALENs (transcription activator-like effectors nucleases) and CRISPR/Cas (cluster regularly interspaced short palindromic repeats/CRISPR-associated proteins) for genome editing (GE) has great potential for crop improvement. The SDNs create specific double-stranded breaks (DSBs) at desired locations in the genome and harness the cell’s endogenous mechanisms to repair the DSBs by homologous recombination (HR) or non homologous end-joining (NHEJ). The NHEJ pathway repairs DSB by ligating broken ends without the help of a homologous template, often resulting in insertions or deletions (InDels) and single nucleotide polymorphism (SNP) at the cut site, thus causing mutations such as frameshift or nonsense mutations. On the other hand, HR allows gene replacement by replacing the DNA sequence using template at the break point. Both NHEJ and HR repair pathways are key processes for nuclease-based GE (Pardo et al. 2009). The field of GE has been witnessing considerable advances in terms of precision, efficiency and accuracy in methodology from meganucleases to ZFNs to TALENs till the most recent CRISPR/Cas nucleases which are revolutionizing the field of plant molecular biology.
The use of GE was pioneered by Paszkowski et al. (1988) for the integration of gene in the tobacco genome via homologous recombination. Later, Bibikova et al. (2003) reported targeted modifications using sequence-specific nucleases by direct gene transfer to Nicotiana tabacum protoplasts. Even after these early leads, it took more than 25 years to build up the information and experimental set-up required for successful application of GE in plants (Puchta and Fauser 2013). The historical timeline of GE in plants is provided in Fig. 1. In crop improvement, GE can be used for development of better phenotypes such as high yield, altering plant architecture, enhancing nutritional value and stress tolerance resistance, etc. The ease, low cost and speed of designing GE tools make it the most appropriate and feasible system for plant improvement in the present era of high-throughput technologies. Additionally, GE offers an advantage over traditional mutation breeding as it creates the variations in the selected target site with high frequency. It certainly transforms the transgenic technology as it integrate cis- or trans-genes without the extra burden of foreign genetic elements, thus may reduce public concern, anxiety and regulatory costs of GM crops. An appreciation of the same is reflected by conferment as a method of the year for 2011 by Nature Methods.
Genome editing tools
The meganucleases (MgNs) are naturally occurring unique enzymes having high activity and long recognition sequences known as the homing site (> 14 bp) and were employed as the first GE tool (Curtin et al. 2012) (Fig. 2a). There are about hundreds of MgNs identified in several organisms including bacteria, fungi and some of the plant species. Most commonly utilized nucleases include I-SceI and I-CreI which have copies of the LAGLIDADG motif. In spite of their greater specificity, use of MgNs in GE is restricted because of the presence of single homing site in genomes of many organisms and overlapping cleavage site and DNA binding domains, thus making it difficult to engineer DNA binding domain. Nonetheless, efforts have been made towards the development of MgN-TALE chimera (megaTAL) having greater flexibility as compared to native MgN. As MgN is an early tool of GE, we have not discussed this tool here further.
Types of different SDNs used for GE. a MgNs: schematic representation of naturally occurring I-SecI MgNs. Homing site for I-SecI is 18 bp and it cleaves DNA within the homing site. DNA binding and cleavage domains are not clearly demarcated. b ZFNs: schematic representation of synthetic ZFNs, ZFNs are synthesized by fusion of zinc finger DNA binding domain and FokI cleavage domain. Zinc finger DNA binding domains are typically composed of three zinc finger arrays each capable of recognizing approximately 3 bp. c TALENs: schematic representation of synthetic TALENs, TALENs are synthesized by fusion of TAL DNA binding domain and nonspecific FokI cleavage domain. Each TAL domain recognizes only one base. Binding specificity is manipulated by combining repeats that recognize individual bases in different orders. TALENs also work in dimer form. d CRISPR/Cas: schematic representation of CRISPR/Cas system. gRNA guides Cas9 protein for DNA DSB. gRNA form complex along with Cas9 protein and bind to seed sequence. Cas9 nucleases create DSB. Presence of PAM (NGG) sequence immediately downstream to target site is must for DSB
The limitations of MgNs were overcome by ZFNs which are created by linking zinc finger proteins to the cleavage domain of FokI endonuclease and were first reported in Arabidopsis by Lioyd et al. (2005). The ZFN DNA binding domain is generally composed of three to four zinc finger arrays each capable of recognizing 3-bp long sequence (Fig. 2b). Relative to the start point of the zinc finger α-helix, amino acids present at − 1, + 2, + 3, and + 6 position contribute to specificity for proper dimerization of FokI domain as it is critical to the functioning of ZFNs (Kim et al. 1996). The amino acids act as engineerable sites and can be customized to fit specific target sequences. The two ZFN monomers designed in such way, flank 6-bp long sequence within the DNA target sequence, allowing the FokI monomers to form an active dimer and to digest within that spacer sequence. Presence of sparse target sites, difficulties in engineering zinc finger arrays (Maeder et al. 2008) and low targeting efficiency leading to frequent off-target effects (DeFrancesco 2012) are the major constraints in the use of ZFNs. Several strategies have been reported to overcome these limitations such as nickases (ZFNickases) that take advantage of nicking at the single strand and emulate the HR rather the error-prone NHEJ pathway, ultimately resulting in reduced off-site targeting. Furthermore, an attempt to increase 4–6 zinc finger domains for each ZFN enhanced the activity and specificity of ZFNs (Sood et al. 2013).
Though ZFNs resolved some of the difficulties associated with MgNs, there was ample scope of further improvement in GE. Christian et al. (2010) for the first time suggested that zinc finger arrays could be substituted with the DNA recognition domain of TALEs (transcription activator-like effectors) to create TALENs that recognize and cleave DNA targets. These path-breaking experiments were done using two highly recognized TALENs, AvrBs3 and PthXo1 from the pepper pathogen, Xanthomonas campestris pv. vesicatoria and rice pathogen X. oryzae pv. oryzae, respectively. Like ZFNs, TALENs consists of TALE DNA binding domain having 33–35-long repeats of amino acids followed by 20 amino acids known as ‘half repeat’ and a nonspecific FokI cleavage domain (Fig. 2c). The conserved 12th and 13th positions of the TALE monomer impart specificity to nucleotide recognition and are thus called repeat-variable di-residues (RVDs). Research showed that first RVD involved in forming contact with RVD loop backbone whereas the second RVD involved in contact with DNA (Deng et al. 2012). The presence of thymine (T) before recognition site is essential to TALEN engineering (Voytas 2013). Although TALE DNA binding monomers are modular in nature, they still suffer from context-dependent specificity and their repetitive sequences incur high cost and labor for construction of novel TALE arrays (Juillerat et al. 2014). Nevertheless, new techniques such as GoldenGate and Platinum Gate assays are in vogue for efficient construction of TALEN. The wide spectrum of engineered TALE for target binding makes it a more robust GE tool in plants (Zhang et al. 2013a, b).
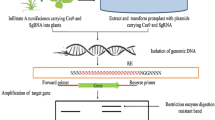
The above three tools of GE described so far are based on recognition of specific DNA sequences by a protein molecule having DNA binding motif. However, engineering DNA binding motif according to the target sequence requires deep knowledge of protein biochemistry and protein engineering. This limitation has been resolved using small RNA sequences as DNA recognition molecules. To attain GE through small RNAs, a prokaryotic immune system called CRISPR/Cas that provides a form of acquired immunity is being deployed (Marraffini and Sontheimer 2008). CRISPRs are often associated with Cas genes that code for proteins related to CRISPRs and three groups of eleven such systems have been reported. The Type II CRISPR/Cas system has been adapted for GE owing to the presence of protospacer adjacent motif (PAM) sequence and a second RNA, called trans-acting CRISPR RNA (tracrRNA). The tracrRNA teams up with CRISPR RNA (crRNA) to assist crRNA maturation and recruit the Cas9 nuclease to DNA. To make this system more promising, the natural three-component system is further simplified by fusing together crRNA and tracrRNA, resulting in the creation of a single synthetic chimeric ‘guide’ RNA (sgRNA or gRNA) (Jinek et al. 2012) (Fig. 2d). For additional information about native CRISPR system of bacteria and archaea, readers are redirected to refer to a review by Bhaya et al. (2011).
A considerable advantage of the CRISPR/Cas system over other SDNs lies with the utilization of an RNA molecule to guide the nuclease to a specific nucleic acid target. RNA is easier and cheaper to synthesize than the protein domains used with ZFN and TALEN approaches. This makes CRISPR/Cas system simple, easy and highly effective (Kim et al. 2017, 2018; Sun et al. 2017). This system also has an advantage with reference to introducing mutations in multiple genes at the same time by introducing multiple gRNAs. However, availability of PAM sequence in adjacent to target site limits target site selection. To overcome such limitations, extensive research is being done especially in mammalian cells to establish proof of concept (Fu et al. 2013). Moreover, a plethora of publications appeared on successful applications of CRISPR/Cas system to engineer desired traits in plants which reflect that it is the most robust tool for crop improvement. A comparative account of GE tools has been summarized in Table 1.
