Abstract
Eukaryotic initiation factor 2B (eIF2B) controls the first step of translation by catalyzing guanine nucleotide exchange on eukaryotic initiation factor 2 (eIF2). Mutations in the genes encoding eIF2B subunits inhibit the nucleotide exchange and eventually slow down the process of translation, causing vanishing white matter disease. We constructed a Saccharomyces cerevisiae genomic DNA library in YEp24 vector and screened it for the identification of extragenic suppressors of eIF2B mutations, corresponding to human eIF2B mutations. We found a suppressor-II (Sup-II) genomic clone, as suppressor of eIF2Bβ (gcd7-201) mutation. Identification of Sup-II reveals the presence of truncated SEC15, full-length TAN1 (tRNA acetyltransferase), full-length EMC4, full-length YGL230C (putative protein) and truncated SAP4 genes. Full-length TAN1 (tRNA acetyltransferase) gene, subcloned into pEG(KG) vector and overexpressed in gcd7-201 gcn2∆ strain, suppresses the slow-growth (Slg−) and general control derepression (Gcd−) phenotype of gcd7-201 gcn2∆ mutation, but YGL230C did not show any effect. A GST-Tan1p fusion protein of 60 kDa was detected by western blotting using α-GST antibodies. Interestingly, Tan1p overexpression also suppresses the temperature-sensitive (Ts−), Slg− and Gcd− phenotype of eIF2Bγ (gcd1-502) mutant. Role of Tan1p protein in eIF2B-mediated translation regulation was also studied. Results revealed that Tan1p overexpression confers resistance to GCD7 GCN2, gcd7-201 gcn2∆, GCD7 gcn2∆ growth defect under ethanol, H2O2 and caffeine stress. No resistance to DMSO-, NaCl- and DTT-mediated growth defect upon GCD7 gcn2∆, GCD7 GCN2, gcd7-201 gcn2∆ was observed by overexpression of TAN1. Hence, we proposed that Tan1p is involved directly or indirectly in regulating eIF2B-mediated translation.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Eukaryotic initiation factor 2B (eIF2B) is a decameric complex containing two copies each of α, β, γ, δ and ε subunits encoded by GCN3, GCD7, GCD1, GCD2 and GCD6, respectively, in yeast (Wortham and Proud 2015; Dever et al. 2016). The α, β and δ subunits of eIF2B constitute a regulatory sub-complex, while the γ and ε subunits form catalytic sub-complex, the latter playing a role in catalyzing the GDP–GTP exchange on its substrate eukaryotic initiation factor 2 (eIF2) (Pavitt et al. 1998). Global protein synthesis is reduced upon phosphorylation of eIF2α at serine 51 (S51) position under stress condition. eIF2B binds tightly to the phosphorylated eIF2 and slows down the process of translation (Krishnamoorthy et al. 2001; Pavitt et al. 1998; Rowlands et al. 1988). Saccharomyces cerevisiae has developed a well-defined mechanism to overcome stress by translation of general control nonderepressible (GCN4) mRNA, stimulated under stress conditions, resulting in transcription of numerous stress-related genes (Natarajan et al. 2001). In S. cerevisiae, Gcn2p is the only enzyme that phosphorylates eIF2 in response to nutrient starvation and sodium or rapamycin exposure (Shenton et al. 2006). Volatile anesthetics inhibit protein synthesis via eIF2B in both S. cerevisiae and higher cells (Palmer et al. 2005). Fusel alcohols also inhibit translation initiation via eIF2B (Ashe et al. 2001). A neurodegenerative disease named differently CACH (childhood ataxia with central nervous system hypomyelination), VWM (leukoencephalopathy with vanishing white matter) or eRDs (eIF2B related disorders) vanishing white matter disease (Fogli and Boespflug-Tanguy 2006; Pronk et al. 2006; Schiffmann and Elroy-Stein 2006) is a rare leukoencephalopathy caused by mutations in the genes encoding different subunits of eIF2B (Hannig et al. 1990; Dever et al. 1993; De Aldana and Hinnebusch 1994; Pavitt et al. 1997). Mutations in all the genes encoding five eIF2B subunits are reported to derepress GCN4 translation independent of eIF2 phosphorylation (Gcd− phenotype) (Fogli and Boespflug-Tanguy 2006; Leegwater et al. 2001; Richardson et al. 2004; van der Knaap et al. 2002). eIF2B guanine nucleotide exchange activity in VWM patients is generally lower than normal cells (Horzinski et al. 2009). In some cases, cataracts, ovaries, kidneys and pancreas are also affected in vanishing white matter disease (van der Knaap et al. 2003; Fogli et al. 2003), but glial cells are severely affected. Function of different subunits of eIF2B in translation regulation in humans is not well understood; therefore, there is the unavailability of proper treatment for such a rare leukoencephalopathy. Identifying target suppressor proteins of eIF2B mutations can be a better approach to understand the complexity of the disease and might be useful in curing VWM disease.
Mutations in eIF2B genes corresponding to VWM disease make S. cerevisiae sensitive to amino acid starvation (Gcd− phenotype) and show slow-growth phenotype (Slg−) and many time temperature-sensitive (Ts−) phenotype. eIF2B mutations impair the function of eukaryotic initiation factor 2B complex in many ways, such as by affecting the stability or integrity of the eIF2B complex, and altering eIF2–eIF2B interactions (Li et al. 2004; Richardson et al. 2004). However, some mutations do not affect eIF2–eIF2B interactions or GEF activity, but can still cause the VWM disease (Liu et al. 2011). In the present work, TAN1 was identified as the extragenic suppressor of gcd7-201 gcn2∆ and gcd1-502 gcn2-101 mutations of S. cerevisiae eIF2B subunits. Defect in protein folding is a major cause of many diseases, including cystic fibrosis and neurodegenerative disorders like Huntington’s, Alzheimer’s and prion diseases. eIF2B protein complex containing eIF2BβV341D (human eIF2BβV316D) mutation is unstable with reduced GEF activity, as a result of which eIF2BβV341D exhibits a reduced growth rate and a defect in global translation (Gcd−) in S. cerevisiae. Moreover, eIF2Bδ is excluded from eIF2BβV341D complexes (Richardson et al. 2004). The function of eIF2Bβv341D can be rescued by overexpression of eIF2Bδ alone (Richardson et al. 2004). In addition to GEF activity, eIF2B is also characterized by possessing GDI displacement factor (GDF) activity. GDF function of eIF2B depends upon the eIF2Bɛ and eIF2Bγ subunits (Jennings et al. 2013). gcd1-502 mutant has defective GCD1 subunit and has a GDF defect, conferring slow growth. Moreover, previous studies have reported that the slow-growth (Slg−) phenotype of eIF2Bγ mutants (G12V) and gcd1-502 (L480Q) strains analogous to a human eIF2B mutation (EIF2B3-G11V) is suppressed by overexpression of the three subunits of eIF2 (Dever et al. 1995). It has been shown that depletion of GCD7 in gcd7-td (degron) leads to codepletion of GCD2 (Dev et al. 2010), thus suggesting that GCD7 provides a platform for GCD2 binding in a pentameric eIF2B complex. Met-tRNAiMet binding stabilizes GTP binding to eIF2 (Kapp and Lorsch 2004), and thus, Met-tRNAiMet overexpression could provide insights into translation defects due to eIF2B mutations. One such example is overexpressing Met-tRNAiMet suppresses the lethality of Gcd2 depletion (gcd2-td mutant) (Dev et al. 2010). Tan1p is required for the formation of modified nucleoside N 4-acetylcytidine (ac4C) in tRNA specific for leucine and serine (Johansson and Bystrom 2004). The role of TAN1 overexpression in eIF2B-mediated translational regulation under H2O2-, alcohol-, caffeine-, DMSO-, DTT- and salt-mediated stress was also studied. Our results suggested that TAN1 overexpression imparts resistance to the H2O2, ethanol and caffeine stress. Thus, TAN1 regulates eIF2B-mediated translation regulation by an unrecognized mechanism.
