Abstract
The genetic diversity of bacterial populations’ nodulating Retama sphaerocarpa grown in the soils of Maamora cork forest (Morocco) was examined. ERIC-PCR fingerprinting of 30 strains distributed them in 2 groups, of which a representative strain from each group was studied by multilocus sequence analysis of the 16S rRNA, atpD, and gyrB genes. The two representative strains RSM25 and RSM32, grouped with “Microvirga tunisiensis”. This is the first description of Retama nodule bacteria as members of the genus Microvirga. A nodC-based phylogeny confirmed that the two representative strains RSM25 and RSM32 are affiliated with symbiovar mediterranense. The 2 strains were capable of nodulating not only R. sphaerocarpa but also R. monosperma, R. dasycarpa and L. luteus, and unable to nodulate Phaseolus vulgaris, Vachellia gummifera, Cicer arietinum, Vigna unguiculata and Glycine max. The inoculation of R. sphaerocarpa with RSM25 or RSM32 produced a 1.22-, and 1.36-fold increase in the dry weight of the plants compared to those grown in the presence of 0.05% KNO3. The 2 strains used monosaccharides and disaccharides as the sole C source, but fructose, glucose, galactose, arabinose and starch were not used. They were unable to grow on glycine as a N source. Phosphate solubilization and siderophore production was not detected, but IAA or IAA-related compounds were produced. The alkaline pH of the sampling site soil in the Maamora forest where Retama grows could explain why Microvirga was the dominant species in the root nodules of the plants.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
Plants of the genus Retama are wild shrubby legumes distributed throughout the Mediterranean area, more precisely in southwest Europe and northwest Africa (Zohary 1959; León-González et al. 2018). The genus Retama belongs to the Genisteae tribe of the Leguminosae (Fabaceae) family and contains four described species, R. monosperma, R. sphaerocarpa, R. raetam, and R. dasycarpa. The latter is endemic to Morocco, and R. sphaerocarpa is widespread in Southern Spain, Morocco, Algeria and Tunisia. The distribution of R. monosperma is limited to Morocco and Algeria, and R. raetam is found only in the arid areas of Southern Mediterranean, from Morocco to Libya (Hannane et al. 2014).
R. sphaerocarpa (yellow retama) has photosynthetic stems and a dimorphic shallow lateral roots that can reach down to a depth of more than 20 m, which enables the plant to lift water from deep, wet layers and release it to shallow, dry soils (Prieto et al. 2010). The hydraulic lift and the open canopy of the plants allow amelioration of the microclimatic conditions and the increase in nutrient content and mineralization rate under the canopy (Rodríguez-Echeverría and Pérez-Fernández 2003). The establishment of effective dinitrogen (N2)-fixing symbioses with soil bacteria, collectively referred to as rhizobia, allows R. sphaerocarpa to grow in N-deficient, poor soils, which contributes to the increase in nitrogen under the canopy (Rodríguez-Echeverría and Pérez-Fernández 2003). Thus, R. sphaerocarpa plays an important ecological role in arid and semiarid areas by maintaining plant diversity and ecosystem functioning and is a valuable species for revegetation and restoration programs, dune stabilization and soil fixation (Rodríguez-Echeverría and Pérez-Fernández 2005).
Rhizobia include genera Rhizobium, Ensifer, Allorhizobium, Pararhizobium, Neorhizobium, Shinella, Mesorhizobium, Aminobacter, Phyllobacterium, Ochrobactrum, Methylobacterium, Microvirga, Bradyrhizobium, Azorhizobium and Devosia from the alpha-Proteobacteria subclass; as well as Paraburkholderia, Cupriavidus and Trinickia from the beta-Proteobacteria subclass (Estrada-de los Santos et al. 2018; de Lajudie et al. 2019). Despite its ecological role, root-nodulating bacteria of R. sphaerocarpa have been studied only in a few sites from Algeria, Morocco and Spain. Rhizobia isolated from R. sphaerocarpa in central-western Spain affiliate with Bradyrhizobium and were later identified as B. canariense (Ruiz-Díez et al. 2009). Similarly, the rhizobial strains isolated from wild-grown R. sphaerocarpa in different ecological-climatic areas of northeastern Algeria formed a phylogenetic clade within the Bradyrhizobium genus (Boulila et al. 2009). B. retamae, originally isolated from R. monosperma grown in the subhumid area of Saidia in Northern Morocco was also able to infect R. sphaerocarpa (Guerrouj et al. 2013). All of the 31 strains infecting R. sphaerocarpa growing in eight different sites with different soil and environmental conditions in the Iberian Peninsula clustered with different members of the B. japonicum and B. elkanii superclades described by Menna et al. (2009) or with the species B. retamae (Rodríguez-Echeverría et al., 2014). Although these studies show that Bradyrhizobium is the dominant microsymbiont isolated from nodules of R. sphaerocarpa (Boulila et al. 2009; Guerrouj et al. 2013), the endophyte Phyllobacterium myrsinacearum has also been isolated from nodules of R. sphaerocarpa plants from central Spain (Ruíz-Díez et al. 2009, 2012). More recently, Ahnia et al. (2018) isolated B. algeriense from N2-fixing nodules of R. sphaerocarpa in Northern Algeria, and Lamin et al. (2019) reported that some strains related to “Ensifer aridi” obtained from the root nodules of R. monosperma, were also able to nodulate R. sphaerocarpa.