Practical considerations
For successful GE in plants, four major steps are involved, viz., identification of target gene for desired trait (fully functionally characterized), construction of suitable vector/s, transformation and screening the transformants for intended mutation. The target gene should be unique in function and the mutation at particular site should either activate or repress its function. The pleiotropic effects and presence of multiple target sites should also be checked for avoiding the off-target effects. Additionally, for CRISPR/Cas system, the presence of PAM sequence at the 3′ end of the target sequence is essential. If there are no PAM sequences for S. pyogenes Cas9 (i.e., NGG) within the desired sequence, Cas enzyme of different species or S. pyogenes Cas9 variants that bind other PAM sequences present in the desired target can be selected as well.
Next crucial step after selection of appropriate target sequence is the selection of a GE strategy. For engineering highly specific ZFNs, several methods such as modular assembly (MA), Oligomerized Pool Engineering (OPEN), Context-Dependent Assembly (CoDA), etc., are available for choice (Maeder et al. 2008). In case of TALEN, the GoldenGate assay is one of the most powerful tools for generating custom-made TALENs (Dahlem et al. 2012). Platinum TALEN and Platinum Gate system are some of the recent systems to design highly efficient TALE repeat assembly (Sakuma et al. 2013). Online and offline tools such as TALEN-NT, idTALE and EENdb furnishes all the relevant information for engineering TALEN. Accurate selection of target site and designing of gRNA is the most critical step in CRISPR/Cas experiment. Sometimes, gRNA can recognize non-target sequences within genome which show partial homology called off-targets. Removal of off-target is an important criterion for designing a gRNA. In addition to “off-target activity”, specific nucleotides within the target sequence should be carefully selected to maximize cleavage of the desired target sequence (on-target activity). Therefore, close examination of predicted on-target and off-target activity of each potential gRNA is necessary (Wolt et al. 2016a). Many softwares and online tools are available for locating potential PAM, target sequences, and ranking of the associated gRNAs based on their predicted on-target and off-target activity (Table 2). Some variants of gRNA are available commercially viz., truncated gRNA, ribozyme gRNA, polycistronic-tRNA–gRNA, which have improved the native CRISPR system for utilization in broader applicability (Khatodia et al. 2016).
Next step is to transform the GE vectors into the plant. Transformation vector possesses one or two nuclear localization signal (NLS), a suitable promoter and other regulatory elements. Generally, constitutive promoters such as CaMV35S, OsUBQ1 and Actin1 are used in constructs. With respect to CRISPR/Cas system, the use of appropriate RNA pol II promoters such as CaMV35s, AtUBQ, 35sPPDK, OsAct1, etc., is equally critical for expression of Cas9 gene (Mao et al. 2013; Li et al. 2013; Wang et al. 2016). Depending on the objectives of the experiment, variants of Cas9 can be selected (Table 3). For expression of target sequence along with gRNA, pol III promoters (mostly U6 and U3) are attached at 5´end. The 5´G is required for transcription initiation from the U6 promoter. The CRISPR/Cas system leverages the use of a single gene construct by combining all three components, viz., Cas gene, sgRNA and tracrRNA. These three components can be constructed separately in different vectors also. Vectors are available according to the objectives of the experiment such as to generate mutation (to create cut), for activation of gene expression or for generation of nick, etc. (Table 4).
After construction of suitable GE vector, it is transformed into plant genome to carry out the intended mutation. Unavailability of reliable transformation methods is a bottleneck for application of GE in plants as compared to animal transformation. Till now, Agrobacterium-mediated transformation and particle bombardment are the most successful transformation methods in plants. However, these methods remain inefficient for many crops due to some of the limitations such as: (1) longer tissue culture periods obligatory to achieve transgenic plants from transformed cells and tissues, (2) low frequency of stably transformed events, (3) small DNA insert delivered by Agrobacterium-mediated gene transfer and (4) low precision of bombardment-mediated gene transfer. Moreover, transformation by particle bombardment has been standardized for certain staple food crops like wheat and maize and is expected to work efficiently for other crops which are difficult to regenerate from protoplast culture. The transformed plants are screened for desired mutations using several methods like restriction-based assays, viz., cel1 endonuclease, PCR-based assays, whole-genome re-sequencing, surveyor assays (Stoddard 2011) and target gene sequencing. Pre-engineered and customized ZFNs, TALENs, CRISPR/Cas are also available from commercial suppliers such as Cellectis (http://www.cellectis.com), Sigma-Aldrich (https://www.sigmaaldrich.com/), Bioresearch and Life Technologies (https://www.biosearchtech.com), etc., as per the specifications of the experiment. For more information readers are directed to refer latest reviews on practical considerations of efficient CRISPR/Cas experiment (Liang et al. 2016).
Strategies to overcome the challenges of off-site targeting
Binding and cleavage at nonspecific loci leads to unintended editing in genome called off-site effect and needs to be addressed for efficient utilization of SDNs. Sometimes, off-site targeting may result in cell toxicity. ZFNs recognize smaller sequence as compared to TALENs and as a result, ZFNs shows more off-site targeting. There is evidence that 4–6 zinc finger domains for each ZFN half-enzyme significantly enhance activity and specificity of ZFN (Sood et al. 2013). Another important consideration is the length of spacer sequence, i.e., sequence separating two target sites. Specificity of ZFNs decreases as target spacer sequence increases more than 7 bp and more length of spacer sequence hinders dimerization of ZFN leading to off-site cleavage (Pattanayak et al. 2014). Zinc finger nickases (ZFNickases) have been developed to increase the specificity of ZFNs by inducing nick in to a single strand of DNA which stimulates HR without activating the error-prone NHEJ repair pathway (Ramirez et al. 2012). Similarly, optimizing spacer length is important in designing TALENs wherein longer binding site ensures increased specificity (Pattanayak et al. 2014). Although CRISPR/Cas system is effective in introducing mutations, it is more subjected to off-target effects than the other tools. To overcome the potential off-target effects of CRISPR/Cas system, Ran et al. (2013) developed a proof of concept in human cells by introducing a truncated version of Cas9 which induced nicks (SSBs) in the target sequence. Modified with less than 20 nucleotide target sequences, the gRNAs truncated at 5′ end of their complementary sequences have been reported to decrease off-target effects in animals; use of these truncated gRNAs is an alternate approach to minimize off-site targeting (Fu et al. 2014; Pattanayak et al. 2014).
To predict specific gRNA spacers which are expected to have little or no off-target risk in RNA guided GE, Xie et al. (2014) developed CRISPR-PLANT database by assembling the genomic sequences of plants, viz., Arabidopsis, soybean, Medicago, tomato, Brachypodium, rice, Sorghum, and maize to access genome-wide predictions of specific gRNAs. Recently, Liang et al. (2017) and Svitashev et al. (2016) have demonstrated the efficiency of ribonucleoprotein-mediated GE in wheat and maize, wherein the off-target mutations were much lower than CRISPR/Cas9 DNA. In addition to that, efficiency of mutation was also higher in ribonucleoprotein-mediated GE.
Latest technological developments in CRISPR/Cas system
Advances have been made in native CRISPR/Cas system to improve its applications in plants. A new facet is being added regularly in the CRISPR technique. In this section technological development pertaining to CRISPR is discussed.
Vector construction There are two types of vector systems available for CRISPR namely binary vectors and single vector system. Binary vector system is an old one and has the advantage of rapid initial testing of CRISPR/Cas system. A vector containing different gRNA can be used for transformation of the plant which is already expressing the Cas9 protein. One more advantage associated with binary vector system is inclusion of different combinations of Cas proteins for specific gRNA, which gives more flexibility in designing the experiment with more targeted efficiency. A single vector containing Cas protein as well as gRNA is becoming more popular among researchers. In most of the single vector system, RNA polymerase II-based promoters such as CaMV35S, ubiquitin are used for expression of Cas9 gene whereas, RNA polymerase III-based promoters such as U6, U3 are used for expression of gRNA. Such type of vector system exploits mixed dual promoters. Dual polymerase II promoter-based vectors use two different RNA polymerase II-based promoters to drive expression of Cas gene and gRNA whereas, single polymerase II promoter-based vectors uses only one RNA polymerase II-based promoters to drive expression of both Cas and gRNA. All these modifications help to reduce vector size which ultimately results in increased transformation efficiency (Lowder et al. 2015).
Delivery methods Transformation methods such as floral dip method, Agrobacterium-mediated in planta inoculation and regeneration of explants, particle bombardments, virus-mediated delivery, plasmid delivery, ribonucleotide protein complex delivery, RNA delivery, etc., are some of the recent advances in transformation methods for CRISPR in plants. Selection of viral delivery is limited, due to low editing efficiency in germline cells. Nevertheless, many reports are available for the virus-mediated delivery of CRISPR components in plants (Ali et al. 2015). Transformation of either Cas9 protein and gRNA complex (ribonucleotide protein) or Cas9 and gRNA alone through gene gun and PEG-mediated methods also reported the success (Wolter and Puchta 2017), but regeneration of protoplast still remains a major challenge associated with these types of methods.
Cpf1-an alternative to Cas9
The cpf1 (also known as Cas12a) was identified by Schunder and colleagues (2013), in Francisella spp. Subsequently, Makarova et al. (2015), proposed a new classification for CRISPR/Cas systems, i.e., type V which is characterized by the Cpf1 ‘signature’ protein. Zetsche et al. (2015), cloned CPf1 from the Francisella novicida (FnCpf1) and tested its function in Escherichia coli. The PAM requirement for FnCpf1 is TTN and CTA (Zetsche et al. 2015). This system is also found in some of the bacteria like Primotella, Acidaminococcus, Francisella, Lachnospiraceae, etc.