Materials and methods
All the chemicals and reagents used were of molecular biology grade and were procured from Thermo Scientific, Himedia Labs, India; MP Biomedicals, USA; Fermentas Inc. USA; and Bio-Rad. USA.
S. cerevisiae strains and plasmids
S. cerevisiae strains used in this study are listed in Table 1. Standard laboratory strain E. coli (DH5α) used for plasmid isolation was cultured in nutrient broth (NB) and grown at 37 °C. YEp24 vector was used for the construction of genomic DNA library, and pEG(KG) was used for cloning and expression of S. cerevisiae genes. S. cerevisiae strains were cultured in YPD agar (1% yeast extract, 2% peptone, 2% dextrose, 2% agar) or liquid media (without agar). Nutrient broth (NB, Himedia Labs, Mumbai) with 100 µg/ml ampicillin was used to culture the bacterial strains of E. coli (DH5α) harboring YEp24 or pEG(KG) at 37 °C. Plasmid DNA of YEp24 and pEG(KG) was isolated and used in transformations of yeast strains (Sambrook et al. 2009; Elble 1992). Transformants were selected on synthetic complete (SC) medium without uracil supplementation at 30 °C.
Construction and screening of S. cerevisiae genomic DNA library
High molecular weight genomic DNA was isolated as described in Sambrook et al. (2009). Genomic DNA was partially digested with three different concentrations of Sau3AI enzyme and was gel purified by using a gel purification kit (Thermo Scientific). A total of 50 ng of purified genomic DNA fragments resulting from restriction digestion was pooled and ligated with 20 µg of shrimp alkaline phosphatase (1U)-treated plasmid YEp24 at BamHI site using T4 DNA ligase (Rose et al. 1987). The ligation reaction was performed at 16 °C for 16 h. DH5α strain was transformed by the heat-shock method with the ligation mix (Sambrook et al. 1989) and plated on NA medium containing ampicillin (100 µg/ml). Transformations were pooled into three groups, each representing ~13,575 cfu. All the three pools were grown independently and plasmid DNA was isolated (Sambrook et al. 1989). Transformation of S. cerevisiae eIF2B mutant strains (Table 1) showing slow-growth (Slg−) phenotype was performed independently with all the three pools of plasmids. Wild-type strains GCD7 GCN2 and GCD7 gcn2∆ GCD6 gcn2∆ strains (Table 2) were transformed with YEp24 vector using LiAc method (Elble 1992). Transformants were plated on SC medium with 2% glucose and lacking uracil. Synthetic complete supplement mixture (SC) without uracil is used as a dropout supplement to select transformants containing uracil-based plasmid. The transformants showing colony size to that of vector transformed GCD7 gcn2∆ were compared by streaking and spot assay (Gunde and Barberis 2005) to that of vector transformed gcd7-201 gcn2∆ mutant strain. The transformants showing Slg+ and Gcd+ phenotype were further screened by spot assay of tenfold serially diluted culture and by streaking. The plasmid DNA was rescued from the potential gcd7-201 gcn2∆ transformant (Slg+, Gcd+) as given by (Hoffman and Winston 1987). Rescued plasmids were transformed back into gcd7-201 gcn2∆ mutant yeast strain in order to reconfirm the Slg+ and Gcd+ phenotype. The insert present in the plasmid was sequenced on both the strands at Eurofins, Bangalore (http://www.eurofins.in/) using vector-specific primers (Table 3).
Functional characterization of suppressor protein
Full-length TAN1 gene and YGL230C gene present in the plasmids were amplified from rescued plasmids using gene-specific primers (Table 3) followed by sub-cloning into pEG(KG) vector with a GAL1 promoter and protease cleavable N-terminal GST tag at the SalI/HindIII and HindIII/BamHI restriction sites, respectively. The resulting recombinant plasmid DNA (100 ng) was transformed into E. coli DH5α by heat-shock method (Sambrook et al. 1989). Plasmid DNA was isolated from transformants and sequences were ensured by DNA sequencing. In order to verify the potential gene responsible for Slg+ and Gcd+ phenotype, pEG(KG)/TAN1 plasmid and pEG(KG)/YGL230C plasmid were transformed into gcd7-201 gcn2∆ strain by LiAc method (Elble 1992) and grown on SC medium supplemented with 2% galactose and lacking uracil. gcd7-201 gcn2∆ strain and GCD7 gcn2∆ were transformed with pEG(KG) vector alone, grown on SC-Ura/Gal medium and were used as controls. pEG(KG) has a galactose-regulated promoter. Gal promoter is repressed in the presence of raffinose, but is induced by addition of galactose. So the gene cloned under Gal1 promoter can be switched on or off depending on the presence of either raffinose or galactose. The transformants were analyzed by isolation of plasmid DNA by HiPurATM yeast plasmid DNA purification kit. The TAN1 transformants were screened for Slg+ and Gcd+ phenotype by spot assay. Gcd phenotype was also tested on SC medium containing 30 mM 3-AT (3-amino-triazole). 3-AT is a histidine analog. Amino acid starvation is commonly mimicked in yeast using 3‐amino‐triazole (3‐AT). Addition of 3‐AT to wild‐type strains causes histidine starvation, which activates Gcn2p kinase, ultimately increasing GCN4 expression. Mutations in eIF2B subunits in gcn2Δ strains inhibit eIF2B activity. Reduction in eIF2B activity in gcn2Δ strains allows growth on media containing 3‐AT. pEG(KG), a uracil-based plasmid containing TAN1, was evicted on 5-fluoroorotic acid (FOA)-containing medium. Colonies from FOA plate were replica plated on SC medium without uracil and containing 2% galactose. URA3 gene product converts 5-fluoroorotic acid (5-FOA) to a toxic product, 5-fluorouracil. Thus, yeast cells containing URA3 marker cannot grow on medium containing 5-FOA, but can grow on medium lacking uracil. This property is used to select for the loss of vectors carrying the wild-type URA marker (Zhang et al. 1997). Plates were incubated at 30 °C for 2 days and were checked for growth.