The Cpf1 has some of the distinct characteristics as compared to Cas9 as it requires only a crRNA and does not utilize tracrRNA. The Cpf1 crRNAs are significantly shorter than ~ 100-nucleotide engineered sgRNAs required by Cas9, thereby offering cheaper and simpler guide RNA production. Small protein size and small gRNA (nearly half of the gRNA of Cas9) facilitate easy and efficient delivery of ribonucleoproteins to plant cell. Furthermore, the different sgRNA and crRNA requirements of Cas9 and Cpf1 allow both systems to be combined when multiplexing of different targets is desired. Although both Cas9 and Cpf1 make DSBs, Cas9 uses its RuvC- and HNH-like domains to make blunt-ended cuts within the seed sequence, whereas Cpf1 uses a RuvC-like domain to produce staggered cuts outside the seed sequence. Cpf1 produce staggered cuts which helps for directional cloning of the gene of interest. Different PAM sequence of Cpf1 gives increased possibilities to a target gene of interest at more than one place. One more advantage associated with the use of Cpf1 is that it cuts distal to PAM sequence and the targeted sequence may be susceptible for repeated cleavage. The publications on applications of Cpf1 in crop plants is on rise (Begemann et al. 2017; Kim et al. 2018; Liu and Wang 2017; Tang et al. 2017; Zaidi et al. 2017).
Screening assays Apart from the methods described in the section, companies like Applied Biological Materials (abm) (https://www.abmgood.com), TaKaRa (http://www.clontech.com), Thermo Fisher (https://www.thermofisher.com) are providing ready-to-use kits to check the efficiency of GE as well as monoallelic and biallelic mutations at target sites.
Various genetic modifications through genome editing in plants
Intervention in crop improvement using GE is increasingly becoming popular, widening its horizon from model plants to economically important cereals, legumes, fruits and vegetable crops. The successful applications of GE are described below under four broad sections.
Targeted mutagenesis
Deliberate changes, viz., addition/deletion/substitution intended at a specific site in the genome are termed as targeted mutagenesis. Zhang et al. (2010) reported an efficient method for targeted mutagenesis of Arabidopsis ADH1 and TT4 genes through regulated expression of ZFNs. In the same year, Osakabe et al. (2010) engineered ZFNs to target gene inactivation of a stress-response regulator gene, ABA-insensitive 4 (ABI4) from Arabidopsis. Curtin et al. (2011) used CoDA method to engineer ZFNs and targeted mutagenesis of soybean Dicer-like (DCL) genes, RNA-dependent RNA polymerase (RDR), and HUA ENHANCER1 (HEN1) family members involved in RNA silencing. To achieve herbicide resistance, Pater et al. 2013, targeted Arabidopsis polyphenol oxidase gene (PPO) involved in heme and chlorophyll synthesis by designing specific ZFNs to create DSB in PPO and achieved butafenacil insensitive PPO. Christian et al. (2013) demonstrated that genome modification using TALENs can be efficiently transmitted to next generation by engineering TALENs which targeted ADH1, MAPKKK1, TT4, NATA2 and DSK2B genes of Arabidopsis. Zhu’s lab fused modified dHax3 DBD domain sequence to C terminal of a FokI cleavage domain and developed a hybrid TALEN (dHax3N) (Mahfouz et al. 2011). In vivo transient expression in tobacco leaves showed desired break using modified Hax3N in its artificial target sequence. Recently, four yield-related genes of rice, viz., Gn1a, DEP1, GS3, and IPA1 were targeted using CRISPR/Cas9 for improving yield traits (Li et al. 2016).
Development of bacterial leaf blight (BLB)-resistant rice by high-efficiency TALE-based gene editing has been a landmark GE application in plants (Li et al. 2012). TALEN was used to edit a specific susceptibility gene, the sucrose-efflux transporter gene (Os11N3 aka OsSWEET14), by disrupting the effector-binding element in its promoter region without changing its expression. The resulting mutant lines displayed resistance to AvrXa7 and PthXo3 with morphologically normal phenotype. Recently, Blanvillain-Baufumé et al. (2017) used TALENs to target promoter of OsSWEET14 to achieve resistance against bacterial blight. Shan et al. (2015) reported the mutation in the OsBADH2 gene using specially designed TALENs resulting in increased aroma. Wang et al. (2016) used CRISPR/Cas system to achieve targeted mutations in OsERF922, ethylene responsive factor (ERF) which act as negative regulator of blast disease in rice (Liu et al. 2012) generating blast resistant rice lines.
Large-scale highly efficient targeted gene knockouts using TALENs were reported in rice (OsDEP1, OsBADH2, OsCKX2, and OsSD1) and Brachypodium (BdABA1, BdCKX2, BdSMC6, BdSPL, BdSBP, BdCOI1, BdRHT and BdHTA1) with mutation rates reaching > 30% (Shan et al. 2013a). Liang et al. (2014), compared the mutagenesis efficiency using TALENs as well as CRISPR/Cas system in maize by targeting genes, viz., ZmIPK1A, ZmPDS, ZmMRP4 and ZmIPK and obtained 23.1% efficiency in protoplasts and up to 13.3–39.1% efficiency in somatic mutations. Anderson et al. (2014), delineated that ZFNs can be effectively used for creating mutations in somatic as well as germline cells in polyploidy genome like soybean. High oleic soybean varieties were developed by targeted mutagenesis in fatty acid desaturase genes, FAD2-1A and FAD2-1B gene using TALENs (Haun et al. 2014). Lor et al. (2014) reported the successful use of TALENs in tomato for targeted mutagenesis of PROCERA (PRO), which is a negative regulator of gibberellin signaling and successfully created new PRO allele. Wendt et al. (2013), reported the assembly of several TALENs for editing an anti-nutritional factor gene, phytase, HvPAPhy in barley and were able reduced its content in grain/seed. Another interesting study by Clasen et al. (2016) demonstrated the use of TALENs to knockout the vacuolar invertase gene (VInv) in potato, making its quality desirable for processing.
The CRISPR/Cas system has been used for targeted mutagenesis and was exploited in Arabidopsis, tobacco, rice and wheat. Zhang et al. (2014) tested 11 target genes in two rice sub-species for their amenability to CRISPR/Cas9-induced editing and determined the patterns, specificity and heritability of the gene modifications. Another study by Fauser et al. (2014), showed that only the nuclease, but not the nickase is an efficient tool for NHEJ-mediated mutagenesis in plants while targeting ADH1 and TT4 genes of Arabidopsis. Highly efficient site-specific modifications in rice and wheat using codon-optimized Cas9 of Streptococcus pyogenes and tailored gRNAs were reported by Shan et al. (2013b). Li et al. (2013), successfully achieved mutagenesis in the phytoene desaturase gene of Arabidopsis and tobacco using CRISPR/Cas9 with 2.7–4.8% frequency. A CRISPR/Cas construct containing two different cassettes of gRNA was successfully used to achieve mutations in two genes viz. LAZY1 and CHLOROPHYLL A OXYGENASE1 in Arabidopsis (Mao et al. 2013). Interestingly, CRISPR/Cas9 has been using in polyploid crop like wheat wherein only one homeoallele of MLO-A1gene was mutated to achieve resistance to powdery mildew (Wang et al. 2014). Jia and Wang (2014), for the first time reported the editing phytoene desaturase gene (CsPDS) in sweet orange and achieved mutation rate of approximately 3.2–3.9%. Zhang et al. (2016), demonstrated that transient expression of CRISPR/Cas9 DNA in wheat callus cells efficiently induced targeted and transgene-free mutants. Homozygous mutants with no detectable transgenes for TaGASR7, TaGW2 and TaLOX2 in hexaploid bread wheat and TdGASR7 in tetraploid durum wheat were generated. Osakabe et al. (2016) used truncated gRNA (tru-gRNA)/Cas9 combination for GE in Arabidopsis to generate new alleles for OST2 gene, a proton pump, with no off-target effects and high average mutation rates (up to 32.8%). The new mutant alleles for OST2 exhibited altered stomata closing in response to environmental conditions which is a highly desirable trait for abiotic stress tolerance. Woo et al. (2015), demonstrated the potential of CRISPR/Cas in targeted mutagenesis by protoplasts transformation in Arabidopsis thaliana, tobacco, lettuce and rice and achieved efficiency up to 46% with small insertions or deletions indistinguishable from naturally occurring genetic variation. Svitashev et al. (2016), also reported targeted mutagenesis in maize using the CRISPR/Cas9 ribonucleoprotein complexes. Ueta et al. (2017) optimized the CRISPR/Cas9 system to introduce somatic mutations selectively into a gene controlling parthenocarpy, SlIAA9 and achieved 100% mutation rates in the T0 generation. Kim et al. (2018), targeted wheat dehydration responsive element binding protein 2 (TaDREB2) and wheat ethylene responsive factor 3 (TaERF3) using CRISPR.
Targeted gene insertion/replacement
Gene replacement differs with targeted gene insertion, in which particular endogenous gene(s) of an organism is replaced with a new version of the same gene. Shukla et al. (2009) selectively targeted IPK1 gene of maize (ZmIPK1) using specifically designed ZFNs to alter phytate biosynthesis in maize seeds. Further, this optimized method was used for site-specific insertion of a gene conferring tolerance to the herbicide bialaphos, phosphinothricin acetyl transferase (PAT). Zhang et al. (2013a, b) developed targeted insertion of herbicide resistance acetolactate synthase gene (ALS) gene in tobacco using TALENs. In another such study by Townsend et al. (2009), successful gene replacement for SurA and SurB genes in tobacco was achieved using ZFN to achieve resistance to imidazolione and sulphonyl urea with the maximum gene replacement frequency of 4%. Ainley et al. (2013), reported sequential stacking of two herbicide resistance genes ‘pat’ and ‘aad1’ using ZFNs with modular trait landing pads into the maize genome and demonstrated co-segregation of traits in subsequent generations. Recently, van de Wiel et al. (2017) discussed various aspects of GE in the presence of oligonucleotide to assist the repair of the DSB.
Targeted gene excision
The intended gene can be deleted or excised from the genomic region using GE. Petolino et al. (2010) demonstrated the excision of GUS by transforming transgene flanked by ZFN cleavage sites and crossing the transformants with the plant expressing a corresponding ZFN gene. Evidence for complete deletion of a 4.3 kb sequence comprising the GUS gene was shown. Antunes et al. (2012) demonstrated the excision of DNA segment from Arabidopsis genome using synthetic homing endonuclease, viz., PB1 which excised unwanted transgenic DNA from the genome. Such capacity of removal of undesired DNA segment may play a very important role in the development of marker free-transgenic plants (Curtin et al. 2012). This will be very much useful to excise antibiotic markers from already developed transgenic events which face biosafety hurdles due to the presence of antibiotic or any other marker genes. It can also be extended to delete highly repetitive DNA to generate new loci or to eliminate undesired loci.
Targeted structural changes of genome
Targeted structural changes include large-scale addition, deletion, inversion, duplication or translocation of DNA at intended site in the selected genome. There are few examples which exploit SDNs for large targeted structural changes in plant species. Zhou et al. (2014) reported large chromosomal segment deletions through Cas9/sgRNA which were inherited in multiple generations in rice. The promoters of rice susceptibility genes, viz., SWEET11 and SWEET14 were edited at specific sites using Cas9/sgRNA resulting into heritable large chromosomal deletions (> 100 kb).
Potential target genes for GE for crop improvement
Though various economically important genes have been modified by GE, there is huge scope to exploit these tools to modify many useful genes in important food crops such as rice, wheat, maize, legumes, etc. Well-characterized genes present in single copy and governing qualitative traits can be the ideal choice for GE. Nevertheless, there are many examples where genes involved in quantitative traits are also targeted. Here, we have provided a list of some well-characterized genes from important crop plants which are highly potential targets for GE (Table 5). Practical considerations mentioned in section “Practical considerations” can be applied for both structural as well as regulatory genes and to target noncoding RNAs such as microRNAs (miRNAs). In case of miRNA, it is essential to understand the role of particular miRNA in the regulation of stress or development. The miRNAs being master regulators of biological pathways, need to be carefully picked for genome editing. It is important to understand the networking of miRNAs in the regulation of important biological processes. In a recent review, Mangrauthia et al. (2017a), have described the possibilities and challenges of miRNA editing for crop improvement.
Editing of quantitative trait nucleotide (QTN)
Quantitative trait locus (QTL) mapping is frequently used to identify genomic regions associated with a complex phenotypic trait of interest. Most of the QTL studies are unable to decipher how multiple genetic factors influence a particular phenotype. The QTLs can act through combined action of multiple sites within a gene or across multiple genes acting in the same gene set. Quantitative trait nucleotide (QTN) is a set of SNPs (single nucleotide polymorphism) in a gene or multiple genes of the same set that act together for expression of a particular trait. Till date, many QTLs have been identified in various crop plants governing agronomically important traits for tolerance/resistance to drought, salinity, cold, heat, disease and insect, etc., but, only a few QTLs are exploited for crop improvement. QTN can facilitate the use of QTLs in crop improvement more effectively (Lee et al. 2014). Identification of QTNs has become easier since the advent of several genome sequence information as well as rapid developments in SNP genotyping techniques including whole-genome association studies, but, the functional validation of these QTNs remains the biggest challenge for the successful application of QTNs in crop improvement. GE would be highly useful for functional analysis of QTN since it allows precise genome modifications at intended multiple genomic regions with ease and cost-effective way.
Introgression of the agronomically important QTLs from wild species or landraces into elite cultivars is one of the widely used practices by plant breeders. Functional analysis of QTN can facilitate the introduction of these QTLs in popular elite cultivars without affecting their original characteristics. GE can help in the creation of the desired allele at an intended locus which can replace marker-assisted introgression of genes and QTLs. This will not only solve the problem of linkage drag, but also would be less laborious and time-consuming. Novel alleles of important genes can also be created and examined for their phenotypic effects which can serve as new source of variation for plant breeding.
With respect to efficient use of QTNs in plant breeding, promotion of alleles by GE (PAGE) is gaining importance in recent times. Variants controlling the quantitative traits should be known to improve the trait using GE. Generally, as compared to small-effect QTNs, large-effect QTNs are easier to detect which is the reason why GE has concentrated more on large-effect QTNs. A large number of QTNs need to be edited for understanding the complex QTLs. Recently, Jenko et al. (2015) has put forth the idea for use of PAGE in association with genomic selection (GS) in livestock breeding program. The potential of PAGE for improvement of quantitative traits has been checked by different strategies. It has been established that GS complemented by PAGE is more useful for selection due to the ability to increase favorable alleles at intended QTNs. Such studies are very much needed in plants to facilitate plant breeding for accurate improvement of quantitative traits.
Potential applications of genome editing for understanding plant genome
Targeted transcriptional regulation
Regulation of gene expression includes a wide range of mechanisms that are used by cells to increase/decrease/maintain level of specific gene products (protein or RNA). Plants are confronted continuously by challenges of surrounding environment and respond accordingly. The transcription factors, activators, enhancers and suppressors are key players in regulating genes at the transcriptional level. The nucleases can be potentially used to target transcriptional regulation of endogenous genes. By engineering the regulatory element binding sites by SDNs, the regulation of endogenous gene can be altered (Fig. 3a). In another approach, binding of SDNs to particular sequences blocks regulatory element to bind to those sequences which in turn attenuate its further regulatory function (Fig. 3b). The role of SDNs in targeted transcriptional regulation is quite new and needs to be explored further. Recent research in animal cells has shown SDNs in the targeted reprogramming of endogenous genes (Dominguez et al. 2015; Braun et al. 2016). Such studies have potential outcomes in plants as well. Reviews by Katayama et al. (2016) and Piatek and Mahfouz (2017) provide an update on programmable transcriptional regulation.
Overview of various CRISPR/Cas 9-based applications. a Cas9 nucleases fused with activation domain can be used for transcriptional activation of targeted gene. b Cas9 nucleases fused with suppression domain can be used for transcriptional suppression of targeted gene. c Cas9 nucleases fused with chromatin modification enzyme DNMT (DNA methyl transferase) domain can be used for epigenetic modifications of DNA or histone. d Cas9 nucleases fused with GFP (green fluorescent protein) can be used to enable imaging of specific genomic locus
One of the interesting targets in plants is the noncoding miRNAs involved in gene regulation of various metabolic activities (Mangrauthia et al. 2017b). The expression of miRNAs can be altered suitably not only to understand their precise function, but also to engineer the trait under regulation. Fine tuning the expression of transcription factors and miRNAs would be highly useful to improve complex or quantitative traits such as drought stress, salt stress, and temperature stress tolerance. This has immense significance in developing resilient plants for combating the challenges of climate change. In future, there lies an expectant scope for plant scientists to exploit SDNs for functional genomic studies and crop improvement through modulation of key transcription factors or regulatory molecules such as miRNAs (Sailaja et al. 2014; Mangrauthia et al. 2017a). Thus, the known microRNA can be an ideal target for regulating the gene expression. Similarly, the roles of unknown miRNA can also be exploited using GE for the desirable trait of interest (Zhou et al. 2017).
Epigenome modifications
The biological code of life lies in the genome as well as in epigenome which has a profound effect on expression and regulation. The role of epigenome has been very well documented in case of stress tolerance, yield, heterosis, etc., in plants (Springer 2013). Engineering the regulators of DNA, histone methylation and other epigenetic modifications in plants gives new directions towards achieving desired phenotypes (Fig. 3c). Chromatin modification regulators such as HAT (histone acetyl transferase), DNMTs (DNA methyl transferase), HDAC (histone deacetyl transferase), IDM1 (increased DNA methylation 1), ROS1 (repressor of silencing 1), KYP3 (Kryptonite 3), DNA methyltransferases (MET1, CMT3, and DRM2), IBM1 (increase in bonsai methylation 1), MBD7 (methylation binding domain 7), ASI1 (Ant-silencing 1), etc., can be recruited at specific site of DNA for altering gene expression. As some epigenetic marks can be transmitted to offspring, epigenetic mechanisms may provide plasticity for the dynamic control of various agronomically important traits such as drought, heat, salinity, cold tolerance, etc., without the generation of genomic lesions. Thus, GE can be employed to create a novel source of epigenetic variation for trait improvement. Efforts have been made for epigenetic modifications in animal cells by targeting enzymes involved in histone modification or chromatin remodeling (Zentner and Henikoff 2015). Vora et al. (2016) have reviewed various aspects of epigenome modifications with CRISPR/Cas9 system.