Western blot analysis
Whole-cell extracts (WCE) from induced and uninduced cultures of S. cerevisiae gcd7-201 gcn2∆ were prepared by a glass bead lysis method using Fast Prep (MP Biomedicals). For this, S. cerevisiae transformants were inoculated into SC medium (5 ml) supplemented with 2% raffinose (w/v) and incubated at 30 °C for 18 h. One percent of overnight grown primary culture was used to inoculate 10 ml SC medium supplemented with 2% raffinose (w/v) and incubated at 30 °C. At an A 600 of ~0.5, an aliquot was taken as the uninduced control, and the remaining culture was induced by the addition of 2% galactose (w/v) followed by continued incubation for an additional 3 h at 30 °C. The cells were harvested by centrifugation (6000 rpm for 10 min) and protein extracts were prepared using 20% trichloroacetic acid (TCA). Twenty micrograms of total protein was resolved by SDS–PAGE transferred to the nitrocellulose membrane (Millipore, Immobilon P 0.45 µm) by electroblotting and incubated at 4 °C for 1 h in blocking solution containing 5% nonfat dried milk. The membrane was incubated with anti-GST antibodies (1:5000, Abcam) for overnight at 4 °C. Immunoreactive proteins were detected by using primary antibody (α-GST for fusion protein and α-GCD6 for GCD6) and secondary antibody anti-rabbit IgG conjugated to horseradish peroxidase (1:10000, Abcam) for 1 h and after three successive washes using PBST (phosphate-buffered saline containing Triton X-100) buffer, blots were developed using an enhanced chemiluminescence kit (ECL, Bio-Rad, Inc., USA).
Expression of TAN1 in eIF2Bε, eIF2Bγ, eIF2Bδ and GCN2 mutants
Transformation of eIF2ε (H1792), eIF2Bγ (H70), eIF2Bδ (H750) and GCN2 (H591) mutants (Table 1) with pEG(KG) or pEG(KG)/TAN1 was done, and they were grown in SC medium lacking uracil and containing 2% galactose for 2 days at 30 °C. Transformants were selected and analyzed for Slg+ and Gcd+ phenotype by streaking and spot assays on SC medium lacking uracil and containing 2% galactose or SC medium lacking uracil and containing 2% galactose and 30 mM 3AT, respectively. Plates were incubated for 2 days at 30 °C. Temperature-sensitive (Ts−) phenotype of eIF2Bγ (H70) was also checked by incubation at 37 °C.
Effect of H2O2, ethanol and caffeine on the growth S. cerevisiae strains overexpressing TAN1
To determine the effect of H2O2, ethanol, caffeine, NaCl, DMSO and dithiothreitol (DTT), wild-type GCD7 GCN2, GCD7 gcn2∆ and gcd7-201 gcn2∆ strains of S. cerevisiae containing pEG(KG) or pEG(KG)/TAN1 were grown for 16 h at 30 °C with shaking in SC medium supplemented with either 2% galactose or raffinose (lacking uracil). SC medium was also supplemented with 4 mM H2O2 (Ring et al. 2008), 10% ethanol (Yamauchi and Izawa 2016), 20 mM caffeine (Wanke et al. 2008), 1.6% DMSO (Motlekar et al. 2009), 35 mM dithiothreitol (DTT) (Ring et al. 2008) and 1 M NaCl (Melamed et al. 2008) separately. Cell density at A 600 nm was measured by using UV visible spectrophotometer. S. cerevisiae transformants were also tested by spot and halo assays (Flower et al. 2005; Yang et al. 2006). Slg− and Gcd− phenotype was tested by spotting tenfold serially diluted cultures on SC medium supplemented with either 2% galactose or raffinose (lacking uracil) and 3-AT (30 mM) for Gcd− phenotype or containing H2O2 (4 mM), ethanol (10%), caffeine (30 mM), DMSO (1.6%), dithiothreitol (DTT) (35 mM) and NaCl (1 M). The plates were incubated at 30 °C for two days. In halo assay, filter disks of H2O2, ethanol, caffeine, DMSO, dithiothreitol (DTT) and NaCl were placed on SC agar medium containing either 2% galactose or raffinose (lacking uracil) with uniformly spread culture of S. cerevisiae mutant or wild-type strains containing pEG(KG) or pEG(KG)/TAN1. Petri plates were incubated at 30 °C for 2 days and observed for zone of inhibition of yeast cell growth. The S. cerevisiae GCD7 GCN2, GCD7 gcn2∆ and gcd7-201 gcn2∆ strains were streaked on YPD plates with or without H2O2, ethanol, caffeine, DMSO, dithiothreitol (DTT) and NaCl and were used as controls.
Results
Construction of S. cerevisiae genomic DNA library in YEp24 plasmid
Genomic DNA of the S. cerevisiae strain H4 (Table 1) and YEp24 plasmid DNA was isolated (Sambrook et al. 2009). DNA quality was checked by agarose gel electrophoresis (data not shown). For construction of genomic DNA library, restriction digestion of genomic DNA was done by using Sau3AI enzyme and YEp24 plasmid was digested with BamHI enzyme. Genomic DNA fragments of 0.5–6 kb were used for ligation reaction in 7.7 kb YEp24 plasmid.