Understanding plant–pathogen interactions
Generally, in compatible plant–pathogen interactions, the pathogen invades plant tissues and diverts its nutrition for its growth by recognizing specific binding sites of nutrient transporters. The pathogen can also inactivate or alter the resistance-associated genes to make plant susceptible, for example, Arabidopsis MPK3 and MPK6 (mitogen-activated protein kinases) genes are inactivated by the HopAI1 effectors of Pseudomonas syringae, resulting in suppression of pathogen-associated molecular pattern-triggered immunity (PTI). The recognition sequences of such plant genes can be altered using SDNs without losing their primary function. Another approach involves knocking out of host susceptibility genes whose products are utilized by the pathogen for its own survival and growth and for negative regulation of plant defense responses. Many susceptible genes like Mlo in barley and Arabidopsis (negative regulator of PEN gene-associated disease resistance to powdery mildews) (Humphry et al. 2006; Piffanelli et al. 2004), tomato recessive allele SlMlo2 and SlMlo1 (Pavan et al. 2008), Arabidopsis LOV1 gene (Sweat et al. 2008) and rice HDT701 gene (Ding et al. 2010) can be targeted for achieving biotic stress resistance. Modifications of such genes using suitable engineered nuclease have brighter prospects for developing next-generation disease/insect-resistant crops (Liu et al. 2017).
GE can be used very effectively to understand the functional genomics of plant pathogenic fungi. Such efforts are being made to annotate the function of fungal elicitors and studying the plant–fungus interaction (Selin et al. 2016). Information generated from such study will be a valuable resource for plant resistance breeding. In a recent review, Barakate and Stephens, (2016) have discussed in depth the potential role of CRISPR/Cas9 system in understanding plant–pathogen interaction.
Functional genomics
Reverse genetics plays an important role in deciphering the function of a gene(s) by analyzing their phenotypic effects (Gilchrist and Haughn 2010). Several techniques of functional genomics like RNA interference (RNAi), Target Induced Local Lesions In Genome (TILLING) and Virus-Induced Gene Silencing (VIGS) have been extensively used in crop plants for deciphering the function of genes. A large number of genes need to be functionally characterized for improving agronomically important traits in various plants. Due to the advent of high-throughput sequencing technologies like next-generation whole-genome sequencing, transcriptome sequencing (RNA-Seq), metagenomics etc., a large amount of nucleic acid data of several plants and microbial species is available in public databases. In spite of the voluminous data, there has been very slow progress in determining the functions of genes (Zimin et al. 2014). Moreover, lack of functional annotation has been a major bottleneck in utilizing the information on genomics in crop improvement through breeding or transgenic approaches. GE has an advantage in analyzing gene function as it mutates either one gene/allele or multiple genes depending on the study. Tools used for GE can target more than one gene at a time which is highly useful in the functional analysis of quantitative traits governed by several genes as against other techniques targeting only one or a couple of genes at a time (Jao et al. 2013). Furthermore, it will also allow understanding the gene/protein interactions and the network of genes involved in several interconnected biological pathways as well as that of repetitive elements. It is also possible to modify a particular domain/motif or individual amino acid of the protein without changing its structure or confirmation which is highly desirable in functional analysis of genes.
Precision plant breeding
Traditional breeding exploits the existing diversity in the gene pool. Lack of required diversity for traits is a bottleneck in present-day plant breeding. GE is one of the effective tools to create diversity in given plant species/germplasm which can be ultimately exploited for breeding (Rani et al. 2016). The genome edited plant can be used as a donor parent in conventional plant breeding to improve the desired trait. This is the fastest way to improve local varieties for desired traits. Cross-incompatibility and hybrid sterility are some of the major limitations of traditional plant breeding which affects exploitation of available allelic and genetic diversity. Knock out of genes involved in cross compatibility and hybrid sterility by GE can help to overcome these problems. Targeting genes involved in cell division and replication can be used to obtain true-to-type plants through apomixes in crop species where maintenance of purity is difficult, e.g., cross-pollinating plants. Development of haploid plant also can be achieved with the help GE by targeting genes involved in spindle fiber formation and cell division. Development of male sterile plants by targeting genes involved in maintaining pollen fertility is one of the interesting and emerging areas to hasten hybrid development (Chang et al. 2016; Zhou et al. 2016). These applications hold a golden promise for future generation crops.
Genome editing and regulatory considerations
The regulatory considerations in case of GE differ from that for genetically modified crops. Regulations can be drafted in a case dependent manner to handle genome edited crop plants. The changes brought through GE are very similar to natural mutations and may be put outside the purview of biosafety regulations. Worldwide reforms are made for regulations governing GM crops to accommodate the latest advances. Many researchers have opinion that GE is likely to be less controversial than GM because of its precision. Additionally, such plants are free from any selectable marker. Although this technology offers a better alternative to GM that could be much more acceptable to consumers, there are issues to be solved. As GE is liable to off-site targeting, its probable side-effects need to be analyzed. Some off-targets can cause unknown nucleotide changes associated with unknown phenotypes. It is imperative to create awareness regarding the genome edited plants, better known to be NBT plants, among the consumers for facilitating its rational discourse in markets. Regulatory agencies need to consider how best to foster responsible use of GE without inhibiting research and development. Further, licensing of technology and issues of intellectual property rights (IPRs) need to be taken care in case of commercial applications of GE in crop plants. For more details regarding regulations, readers are directed to read review by Wolt et al. (2016b).
Challenges
The improvements in GE and their applications are ever expanding in the field of plant science. Although success has been witnessed in the specificity and ease of genome engineering in the last few years, challenges still exist. To achieve the full potentiality of this technology, major challenges must be addressed. Probability of getting DSBs and its further repair at both locus of a targeted region is less in diploid plant species. In case of polyploid plants, the targeted locus is present in more than two copies, in such condition to get a homozygous plant for a modified locus is more difficult. To achieve homozygous plant for all targeted loci, one needs to screen a large number of plant population after GE experiment. There is a demand to develop facilities like high-throughput phenotyping to evaluate the phenotypes of genome edited lines.
To boost applications of GE in crop plants, there is need to improve recombination frequency in several folds. The limitations associated with polyploid plants can be overcomed by targeting a specific gene from the gene family by proper selection of target sequence (Anderson et al. 2014; Sánchez-León et al. 2017; Wang et al. 2017). Selection of unique gene sequence and avoidance of conserved sequence is the key to success for editing member of multigenes family.
Unavailability of highly efficient and sophisticated transformation methods for some important crops also poses a challenge. Nano-particle (gold or tungsten) delivery by gene gun, liposome-mediated transformation, macro-injection and WHISKR transformation methods should be validated for its efficiency. These methods can be used for better and effective transformation. There is need to develop well-standardized regeneration protocol for major food crops, vegetables, and horticultural crops. Another challenge pertains in context to toxicity to cells caused by off-target DSBs, which prevent widespread use of these nuclease targeting systems in plants. Some of the possible strategies to overcome off-site targeting of SDNs have already been discussed. A better understanding of homology-directed endogenous DNA repair mechanisms that follow nuclease-mediated DNA cleavage will help to increase specificity and accuracy of an experiment by reducing off-site targeting and also help to develop new tools of GE. High-throughput sequencing platforms facilitate de novo sequencing and re-sequencing of genomes of many plant species which can generate vast data of SNPs. Most of the available SNPs are not phenotypically characterized. Phenotypically well-characterized SNPs can be good candidates for GE for crop improvements. So, there is need to initiate HapMap projects in important food crops which will provide a phenotypic characterization of SNPs.
Availability of pooled CRISPR libraries is one of the latest advancements. Addgene (http://www.addgene.org) provides such CRISPR-pooled libraries for different organisms. Such pooled libraries consist of thousands of plasmids, each plasmid containing a gRNA for a different target gene. Target cell should be treated with the pooled library to create a mutant population of cells, which can be phenotyped for the desired trait. To date, such pooled CRISPR libraries are not available for plant species. Availability of such libraries at least for model plants will be definitely a valuable resource for functional genomics and ultimately for crop improvement. Besides the coding region, libraries designed for the noncoding genomic region DNA will help us for functional characterization.
Conclusions
In the last few years, there has been an enormous acceleration in the development of GE tools and its applications for trait improvement. CRISPR/Cas is the most flexible and versatile version with tremendous potential for crop improvement. Biosafety rules and regulations for GE crops may not be much stringent as for transgenics because of its accuracy, specificity, and efficiency in the mode of action. GE not only helps in improving crops by altering traits, but also has been playing a tremendous role in functional genomics. Till date, GE is employed for improvement of single trait, but there is a great scope for improving multiple traits in a way to develop designer crops. There is a need to put more concerted efforts to address concerns like minimizing the off-site targeting, increasing rate of recombination frequency and developing high-throughput protocols for commercial crops. There is a need to formulate an international consortium to develop genome edited lines of important crops (e.g., rice, maize, wheat) covering the whole genome. Huge data of SNPs can be used in defining the QTNs, and their use in crop improvement by GE. Overall, GE possesses the ability to boost next-generation plant breeding and plant genomic research.