Screening of genomic DNA library clones for rescuing slow-growth phenotype of eIF2B mutant strains
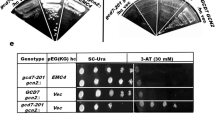
gcd7-201 gcn2∆ transformed with genomic DNA library having colony size equivalent to that of isogenic wild-type GCD7 gcn2∆ transformed with vector alone (Fig. 1a, c), but unlike vector transformed gcd7-201 gcn2∆ were observed (Fig. 1b). Comparing with the vector transformed mutant strains, we observed approximately 20 transformants showing growth comparable to the wild type. We only indicated eight of the transformants as representative colonies by pointing arrows. We screened all the 20 transformants for rescuing Slg+ and Gcd+ phenotype associated with mutations in different subunits of eIF2B. Further, we found one transformant which suppressed the Slg+ and Gcd+ phenotype of gcd7-201 gcn2∆. A transformant named as Sup-II (suppressor clone II) restored growth (Fig. 1d) as well as Gcd− phenotype of gcd7-201 gcn2∆ (Fig. 1e). Sup-II showed very similar growth pattern to that of isogenic wild-type GCD7 gcn2∆ transformed with empty vector (Fig. 1d). In order to reconfirm the slow-growth suppression and Gcd+ phenotype, Sup-II plasmid DNA was rescued from gcd7-201 gcn2∆ transformants (Fig. 1f) and transformed into gcd7-201 gcn2∆ strain. The transformants were restreaked to compare the growth. All the gcd7-201 gcn2∆ transformants were of uniform size and showed Slg+ phenotype (Fig. 1g, h). The results suggested that a genomic clone (Sup-II) suppresses Slg− and Gcd− phenotype of gcd7-201 gcn2∆ mutant strain. The Sup-II clone was sequenced using YEp24-specific primers (Table 3). Further, only EMC4 (~573 bp), TAN1 (~928 bp) and YGL230C (~444 bp) gene encoding for chaperone, tRNA acetyltransferase and putative protein, respectively, showed the complete ORF, whereas SEC15 and SAP4 were truncated. These results suggest that Sup-II harbors full-length EMC4, TAN1 and YGL230C that rescue the slow-growth and Gcd− phenotype of gcd7-201 gcn2∆ (Fig. 2a).
Screening for extragenic suppressors of gcd7-201 gcn2∆ mutant strain transformed with a genomic DNA library. b (YEp24) vector and c GCD7 gcn2∆ transformed with (YEp24) vector. S. cerevisiae strains were plated on SC medium lacking uracil and incubated at 30 °C for 2 days. d gcd7-201 gcn2 ∆ transformant showing Slg+ phenotype (Sup-II) was streaked along with gcd7-201 gcn2∆ and GCD7 gcn2∆ transformed with YEp24. e Serial dilutions of the transformant (Sup-II) showing Slg+ phenotype were spotted on SC-Ura and SC medium containing 30 mM-3AT. GCD7 gcn2∆ and GCD7 GCN2∆ transformed with vector were spotted as control. f The transformants (Sup-II) showing Slg+ and Gcd + phenotype were used for rescuing plasmid as indicated in lane 2. g The rescued plasmid was retransformed into gcd7-201 gcn2∆ mutant strain and h further verified by streaking on SC-Ura medium as indicated
PCR amplification of TAN1 gene. a Schematic representation of Sup-II clone showing complete ORF of EMC4, TAN1, YGL230C and truncated SEC15 and SAP4 genes on chromosome VII, b PCR amplification of the TAN1 gene from genomic clone (Sup-II) and separated on 1% agarose gel, c pEG(KG) plasmid was isolated (lane 3) and digested with XbaI and SalI (lane 1). b, c Lane 1 represents molecular size marker (kb)
Sub-cloning of potential suppressors into pEG(KG) (yeast expression vector)
TAN1 and YGL230C genes were amplified using gene-specific primers (Table 3) containing SalI restriction site in the forward primer and HindIII in the reverse primer for TAN1 and HindIII and BamHI for YGL230C followed by cloning under GST expression vector at respective restriction sites. A PCR product of ~ 950 bp was observed on agarose gel as expected for TAN1 gene (Fig. 2b). A PCR product of ~ 450 bp was also observed on agarose gel as expected for YGL230C (data not shown). TAN1 gene is present on chromosome VII of S. cerevisiae genome (http://www.yeastgenome.org/) (Fig. 2a). pEG(KG) vector of 9.3 kb containing GST tag under GAL1 promoter was used for sub-cloning of TAN1 and YGL230C (Fig. 2c). TAN1 and YGL230C gene sequences were verified by sequencing, and complete and error-free sequences of 928bp and 444 bp were observed. Data are not shown for YGL230C.
TAN1 overexpression rescued slow-growth and Gcd− phenotype of gcd7-201 gcn2∆
The gcd7-201 gcn2∆ strain was transformed with vector alone or with pEG(KG)/TAN1 or pEG(KG)/YGL230C expression constructs and transformants were streaked on SC medium without uracil, but containing either raffinose (Fig. 3a) or galactose (Fig. 3b). As expected, gcd7-201 gcn2∆ transformed with vector alone or TAN1 construct showed slow-growth phenotype on SC medium containing raffinose. Interestingly, TAN1 overexpression rescued the slow growth of gcd7-201 gcn2∆, when streaked on SC medium containing galactose (Fig. 3b). In contrast, gcd7-201 gcn2∆ transferred with pEG(KG)/YGL230C did not rescue the slow-growth phenotype (data not shown). In order to verify the results, pEG(KG)/TAN1 a uracil-based plasmid was evicted on FOA containing medium that showed the original slow-growth phenotype of gcd7-201 gcn2∆ (Fig. 3c) and did not grow on SC medium without uracil supplementation (Fig. 3d). These data clearly suggest that overexpression of TAN1 rescued the Slg− of gcd7-201 gcn2∆ mutant. Further TAN1 overexpression was analyzed for rescuing Gcd− phenotype of gcd7-201 gcn2∆ mutant strain (Fig. 3e). As expected, gcd7-201 gcn2∆ transformed with vector alone showed slow growth in SC medium without uracil supplementation as well as Gcd− phenotype on medium containing 3-AT. Interestingly, Gcd− phenotype of gcd7-201 gcn2∆ was also rescued when TAN1 was overexpressed (Fig. 3e). Overexpression of TAN1 suppressed the Slg− and Gcd− phenotype of gcd7-201 gcn2∆. This clearly suggests that Tan1p play a direct or indirect role in eIF2B-mediated amino acid starvation. Expression of GST-Tan1 was verified by western blotting. Whole-cell extracts (WCE) of gcd7-201 gcn2∆ mutant expressing GST-Tan1 or GST alone were analyzed by western blotting using anti-GST antibodies. As expected, a 26-kDa GST protein was detected in extracts expressing GST alone. On the other hand, ~60-kDa protein was detected in WCE of gcd7-201 gcn2∆ mutant transformed with pEG(KG)/TAN1 construct. α-GCD6 antibodies were used as an internal loading control (Fig. 3f).