References
Ainley WM, Sastry-Dent L, Welter ME et al (2013) Trait stacking via targeted genome editing. Plant Biotechnol J 11(9):1126–1134
Akagi A, Fukushima S, Okada K et al (2014) WRKY45-dependent priming of diterpenoid phytoalexin biosynthesis in rice and the role of cytokinin in triggering the reaction. Plant Mol Biol 86(1–2):171–183
Ali Z, Abulfaraj A, Idris A et al (2015) CRISPR/Cas9-mediated viral interference in plants. Genome Biol 16:238
Anderson JE, Kantar MB, Kono TY et al (2014) A roadmap for functional structural variants in the soybean genome. G3 4(7):1307–1318
Antunes MS, Smith JJ, Jantz D, Medford JI et al (2012) Targeted DNA excision in Arabidopsis by a re-engineered homing endonuclease. BMC Biotechnol 12(1):86
Barakate A, Stephens J (2016) An overview of CRISPR-based tools and their improvements: new opportunities in understanding plant–pathogen interactions for better crop protection. Front Plant Sci 7:765
Begemann MB, Gray BN, January E et al (2017) Precise insertion and guided editing of higher plant genomes using Cpf1 CRISPR nucleases. Sci Rep 7(1):11606
Belhaj K, Chaparro-Garcia A, Kamoun S et al (2013) Plant genome editing made easy: targeted mutagenesis in model and crop plants using the CRISPR/Cas system. Plant Methods 9:39
Bhaya D, Davison M, Barrangou R et al (2011) CRISPR-Cas systems in bacteria and archaea: versatile small RNAs for adaptive defence and regulation. Ann Rev Genet 45:273–297
Bibikova M, Beumer K, Trautman JK et al (2003) Enhancing gene targeting with designed zinc finger nucleases. Science 300:764–764
Blanvillain-Baufumé S, Reschke M, Solé M et al (2017) Targeted promoter editing for rice resistance to Xanthomonas oryzae pv. oryzae reveals differential activities for SWEET14-inducing TAL effectors. Plant Biotechnol J 15(3):306–317
Braun CJ, Bruno PM, Horlbeck MA et al (2016) Versatile in vivo regulation of tumor phenotypes by dCas9-mediated transcriptional perturbation. Proc Natl Acad Sci 113(27):E3892–E3900
Butterbach P, Verlaan MG, Dullemans A et al (2014) Tomato yellow leaf curl virus resistance by Ty-1 involves increased cytosine methylation of viral genomes and is compromised by cucumber mosaic virus infection. PNAS 111(35):12942–12947
Chang Z, Chen Z, Wang N et al (2016) Construction of a male sterility system for hybrid rice breeding and seed production using a nuclear male sterility gene. PNAS 113(49):14145–14150
Chawla R, Shakya R, Rommens CM et al (2012) Tuber-specific silencing of asparagine synthetase-1 reduces the acrylamide-forming potential of potatoes grown in the field without affecting tuber shape and yield. Plant Biotechnol Commun 5:3352
Chen X, Lu X, Shu N, Wang S, Wang J, Wang D, Guo L, Ye W (2017) Targeted mutagenesis in cotton (Gossypium hirsutum L.) using the CRISPR/Cas9 system. Sci Rep 7:44304
Chou WL, Huang LF, Fang JC et al (2014) Divergence of the expression and subcellular localization of CCR4-associated factor 1 (CAF1) deadenylase proteins in Oryza sativa. Plant Mol Biol 85:443–458
Christian M, Cermak T, Doyle EL et al (2010) Targeting DNA double-strand breaks with TAL effector nucleases. Genet 186(2):757–761
Christian M, Qi Y, Zhang Y et al (2013) Targeted mutagenesis of Arabidopsis thaliana using engineered TAL effector nucleases. G3: Genes|. Genomes| Genet 3:1697–1705
Clasen BM, Stoddard TJ, Luo S et al (2016) Improving cold storage and processing traits in potato through targeted gene knockout. Plant Biotechnol J 14(1):169–176
Curtin SJ, Zhang F, Sander JD et al (2011) Targeted mutagenesis of duplicated genes in soybean with zinc-finger nucleases. Plant Physiol 156(2):466–473
Curtin SJ, Voytas DF, Stupar RM (2012) Genome engineering of crops with designer nucleases. Plant Genome 5(2):42–50
Dahlem TJ, Hoshijima K, Jurynec MJ et al (2012) Simple methods for generating and detecting locus-specific mutations induced with TALENs in the zebrafish genome. PLoS Genet 8(8):e1002861
Daspute A, Fakrudin B, Bhairappanavar SB et al (2014) Inheritance of pigeonpea sterility mosaic disease resistance in pigeonpea. Plant Pathol J 30(2):188
DeFrancesco L (2012) Erratum: Move over ZFNs. Nat Biotechnol 30(1):112–112
Deng D, Yan C, Pan X et al (2012) Structural basis for sequence-specific recognition of DNA by TAL effectors. Science 335(6069):720–723
Ding SL, Liu W, Iliuk A et al (2010) The tig1 histone deacetylase complex regulates infectious growth in the rice blast fungus Magnaporthe oryzae. Plant Cell 22(7):2495–2508
Dominguez AA, Lim WA, Qi LS (2015) Beyond editing: repurposing CRISPR-Cas9 for precision genome regulation and interrogation. Nat Rev Mol Cell Biol 17(1):5
Dong X, Wang X, Zhang L et al (2013) Identification and characterization of OsEBS, a gene involved in enhanced plant biomass and spikelet number in rice. Plant Biotechnol J 11(9):1044–1057
Dong Y, Yang X, Liu J et al (2014) Pod shattering resistance associated with domestication is mediated by a NAC gene in soybean. Nat commun 5:3352
Esvelt KM, Mali P, Braff JL et al (2013) Orthogonal Cas9 proteins for RNA-guided gene regulation and editing. Nat Methods 10:1116–1121
Fan Y, Yang J, Mathioni SM et al (2016) PMS1T, producing phased small-interfering RNAs, regulates photoperiod-sensitive male sterility in rice. Proc Natl Acad Sci USA 13:15144–15149
Fang Y, Xie K, Xiong L (2014) Conserved miR164-targeted NAC genes negatively regulate drought resistance in rice. J Exp Bot 65:2119–2135
Fauser F, Schiml S, Puchta H (2014) Both CRISPR/Cas-based nucleases and nickases can be used efficiently for genome engineering in Arabidopsis thaliana. The Plant J 79:348–359
Fondevilla S, Carver TLW, Moreno MT, Rubiales D (2006) Macroscopic and histological characterisation of genes er1 and er2 for powdery mildew resistance in pea. Eur J Plant Pathol 115(3):309–321
Fu Y, Foden JA, Khayter C et al (2013) High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat Biotechnol 31(9):822–826
Fu Y, Sander JD, Reyon D et al (2014) Improving CRISPR-Cas nuclease specificity using truncated guide RNAs. Nat Biotechnol 32:279–284
Gilchrist E, Haughn G (2010) Reverse genetics techniques: engineering loss and gain of gene function in plants. Brief Funct Genomics 9(2):103–110
Haun W, Coffman A, Clasen BM et al (2014) Improved soybean oil quality by targeted mutagenesis of the fatty acid desaturase 2 gene family. Plant Biotechnol J 12(7):934–940
Hou Z, Zhang Y, Propson NE et al (2013) Efficient genome engineering in human pluripotent stem cells using Cas9 from Neisseria meningitidis. PNAS USA 110:15644–15649
Hu B, Zhu C, Li F et al (2011) LEAF TIP NECROSIS1 plays a pivotal role in the regulation of multiple phosphate starvation responses in rice. Plant Physiol 156:1101–1115
Hu B, Wang W, Deng K et al (2015) microRNA399 is involved in multiple nutrient responses in rice. Front Plant Sci 6:188
Humphry M, Consonni C, Panstruga R (2006) MLO-based powdery mildew immunity: silver bullet or simply non-host resistance? Mol Plant Pathol 7(6):605–610
Hurni S, Scheuermann D, Krattinger SG et al (2015) The maize disease resistance gene Htn1 against northern corn leaf blight encodes a wall-associated receptor-like kinase. PNAS 112(28):8780–8785
Janila P, Pandey MK, Shasidhar Y et al (2016) Molecular breeding for introgression of fatty acid desaturase mutant alleles (ahFAD2A and ahFAD2B) enhances oil quality in high and low oil containing peanut genotypes. Plant Sci 242:203–213
Jao LE, Wente SR, Chen W (2013) Efficient multiplex biallelic zebrafish genome editing using a CRISPR nuclease system. PNAS 110:13904–13909
Jaskula-Ranga V, Zack DJ (2016) grID: a CRISPR-Cas9 guide RNA database and resource for genome-editing. bioRxiv 097352
Jenko J, Gorjanc G, Cleveland MA (2015) Potential of promotion of alleles by genome editing to improve quantitative traits in livestock breeding programs. Genet Select Evol 47(1):55
Ji H, Kim SR, Kim YH et al (2016) Map-based cloning and characterization of the BPH18 gene from wild rice conferring resistance to brown plant hopper (BPH) insect pest. Sci Rep 6:34376
Jia H, Wang N 2(014) Targeted genome editing of sweet orange using Cas9/sgRNA. PloS one 9:e93806
Jinek M, Chylinski K, Fonfara I et al (2012) A programmable dual-RNA—guided DNA endonuclease in adaptive bacterial immunity. Science 337:816–821
Jo KR, Visser RG, Jacobsen E et al (2015) Characterisation of the late blight resistance in potato differential MaR9 reveals a qualitative resistance gene, R9a, residing in a cluster of Tm-2 2 homologs on chromosome IX. Theor Applied Genet 128(5):931–941
Juillerat A, Dubois G, Valton J et al (2014) Comprehensive analysis of the specificity of transcription activator-like effector nucleases. Nucleic Acids Res 42(8):5390–5402
Katayama S, Moriguchi T, Ohtsu N et al (2016) A powerful CRISPR/Cas9 based method for targeted transcriptional activation. Angew Chem 128:6562–6566
Khatodia S, Bhatotia K, Passricha N et al (2016) The CRISPR/Cas genome-editing tool: application in improvement of crops. Front Plant Sci 7: 506
Kim YG, Cha J, Chandrasegaran S (1996) Hybrid restriction enzymes: zinc finger fusions to Fok I cleavage domain. PNAS 93:1156–1160
Kim H, Kim ST, Ryu J et al (2017) CRISPR/Cpf1-mediated DNA-free plant genome editing. Nat Commun 8:14406
Kim D, Alptekin B, Budak H (2018) CRISPR/Cas9 genome editing in wheat. Funct Integr Genom 18(1):31–41
Kleinstiver BP, Prew MS, Tsai SQ et al (2015) Engineered CRISPR-Cas9 nucleases with altered PAM specificities. Nature 523(7561):481
Kleinstiver BP, Pattanayak V, Prew MS et al (2016) High-fidelity CRISPR-Cas9 nucleases with no detectable genome-wide off-target effects. Nature 529(7587):490
Krattinger SG, Sucher J, Selter LL et al (2016) The wheat durable, multipathogen resistance gene Lr34 confers partial blast resistance in rice. Plant Biotechnol J 14(5):1261–1268
Lee YW, Gould BA, Stinchcombe JR (2014) Identifying the genes underlying quantitative traits: a rationale for the QTN programme. AoB Plants. https://doi.org/10.1093/aobpla/plu004
Li T, Liu B, Spalding MH et al (2012) High-efficiency TALEN-based gene editing produces disease-resistant rice. Nat Biotechnol 30:390–392
Li JF, Norville JE, Aach J et al (2013) Multiplex and homologous recombination-mediated genome editing in Arabidopsis and Nicotiana benthamiana using guide RNA and Cas9. Nat Biotechnol 31(8):688–691
Li M, Li X, Zhou Z et al (2016) Reassessment of the four yield-related genes Gn1a, DEP1, GS3, and IPA1 in rice using a CRISPR/Cas9 system. Front Plant Sci 7:377
Liang Z, Zhang K, Chen K et al (2014) Targeted mutagenesis in Zea mays using TALENs and the CRISPR/Cas system. J Genet Genom 41:63–68
Liang G, Zhang H, Lou D et al (2016) Selection of highly efficient sgRNAs for CRISPR/Cas9-based plant genome editing. Sci Rep 6:21451
Liang Z, Chen K, Li T et al (2017) Efficient DNA-free genome editing of bread wheat using CRISPR/Cas9 ribonucleoprotein complexes. Nat Commun 8:14261
Lioyd A, Plaisier CL, Carroll D et al (2005) Targeted mutagenesis using zinc-finger nucleases in Arabidopsis. PNAS USA 102:2232–2237
Liu XHCWQ, Wang YFK (2017) Targeted mutagenesis in rice using CRISPR-Cpf1 system. J Genet Genomics 44(1):71–73
Liu D, Chen X, Liu J et al (2012) The rice ERF transcription factor OsERF922 negatively regulates resistance to Magnaporthe oryzae and salt tolerance. J Exp Bot 63:3899–3911
Liu H, Soyars C, Li J et al (2017) CRISPR/Cas9-mediated resistance to cauliflower mosaic virus. Plant Direct 2(3):00047
Lor VS, Starker CG, Voytas DF, Weiss D, Olszewski NE (2014) Targeted mutagenesis of the tomato PROCERA gene using transcription activator-like effector nucleases. Plant Physiol 166(3):1288–1291
Lowder LG, Zhang D, Baltes NJ et al (2015) A CRISPR/Cas9 toolbox for multiplexed plant genome editing and transcriptional regulation. Plant Physiol 169(2):971–985
Lusser M, Parisi C, Plan D et al (2012) Deployment of new biotechnologies in plant breeding. Nat Biotechnol 30(3):231
Maeder ML, Thibodeau-Beganny S, Osiak A et al (2008) Rapid “open-source” engineering of customized zinc-finger nucleases for highly efficient gene modification. Mol cell 31(2):294–301
Mahfouz MM, Li L, Shamimuzzaman M et al (2011) De novo-engineered transcription activator-like effector (TALE) hybrid nuclease with novel DNA binding specificity creates double-strand breaks. PNAS 108:2623–2628
Makarova KS, Wolf YI, Alkhnbashi OS et al (2015) An updated evolutionary classification of CRISPR-Cas systems. Nat Rev Microbiol 13:722–736
Mali P, Aach J, Stranges PB et al (2013) CAS9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering. Nat Biotechnol 31:833–838
Mangrauthia SK, Maliha A, Prathi NB et al (2017a) MicroRNAs: potential target for genome editing in plants for traits improvement. Indian J Plant Physiol 22(4):530–548
Mangrauthia SK, Bhogireddy S, Agarwal S et al (2017b) Genome-wide changes in microRNA expression during short and prolonged heat stress and recovery in contrasting rice cultivars. J Exp Bot 68:2399–2412
Mao Y, Zhang H, Xu N et al (2013) Application of the CRISPR–Cas system for efficient genome engineering in plants. Mol Plant 6:2008
Marraffini LA, Sontheimer EJ (2008) CRISPR interference limits horizontal gene transfer in staphylococci by targeting DNA. Science 322:1843–1845
Mertz E, Bates L, Nelson OE et al (1964) Mutant gene that changes protein composition and increases lysine content of maize endosperm. Science 145:279–280
Mojica FJ, Diez-Villasenor C, Garcia-Martinez J et al (2009) Short motif sequences determines the targets of the prokaryotic CRISPR defense system. Microbiology 155:733–740
Nekrasov V, Staskawicz B, Weigel D et al (2013) Targeted mutagenesis in the model plant Nicotiana benthamiana using Cas9 RNA-guided endonuclease. Nat Biotechnol 31:691–693
Osakabe K, Osakabe Y, Toki S (2010) Site-directed mutagenesis in Arabidopsis using custom-designed zinc finger nucleases. PNAS 107:12034–12039
Osakabe Y, Watanabe T, Sugano SS et al (2016) Optimization of CRISPR/Cas9 genome editing to modify abiotic stress responses in plants. Sci Rep 6:26685
Padaria JC, Vishwakarma H, Biswas K et al (2014) Molecular cloning and in silico characterization of high temperature stress responsive pAPX gene isolated from heat tolerant Indian wheat cv. Raj 3765. BMC Rese Notes 7(1):713
Pardo B, Gómez-González B, Aguilera A (2009) DNA repair in mammalian cells. Cell Mol Life Sci 66:1039–1056
Paszkowski J, Baur M, Bogucki et al (1988) Gene targeting in plants. EMBO J 7:4021
Pater S, Pinas JE, Hooykaas PJ et al (2013) ZFN-mediated gene targeting of the Arabidopsis protoporphyrinogen oxidase gene through Agrobacterium-mediated floral dip transformation. Plant Biotechnol J 11:510–515
Pattanayak V, Lin S, Guilinger JP et al (2014) High-throughput profiling of off-target DNA cleavage reveals RNA-programmed Cas9 nuclease specificity. Nat Biotechnol 31:839–843
Pavan S, Zheng Z, Borisova M et al (2008) Map-vs. homology-based cloning for the recessive gene ol-2 conferring resistance to tomato powdery mildew. Euphytica 162(1):91–98
Petolino JF, Worden A, Curlee K et al (2010) Zinc finger nuclease-mediated transgene deletion. Plant Mol Biol 73(6):617–628
Piatek A, Mahfouz MM (2017) Targeted genome regulation via synthetic programmable transcriptional regulators. Crit Rev Biotechnol 37(4):429–440
Piffanelli P, Ramsay L, Waugh R et al (2004) A barley cultivation-associated polymorphism conveys resistance to powdery mildew. Nature 430(7002):887
Puchta H, Fauser F (2013) Gene targeting in plants: 25 years later. Int J Dev Biol 57:629–637
Ramirez CL, Certo MT, Mussolino C et al (2012) Engineered zinc finger nickases induce homology-directed repair with reduced mutagenic effects. Nucleic Acids Res 40(12):5560–5568
Ran FA, Hsu PD, Lin CY et al (2013) Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell 154:1380–1389
Ran FA, Cong L, Yan WX et al (2015) In vivo genome editing using Staphylococcus aureus Cas9. Nature 520(7546):186–191
Rani R, Yadav P, Barbadikar KM et al (2016) CRISPR/Cas9: a promising way to exploit genetic variation in plants. Biotechnol Lett 38(12):1991–2006
Sailaja B, Anjum N, Prasanth VV et al (2014) Comparative study of susceptible and tolerant genotype reveals efficient recovery and root system contributes to heat stress tolerance in rice. Plant Mol Biol Rep 32(6):1228–1240
Sakuma T, Ochiai H, Kaneko T et al (2013) Repeating pattern of non-RVD variations in DNA-binding modules enhances TALEN activity. Sci Rep 3:3379