gcd7-201 gcn2∆ rescued Slg+ and Gcd+ phenotype when transformed with pEG(KG)/TAN1 plasmid. GCD7 gcn2∆ and gcd7-201 Gcn2∆ harboring pEG(KG)/TAN1 or empty vector pEG(KG) were streaked in parallel on SC medium laking uracil, but either containing a raffinose or b galactose. Uracil-based plasmid pEG(KG)/TAN1 was evicted on SC medium containing, c FOA and further streaked on d SC-Ura medium. e Spotting on SC medium containing 2% galactose and lacking uracil or SC medium containing 2% galactose, 30 mM 3-AT and lacking uracil. Plates were incubated at 30 °C for 2 days. f Western analysis of GST-Tan1p expression in gcd7-201 gcn2∆ strain with anti-GST antibody. The whole-cell protein extracts (20 µg) were prepared from uninduced and 2% galactose-induced cultures of the strain harboring pEG(KG)/TAN1. Samples were separated on 10% SDS gel followed by western blotting using anti-GST for GST-Tan1 and anti-Gcd6 antibodies (loading control). UI uninduced, I induced
Tan1p overexpression suppresses specific eIF2B mutations
To test whether Tan1p overexpression is specific, we transferred S. cerevisiae harboring mutations in different subunits which include H70 (gcd1-502), H1792 (gcd6-1), H591 (gcd12-503) and H750 (gcn2::LEU2). Interestingly, the TAN1 overexpression rescued temperature-sensitive (Ts−) phenotype of gcd1-502 at 37 °C (Fig. 4a), but not that of H1792 (gcd6-1), H591 (gcd12-503) and H750 (gcn2::LEU2) (data not shown). TAN1 overexpression also rescued Slg− and Gcd− phenotype of gcd1-502 (Fig. 4b). Thus, Tan1p causes strain-specific suppression of eIF2B mutants.
Tan1p overexpression rescues Slg−, Gcd− and Ts− phenotype of gcd1-502 gcn2-101 strain. a gcd1-502 gcn2-101 transformants showing Slg+ phenotype were streaked along with gcd1-502 gcn2-101 transformed with pEG(KG) vector alone. b Serial dilutions of the gcd1-502 gcn2-101 transformants showing Slg+ phenotype were spotted on SC-Ura or SC medium containing 3-AT (30 mM). gcd1-502 gcn2-101 transformed with pEG(KG) was spotted as control. Plates were incubated for 2 days at 30 °C
pEG(KG)/TAN1 overexpression enhanced stress tolerance in S. cerevisiae
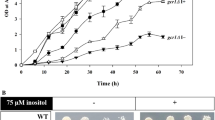
Unlike amino acid stress, TAN1 overexpression protects GCD7 GCN2, GCD7 gcn2∆ and gcd7-201 gcn2∆ against H2O2-, ethanol- and caffeine-mediated stress. GCD7 gcn2∆, gcd7-201 gcn2∆ and GCD7 GCN2∆ were streaked on YPD agar medium containing H2O2, ethanol and caffeine showed Slg− phenotype (Fig. 5a). The GCD7 GCN2, GCD7 gcn2∆ and gcd7-201 gcn2∆ S. cerevisiae strains transformed with empty vector showed reduced growth (twofold) in the presence of 4 mM H2O2, 10% ethanol and 30 mM caffeine in the presence of either raffinose or galactose. S. cerevisiae strains GCD7 GCN2, GCD7 gcn2∆ and gcd7-201 gcn2∆ transformed with pEG(KG)/TAN1 showed normal growth even in the presence of H2O2, ethanol and caffeine only in the presence of galactose (Fig. 5b, c, d), indicating that overexpression of Tan1p protects S. cerevisiae from H2O2-, ethanol- and caffeine-induced cell death. No significant protective effect was observed against DMSO, NaCl and DTT (data not shown). Halo assay shows zone of no growth surrounding the hydrogen peroxide-, caffeine- and alcohol-containing filter disks. Zone of inhibition was significantly reduced in GCD7 GCN2, GCD7 gcn2∆ and gcd7-201 gcn2∆ strains overexpressing TAN1 in the presence of 2% galactose as compared to control cells (Fig. 6a, b, c). Using the cell spot assay, it was observed that different yeast strains (GCD7 GCN2, GCD7 gcn2∆ and gcd7-201 gcn2∆) were protected from H2O2-, ethanol- and caffeine-mediated cell death only when Tan1p was expressed in galactose (Fig. 7a, b, c). These results strongly suggest that overexpression of TAN1 is capable of preventing cell death due to H2O2, alcohol and caffeine. Thus, overexpression of TAN1 repairs GCD7 GCN2-, GCD7 gcn2∆- and gcd7-201 gcn2∆-based defect in translation initiation in some or other way. Based on our results, we have proposed a new model (Fig. 8) explaining the amino acid starvation stress in eIF2B mutant of S. cerevisiae and H2O2, ethanol and caffeine stress-induced (wild-type and eIF2B mutant of S. cerevisiae) derepression of Gcn4p. The H2O2-, ethanol- and caffeine-mediated stress bypasses Gcn2p phosphorylation because similar results were observed in the presence or absence of Gcn2p. Overexpression of Tan1p modulates the eIF2B activity and in turn lowers GCN4 expression. Our model is supported by previous reports such as butanol-mediated derepression of GCN4 in a Gcn2p-independent manner (Ashe et al. 2001). Rapamycin-induced stress has also shown to down regulate translation and GCN4 induction (Barbet et al. 1996; Cherkasova and Hinnebusch 2003; Kubota et al. 2003). Moreover, oxidative stress (H2O2) inhibits general translation independent of Gcn2p or eIF2α phosphorylation (Knutsen et al. 2015).