Sánchez-León S, Gil-Humanes J, Ozuna CV et al (2017) Low-gluten, non-transgenic wheat engineered with CRISPR/Cas9. Plant Biotechnol J 16(4):902–910
Sarrion-Perdigones A, Vazquez-Vilar M, Palací J et al (2013) GoldenBraid 2.0: a comprehensive DNA assembly framework for plant synthetic biology. Plant Physiol 162(3):1618–1631
Schunder E, Rydzewski K, Grunow R et al (2013) First indication for a functional CRISPR/Cas system in Francisella tularensis. Int J Med Microbiol 303:51
Selin C, de Kievit TR, Belmonte MF et al (2016) Elucidating the role of effectors in plant–fungal interactions: progress and challenges. Front Microbiol 7:600
Shan Q, Wang Y, Chen K et al (2013a) Rapid and efficient gene modification in rice and Brachypodium using TALENs. Mol Plant 6(4):1365–1368
Shan Q, Wang Y, Li J et al (2013b) Targeted genome modification of crop plants using a CRISPR-Cas system. Nat Biotechnol 31(8):686–688
Shan Q, Zhang Y, Chen K et al (2015) Creation of fragrant rice by targeted knockout of the OsBADH2 gene using TALEN technology. Plant Biotechnol J 13:791–800
Shukla VK, Doyon Y, Miller JC et al (2009) Precise genome modification in the crop species Zea mays using zinc-finger nucleases. Nature 459(7245):437
Slaymaker IM, Gao L, Zetsche B et al (2016) Rationally engineered Cas9 nucleases with improved specificity. Science 351:84–88
Sood R, Carrington B, Bishop K et al (2013) Efficient methods for targeted mutagenesis in zebrafish using zinc-finger nucleases: data from targeting of nine genes using CompoZr or CoDA ZFNs. PloS one 8(2):e57239
Springer NM (2013) Epigenetics and crop improvement. Trends Genet 29(4):241–247
Stoddard BL (2011) Homing endonucleases: from microbial genetic invaders to reagents for targeted DNA modification. Structure 19:7–15
Sun Y, Jiao G, Liu Z et al (2017) Generation of high-amylose rice through CRISPR/Cas9-mediated targeted mutagenesis of starch branching enzymes. Front Plant Sci 8:298
Svitashev S, Schwartz C, Lenderts B et al (2016) Genome editing in Maize directed by CRISPR-Cas9 ribonucleoprotein complexes. Nat Commun 7:13274
Sweat TA, Lorang JM, Bakker EG et al (2008) Characterization of natural and induced variation in the LOV1 gene, a CC-NB-LRR gene conferring victorin sensitivity and disease susceptibility in Arabidopsis. Mol Plant-Microbe Interact 21:7–19
Tang X, Lowder LG, Zhang T et al (2017) A CRISPR–Cpf1 system for efficient genome editing and transcriptional repression in plants. Nat Plants 3(3):17018
Tian C, Zuo Z, Qiu JL (2015) Identification and characterization of ABA-responsive microRNAs in rice. J Genet Genom 42:393–402
Townsend JA, Wright DA, Winfrey RJ et al (2009) High-frequency modification of plant genes using engineered zinc-finger nucleases. Nature 459:442–445
Tyler AM, Bhandari DG, Poole M et al (2015) Gluten quality of bread wheat is associated with activity of RabD GTPases. Plant Biotechnol J 13(2):63–176
Ueta R, Abe C, Watanabe T, Sugano SS et al (2017) Rapid breeding of parthenocarpic tomato plants using CRISPR/Cas9. Sci Rep 7:507
van de Wiel CCM, Schaart JG, Lotz LAP et al (2017) New traits in crops produced by genome editing techniques based on deletions. Plant Biotechnol Rep 11(1):1–8
Varshney RK, Mir RR, Bhatia S et al (2014) Integrated physical. Genet J 10(8):913–924
Vinoth A, Ravindhran R (2017) Biofortification in millets: a sustainable approach for nutritional security. Front Plant Sci 8:29
Vora S, Tuttle M, Cheng J, Church G (2016) Next stop for the CRISPR revolution: RNA guided epigenetic regulators. FEBS J 283:3181–3193
Voytas DF (2013) Plant genome engineering with sequence-specific nucleases. Plant Biol 64:327
Wang Y, Cheng X, Shan Q et al (2014) Simultaneous editing of three homoeoalleles in hexaploid bread wheat confers heritable resistance to powdery mildew. Nat Biotechnol 32(9):947–951
Wang F, Wang C, Liu P et al (2016) Enhanced rice blast resistance by CRISPR/Cas9-targeted mutagenesis of the ERF transcription factor gene OsERF922. PloS one 11(4):0154027
Wang P, Zhang J, Sun L et al (2017) High efficient multi-sites genome editing in allotetraploid cotton (Gossypium hirsutum) using CRISPR/Cas9 system. Plant Biotechnol J 16(1):137–150
Wendt T, Holm PB, Starker CG et al (2013) TAL effector nucleases induce mutations at a pre-selected location in the genome of primary barley transformants. Plant Mol Biol 83(3):279–285
Wissuwa M, Yano M, Ae N (1998) Mapping of QTLs for phosphorus deficiency tolerance in rice (Oryza sativa L.). Theor Appl Genet 97:777–783
Wissuwa M, Wegner J, Ae N et al (2002) Substitution mapping of Pup1: a major QTL increasing phosphorus uptake of rice from a phosphorus deficient soil. Theor Appl Genet 105:890–897
Wolt JD, Wang K, Yang B (2016a) The regulatory status of genome-edited crops. Plant Biotechnol J 14:510–518
Wolt JD, Wang K, Sashital D et al (2016b) Achieving plant CRISPR targeting that limits off-target effects. Plant Genome 9(3):1–8
Wolter F, Puchta H (2017) Knocking out consumer concerns and regulator’s rules: efficient use of CRISPR/Cas ribonucleoprotein complexes for genome editing in cereals. Genome biol 18(1):43
Woo JW, Kim J, Kwon SI et al (2015) DNA-free genome editing in plants with preassembled CRISPR-Cas9 ribonucleoproteins. Nat Biotechnol 33(11):1162–1164
Xie K, Yang Y (2013) RNA-guided genome editing in plants using a CRISPR–Cas system. Mol Plant 6:1975–1983
Xie K, Zhang J, Yang Y (2014) Genome-wide prediction of highly specific guide RNA spacers for CRISPR–Cas9-mediated genome editing in model plants and major crops. Mol Plant 7(5):923–926
Xing HL, Dong L, Wang ZP et al (2014) A CRISPR/Cas9 toolkit for multiplex genome editing in plants. BMC Plant Biol 14(1):327
Zaidi SSEA, Mahfouz MM, Mansoor S (2017) CRISPR-Cpf1: A new tool for plant genome editing. Trends Plant Sci 22(7):550–553
Zentner GE, Henikoff S (2014) High-resolution digital profiling of the epigenome. Nat Rev Genet 15:814–827
Zetsche B, Gootenberg JS, Abudayyeh OO et al (2015) Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell 163(3):759–771
Zhang F, Maeder ML, Unger-Wallace E et al (2010) High frequency targeted mutagenesis in Arabidopsis thaliana using zinc finger nucleases. PNAS 107(26):12028–12033
Zhang Y, Zhang F, Li X et al (2013a) Transcription activator-like effector nucleases enable efficient plant genome engineering. Plant Physiol 161:20–27
Zhang YC, Yu Y, Wang CY et al (2013b) Overexpression of microRNA OsmiR397 improves rice yield by increasing grain size and promoting panicle branching. Nat Biotechnol 31:848–852
Zhang D, Wang Z, Wang N et al (2014) Tissue culture-induced heritable genomic variation in rice, and their phenotypic implications. PloS one 9(5):96879
Zhou H, Liu B, Weeks DP et al (2014) Large chromosomal deletions and heritable small genetic changes induced by CRISPR/Cas9 in rice. Nucleic Acids Res 42(17):10903–10914
Zhou H, He M, Li J et al (2016) Development of commercial thermo-sensitive genic male sterile rice accelerates hybrid rice breeding using the CRISPR/Cas9-mediated TMS5 editing system. Sci Rep 6:37395
Zhou J, Deng K, Cheng Y et al (2017) CRISPR-Cas9 Based Genome Editing Reveals New Insights into MicroRNA Function and Regulation in Rice. Front Plant Sci 8:1598
Zimin A, Stevens KA, Crepeau (2014) Sequencing and assembly of the 22-Gb loblolly pine genome. Genet 196(3):875–890
Zentner GE, Henikoff S (2015) Epigenome editing made easy. Nat Biotechnol 33(6):606–607
Zhang Y, Liang Z, Zong Y, Wang Y, Liu J, Chen K, Qiu J-L, Gao C (2016) Efficient and transgene-free genome editing in wheat through transient expression of CRISPR/Cas9 DNA or RNA. Nat Commun 7:12617
Acknowledgements
Authors are thankful to Director, ICAR-IIRR for encouragement and support.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
Authors declare that there is no conflict of interests.
Rights and permissions
About this article
Cite this article
Aglawe, S.B., Barbadikar, K.M., Mangrauthia, S.K. et al. New breeding technique “genome editing” for crop improvement: applications, potentials and challenges. 3 Biotech 8, 336 (2018). https://doi.org/10.1007/s13205-018-1355-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s13205-018-1355-3