S. cerevisiae strains overexpressing TAN1 showed enhanced stress tolerance. a gcd7-201 gcn2∆, GCD7 gcn2∆ and GCN2 gcn2∆ S. cerevisiae strains were streaked on YPD agar plates containing 4 mM H2O2, 10% ethanol and 30 mM caffeine. Number of plus signs (+) indicates comparative visible growth. Negative signs (−) indicates no visible growth. SC medium supplemented with b 4 mM H2O2, c 10% ethanol and d 30 mM caffeine in the presence of raffinose or galactose. Cultures were grown for 16 h at 30 °C. Cultures with raffinose supplementation were served as control. The cell density was obsereved by measuring absorbance at 600 nm (A 600). Data of three independent experiments were plotted with standard deviation
Halo assay to test the effect of H2O2, ethanol and caffeine on the growth of eIF2B mutants. Hydrogen peroxide, ethanol and caffeine halo assays were performed with gcd7-201 gcn2∆, GCD7 gcn2∆ and GCD7 GCN2. S. cerevisiae strains harboring either the empty vector or the TAN1 gene-expressing plasmid were spread on SC medium, containing filter disks soaked in a H2O2 (4 mM), b ethanol (10%) and c caffeine (30 mM) in the presence of either raffinose or galactose. The plates were incubated at 30 °C for 2–3 days
Spot assay to test the effect of H2O2, ethanol and caffeine on the growth of eIF2B mutants. Hydrogen peroxide, ethanol and caffeine halo assays were performed with gcd7-201 gcn2∆, GCD7 gcn2∆ and GCD7 GCN2. Tenfold serially diluted transformants harboring empty vector (pEG(KG) or pEG(KG)-TAN1 were spotted on SC-Ura or SC-Ura/3-AT agar plates supplemented with a H2O2 (4 mM), b ethanol (10%) and c caffeine (30 mM) in the presence of either raffinose or galactose. Cultures were grown for 2 days at 30 °C
Schematic model indicating the repression of GCN4 by overexpression of GST-Tan1p via eIF2B. Panel I indicates that in the presence of amino acids (+a.a), Gcn2p is inhibited in wild-type cells that in turn inhibits Gcn4p expression. In contrast, amino acid starvation (−a.a) in wild-type cells leads to eIF2α phosphorylation and Gcn4p induction via activation of Gcn2p kinase. Similarly, the presence of H2O2, ethanol and caffeine in the growth medium inhibits eIF2B activity that in turn activates GCN4p. On the other hand, in panel II, eIF2B mutants grown in the presence or absence of a.a, H2O2, ethanol and caffeine inhibit eIF2B activity that can lead to Gcn4p activation. In panel III, the eIF2B mutants gcd7-201 gcn2∆, GCD7 gcn2∆ and GCD7 GCN2∆ overexpressing Tan1p repress Gcn4p in the presence or absence of a.a, H2O2, ethanol and caffeine and rescue their Slg−, Gcd− and Ts− phenotype
Discussion
Eukaryotic initiation factor 2B (eIF2B) mutations lead to the translation dysregulation (Hannig et al. 1990; Dever et al. 1993; De Aldana and Hinnebusch 1994; Pavitt et al. 1997) causing a rare encephalopathy, vanishing white matter disease (VWM). In this study, we identified the extragenic suppressors of eIF2B mutations in Saccharomyces cerevisiae, corresponding to VWM. By screening the S. cerevisiae genomic DNA library, TAN1 gene was observed to suppress the slow-growth (Slg−) and general control derepression (Gcd−) phenotype of S. cerevisiae, caused by gcd7-201 gcn2∆ and gcd1-502 gcn2-101 mutations. It is also interesting to note that Tan1p overexpression rescued Slg−, Gcd− and Ts− phenotypes of one of the regulatory eIF2B subunits and one of the catalytic eIF2B subunits. It has been already reported that overexpressing Met-tRNAiMet suppresses the lethality of Gcd2 depletion (gcd2-td mutant) (Dev et al. 2010). Tan1p is a putative tRNA acetyltransferase required for the formation of modified nucleoside N 4-acetylcytidine (ac4C) in tRNA and is important for tRNA stability (Johansson and Bystrom 2004). Tan1p protein abundance increases in response to DNA replication stress (Tkach et al. 2012). Thus, we also tested the effect of TAN1 overexpression in Saccharomyces cerevisiae strains under different stress conditions (H2O2, ethanol, caffeine, DMSO, DTT and NaCl). gcd7-201 gcn2∆ GCD7 GCN2 and GCD7 gcn2∆ strains were found to be more sensitive to H2O2, ethanol and caffeine and less sensitive to NaCl-mediated (Goossens et al. 2001), DMSO-mediated and DTT-mediated stress. But the growth was reduced to a greater extent in gcd7-201 gcn2∆ as compared to GCD7 GCN2 cells, thus proposing the role of Tan1p in eIF2B-mediated translation regulation. This is in consistent with the previous reports of Gcn2p protein-dependent/independent Gcn4p protein induction and translation regulation. For example, butanol-mediated induction of GCN4 by a Gcn2p-independent manner has already been reported (Ashe et al. 2001). Oxidative stress (H2O2) causes translation inhibition independent of eIF2α–p mediated by GCN2 (Knutsen et al. 2015). Moreover, rapamycin causes translational inhibition (Barbet et al. 1996) by GCN4-mediated stress responses (Cherkasova and Hinnebusch 2003; Kubota et al. 2003). Thus, H2O2, ethanol and caffeine might activate Gcn4p in the presence of functional wild-type eIF2B. Interestingly, TAN1 overexpression restored the cell growth of GCD7 GCN2, gcd7-201 Gcn2∆ strains under different stress conditions by directly or indirectly modulating eIF2B GEF activity. Thus, H2O2, ethanol and caffeine might be used as chemical modulators to study eIF2B-mediated pathways leading to stress responses (Motlekar et al. 2009). Although the mechanism by which Tan1p protein functions in translation regulation is still unknown, the requirement of more stable tRNA specific for leucine and serine depends upon the expression of Tan1p protein. The stability of tRNA is maintained by various enzymes. For example, tRNA-modifying enzymes such as tRNA(m5U54) methyltransferase Trm2p (Nordlund et al. 2000), pseudouridine (Ψ 55) synthase Pus4p (Becker et al. 1997), tRNA (m 22 G26) dimethyltransferase Trm1p (Ellis et al. 1986) and the putative tRNA (Gm18) 2′-O-ribose methyltransferase Trm3p (Cavaillé et al. 1999) are known for tRNA modification. Overexpression of Tan1p protein suppresses the Slg− and Gcd− phenotype of eIF2B mutants possibly by stabilizing the tRNA by recognizing general tRNA structure. This explanation is supported by the previous study explaining the in vitro binding of E. coli tRNA (m1G37) methyltransferase to the tRNA species that are not methylated by the same enzyme (Redlak et al. 1997). On comparison with previous studies, it can be concluded that Tan1p protein might be involved in eIF2B-mediated translation regulation. More importantly, the resistance to different stress conditions brought upon the overexpression of TAN1 in the wild-type and mutant yeast strains provides a facile model system to begin a detailed structural and functional analysis of its role in eIF2B-mediated translation regulation.
References
Ashe MP, Slaven JW, Susan K, Ibrahimo S, Sachs AB (2001) A novel eIF2B-dependent mechanism of translational control in yeast as a response to fusel alcohols. EMBO J 20:6464–6474
Barbet NC, Schneider U, Helliwell SB, Stansfield I, Tuite MF, Hall MN (1996) TOR controls translation initiation and early G1 progression in yeast. Mol Biol Cell 7:25–42
Becker HF, Motorin Y, Planta RJ, Grosjean H (1997) The yeast gene YNL292w encodes a pseudouridine synthase (Pus4) catalyzing the formation of Ψ55 in both mitochondrial and cytoplasmic tRNAs. Nucleic Acids Res 25:4493–4499
Cavaillé JEROME, Chetouani FARID, Bachellerie JP (1999) The yeast Saccharomyces cerevisiae YDL112w ORF encodes the putative 2′-O-ribose methyltransferase catalyzing the formation of Gm18 in tRNAs. RNA 5:66–81
Cherkasova V, Hinnebusch AG (2003) Translational control by TOR and TAP42 through dephosphorylation of eIF2α kinase GCN2. Genes Dev 17:859–872
De Aldana CV, Hinnebusch AG (1994) Mutations in the GCD7 subunit of yeast guanine nucleotide exchange factor eIF-2B overcome the inhibitory effects of phosphorylated eIF-2 on translation initiation. Mol Cell Biol 14:3208–3222
Dev K, Qiu H, Dong J, Zhang F, Barthlme D, Hinnebusch AG (2010) The β/Gcd7 subunit of eukaryotic translation initiation factor 2B (eIF2B), a guanine nucleotide exchange factor, is crucial for binding eIF2 in vivo. Mol Cell Biol 30(21):5218–5233
Dever TE, Chen JJ, Barber GN, Cigan AM, Feng L, Donahue TF, London IM, Katze MG, Hinnebusch AG (1993) Mammalian eukaryotic initiation factor 2 alpha kinases functionally substitute for GCN2 protein kinase in the GCN4 translational control mechanism of yeast. Proc Natl Acad Sci 90:4616–4620
Dever TE, Yang W, Aström S, Byström AS, Hinnebusch AG (1995) Modulation of tRNA(iMet), eIF-2, and eIF-2B expression shows that GCN4 translation is inversely coupled to the level of eIF-2.GTP.Met-tRNA(iMet) ternary complexes. Mol Cell Biol 15 (11):6351–6363
Dever TE, Kinzy TG, Pavitt GD (2016) Mechanism and regulation of protein synthesis in Saccharomyces cerevisiae. Genetics 203:65–107
Elble R (1992) A simple and efficient procedure for transformation of yeasts. Biotechniques 13:18–20
Ellis SR, Morales MJ, Li JM, Hopper AK, Martin NC (1986) Isolation and characterization of the TRM1 locus, a gene essential for the N2, N2-dimethylguanosine modification of both mitochondrial and cytoplasmic tRNA in Saccharomyces cerevisiae. J Biol Chem 261:9703–9709
Flower TR, Chesnokova LS, Froelich CA, Dixon C, Witt SN (2005) Heat shock prevents alpha-synuclein-induced apoptosis in a yeast model of Parkinson’s disease. J Mol Biol 351:1081–1100
Fogli A, Boespflug-Tanguy O (2006) The large spectrum of eIF2B-related diseases. Biochem Soc Trans 34(1):22–29
Fogli A, Rodriguez D, Eymard-Pierre E, Bouhour F, Labauge P, Meaney BF, Zeesman S, Kaneski CR, Schiffmann R, Boespflug-Tanguy O (2003) Ovarian failure related to eukaryotic initiation factor 2B mutations. Am J Human Genetics 72:1544–1550
Goossens A, Dever TE, Pascual-Ahuir A, Serrano R (2001) The protein kinase Gcn2p mediates sodium toxicity in yeast. J Biol Chem 276:30753–30760
Gunde T, Barberis A (2005) Yeast growth selection system for detecting activity and inhibition of dimerization-dependent receptor tyrosine kinase. Biotechniques 39:541
Hannig EM, Williams NP, Wek RC, Hinnebusch AG (1990) The translational activator GCN3 functions downstream from GCN1 and GCN2 in the regulatory pathway that couples GCN4 expression to amino acid availability in Saccharomyces cerevisiae. Genetics 126:549–562
Hoffman CS, Winston F (1987) A ten-minute DNA preparation from yeast efficiently releases autonomous plasmids for transformation of Escherichia coli. Gene 57:267–272
Horzinski L, Huyghe A, Cardoso MC, Gonthier C, Ouchchane L, Schiffmann R, Blanc P, Boespflug-Tanguy O, Fogli A (2009) Eukaryotic initiation factor 2B (eIF2B) GEF activity as a diagnostic tool for EIF2B-related disorders. PLoS One 4:8318
Jennings MD, Zhou Y, Mohammad-Qureshi SS (2013) eIF2B promotes eIF5 dissociation from eIF2• GDP to facilitate guanine nucleotide exchange for translation initiation. Genes Dev 27(24):2696–2707
Johansson MJ, Bystrom AS (2004) The Saccharomyces cerevisiae TAN1 gene is required for N4-acetylcytidine formation in tRNA. RNA 10:712–719
Kapp LD, Lorsch JR (2004) GTP-dependent recognition of the methionine moiety on initiator tRNA by translation factor eIF2. J Mol Biol 335:923–936
Knutsen JHJ, Rødland GE, Bøe CA, Håland TW, Sunnerhagen P, Grallert B, Boye E (2015) Stress-induced inhibition of translation independently of eIF2α phosphorylation. J Cell Sci 128:4420–4427
Krishnamoorthy T, Pavitt GD, Zhang F, Dever TE, Hinnebusch AG (2001) Tight binding of the phosphorylated α subunit of initiation factor 2 (eIF2α) to the regulatory subunits of guanine nucleotide exchange factor eIF2B is required for inhibition of translation initiation. Mol Cell Biol 21:5018–5030
Kubota H, Obata T, Ota K, Sasaki T, Ito T (2003) Rapamycin-induced translational derepression of GCN4 mRNA involves a novel mechanism for activation of the eIF2α kinase GCN2. J Biol Chem 278:20457–20460
Leegwater PA, Vermeulen G, Könst AA, Naidu S, Mulders J, Visser A, Kersbergen P, Mobach D, Fonds D, van Berkel CG, Lemmers RJ (2001) Subunits of the translation initiation factor eIF2B are mutant in leukoencephalopathy with vanishing white matter. Nat Genet 29:383–388
Li W, Wang X, Van Der Knaap MS, Proud CG (2004) Mutations linked to leukoencephalopathy with vanishing white matter impair the function of the eukaryotic initiation factor 2B complex in diverse ways. Mol Cell Biol 24:3295–3306
Liu R, Van der Lei HD, Wang X, Wortham NC, Tang H, Van Berkel CG, Mufunde TA, Huang W, Van der Knaap MS, Scheper GC et al (2011) Severity of vanishing white matter disease does not correlate with deficits in eIF2B activity or the integrity of eIF2B complexes. Hum Mutat 32:1036–1045
Melamed D, Pnueli L, Arava Y (2008) Yeast translational response to high salinity: global analysis reveals regulation at multiple levels. RNA 14:1337–1351
Motlekar N, de Almeida RA, Pavitt GD, Diamond SL, Napper AD (2009) Discovery of chemical modulators of a conserved translational control pathway by parallel screening in yeast. Assay Drug Dev Technol 7:479–494
Natarajan K, Meyer MR, Jackson BM, Slade D, Roberts C, Hinnebusch AG, Marton MJ (2001) Transcriptional profiling shows that Gcn4p is a master regulator of gene expression during amino acid starvation in yeast. Mol Cell Biol 21:4347–4368
Nordlund ME, Johansson JO, von Pawel-Rammingen UL, BYSTROeM AS (2000) Identification of the TRM2 gene encoding the tRNA (m5U54) methyltransferase of Saccharomyces cerevisiae. Rna 6(6):844–860
Palmer LK, Shoemaker JL, Baptiste BA, Wolfe D, Keil RL (2005) Inhibition of translation initiation by volatile anesthetics involves nutrient-sensitive GCN-independent and-dependent processes in yeast. Mol Biol Cell 16:3727–3739
Pavitt GD, Yang W, Hinnebusch AG (1997) Homologous segments in three subunits of the guanine nucleotide exchange factor eIF2B mediate translational regulation by phosphorylation of eIF2. Mol Cell Biol 17:1298–1313
Pavitt GD, Ramaiah KV, Kimball SR, Hinnebusch AG (1998) eIF2 independently binds two distinct eIF2B subcomplexes that catalyze and regulate guanine–nucleotide exchange. Genes Dev 12:514–526
Pronk JC, Van Kollenburg B, Scheper GC, Van der Knaap MS (2006) Vanishing white matter disease: a review with focus on its genetics. Dev Disabil Res Rev 12:123–128
Redlak M, Andraos-Selim C, Giege R, Florentz C, Holmes WM (1997) Interaction of tRNA with tRNA (guanosine-1) methyltransferase: binding specificity determinants involve the dinucleotide G36pG37 and tertiary structure. Biochemistry 36:8699–8709
Richardson JP, Mohammad SS, Pavitt GD (2004) Mutations causing childhood ataxia with central nervous system hypomyelination reduce eukaryotic initiation factor 2B complex formation and activity. Mol Cell Biol 24:2352–2363
Ring G, Khoury CM, Solar AJ, Yang Z, Mandato CA, Greenwood MT (2008) Transmembrane protein 85 from both human (TMEM85) and yeast (YGL231c) inhibit hydrogen peroxide mediated cell death in yeast. FEBS Lett 582:2637–2642
Rose MD, Novick P, Thomas JH, Botstein D, Fink GR (1987) A Saccharomyces cerevisiae genomic plasmid bank based on a centromere-containing shuttle vector. Gene 60:237–243
Rowlands AG, Panniers R, Henshaw EC (1988) The catalytic mechanism of guanine nucleotide exchange factor action and competitive inhibition by phosphorylated eukaryotic initiation factor 2. J Biol Chem 263:5526–5533
Sambrook J, Fritsch EF, Maniatis T (1989) Molecular cloning: a laboratory manual, 2nd edn. Cold spring harbor laboratory press, New York
Sambrook J, Fritsch EF, Maniatis T (2009) Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, New York
Schiffmann R, Elroy-Stein O (2006) Childhood ataxia with CNS hypomyelination/vanishing white matter disease—a common leukodystrophy caused by abnormal control of protein synthesis. Mol Genet Metab 88:7–15
Shenton D, Smirnova JB, Selley JN, Carroll K, Hubbard SJ, Pavitt GD, Ashe MP, Grant CM (2006) Global translational responses to oxidative stress impact upon multiple levels of protein synthesis. J Biol Chem 281:29011–29021
Tkach JM, Yimit A, Lee AY, Riffle M, Costanzo M, Jaschob D, Hendry JA, Ou J, Moffat J, Boone C, Davis TN (2012) Dissecting DNA damage response pathways by analysing protein localization and abundance changes during DNA replication stress. Nat Cell Biol 14:966–976
Van Der Knaap MS, Leegwater PA, Könst AA, Visser A, Naidu S, Oudejans C, Schutgens RB, Pronk JC (2002) Mutations in each of the five subunits of translation initiation factor eIF2B can cause leukoencephalopathy with vanishing white matter. Ann Neurol 51:264–270
Van der Knaap MS, Van Berkel CG, Herms J, Van Coster R, Baethmann M, Naidu S, Boltshauser E, Willemsen MA, Plecko B, Hoffmann GF, Proud CG (2003) eIF2B-related disorders: antenatal onset and involvement of multiple organs. Am J Human Genetics 73:1199–1207
Wanke V, Cameroni E, Uotila A, Piccolis M, Urban J, Loewith R, De Virgilio C (2008) Caffeine extends yeast lifespan by targeting TORC1. Mol Microbiol 69:277–285
Wortham NC, Proud CG (2015) Biochemical effects of mutations in the gene encoding the alpha subunit of eukaryotic initiation factor (eIF) 2B associated with Vanishing White Matter disease. BMC Med Genet 16:64
Yamauchi Y, Izawa S (2016) Prioritized expression of BTN2 of Saccharomyces cerevisiae under pronounced translation repression induced by severe ethanol stress. Front microbiol 7:1319
Yang Z, Khoury C, Jean-Baptiste G, Greenwood MT (2006) Identification of mouse sphingomyelin synthase 1 as a suppressor of Bax-mediated cell death in yeast. FEMS Yeast Res 6:751–762
Zhang BE, Kraemer B, Sen Gupta D, Fields S, Wickens M (1997) A three-hybrid system to detect and analyze RNA-protein interactions in vivo. In: Bartel P, Fields (eds) The Yeast Two-Hybrid System. Oxford University Press, New York, pp 298–315
Acknowledgements
Authors are thankful to Dr. Alan G. Hinnebusch, NICHD, USA, for providing yeast strains, αGCD6 antibodies and plasmids used in this study. Authors are also thankful to Shoolini University, Bajhol, Solan, Himachal Pradesh, India, for providing financial and infrastructural support for this study and all the members of Yeast Biology Lab for moral support.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors hereby declare that they have no conflict of interest.
Funding and support
Shoolini University.
Rights and permissions
About this article
Cite this article
Sharma, S., Sourirajan, A. & Dev, K. Role of Saccharomyces cerevisiae TAN1 (tRNA acetyltransferase) in eukaryotic initiation factor 2B (eIF2B)-mediated translation control and stress response. 3 Biotech 7, 223 (2017). https://doi.org/10.1007/s13205-017-0857-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s13205-017-0857-8