R. sphaerocarpa grows both in alkaline and acidic soils ranging from sea level to 2000 m, in sites with mean annual precipitation of 200–980 mm (Armas et al. 2011), reasons why we hypothesize that rhizobia associated with the plant could be more diverse than that reported thus far. The Maamora is the largest cork oak forest in the south Mediterranean region, where it plays economic, social and environmental roles (El Boukhari et al. 2016). Legumes occupy the second place in importance after the Poaceae family in Maamora, and Retama is one of the main diverse legume genera (Aafi et al. 2005).
Therefore, the aim of this study was to identify the microsymbionts associated with R. sphaerocarpa grown wild in the Maamora forest.
2 Materials and methods
2.1 Bacterial strains
Bacteria were isolated from root nodules of R. sphaerocarpa grown in soils of the Maamora Forest (34°.02 76 82 N; −6°. 71 22 23 W) located in the vicinity of Rabat (Morocco).
Root segments bearing nodules were collected, washed under running tap water, surface-sterilized by immersion in 5% sodium hypochlorite for 3 min, and finally washed thoroughly with sterile water. Individual nodules were then placed in Petri dishes and crushed with a sterile glass rod. The obtained extracts were then streaked onto Petri dishes containing solid yeast extract mannitol (YEM) medium (Vincent 1970) supplemented with Congo Red dye (0.025 g l−1). After incubation for 10 d at 28 °C, the single colony-forming units (CFUs) appeared on the plates were checked for purity by repeated streaking on the same medium. YEM medium was routinely used for bacterial culture.
2.2 DNA extraction, PCR amplifications and genes sequencing
Essentially, bacterial genomic DNA was isolated after growth of the cells in liquid YEM medium, collected by centrifugation in a microfuge, treated with proteinase K (20 mg ml−1) and extracted as described by Brenner et al. (1982). After recovery, the DNA was kept at −20 °C until use. The quantity of DNA was determined using a Nanodrop spectrophotometer (NanoDrop ND1000). Enterobacterial repetitive intergenic consensus (ERIC)-polymerase chain reactions (ERIC-PCR) were performed using primers ERIC1R and ERIC2 and reactions conditions previously published (Versalovic et al. 1991). PCR amplifications of the 16S rRNA gene fragments were carried out using the two opposing primers fD1 and rD1 as indicated earlier (Weisburg et al. 1991). The primer pairs atpD255F/atpD782R (Vinuesa et al. 2005) and gyrBF/gyrBR (Martens et al. 2007) were used for amplification of the atpD and gyrB genes, respectively. For the amplification of nodC and nodA genes, we used the primers nodCFn/nodCI and nodA1F/nodAb1r as previously described (Laguerre et al. 2001; Chantereuil et al., 2001). All the primers and amplification protocols used in this study are reported in Table 2S.
The amplification products were purified with the Qiaquick PCR purification kit (Qiagen), verified by electrophoresis in agarose gels and subjected to cycle sequencing using the same primers as for PCR amplification with ABI Prism dye chemistry. The products were analyzed with a 3130xl automatic sequencer at the sequencing facilities of UATRS (Unités d’Appui Technique à la Recherche Scientifique, CNRST, Rabat, Morocco) and Estación Experimental del Zaidín, CSIC, Granada, Spain. All the obtained sequences were compared with those from GenBank using the BLASTn program (Altschul et al. 1990) and the EzBiocloud.net (Yoon et al. 2017). Distances calculated according to Kimura’s two-parameter model (Kimura 1980) were used to infer phylogenetic trees with the neighbor-joining analysis (Saitou and Nei 1987) with MEGA7 software (Kumar et al. 2016). Identity values were calculated by pairwise analysis and gaps were not considered. The accession numbers of the nucleotide sequences of the bacterial strains used in this study are shown in the phylogenetic trees.
2.3 Plant nodulation tests
Selected strains were tested for nodulation of R. monosperma, R. dasycarpa, Lupinus luteus, Phaseolus vulgaris, Cicer arietinum, Vachellia gummifera, Glycine max and Vigna unguiculata. Seeds of lupine, V. gummifera, R. dasycarpa, and R. monosperma were surface-sterilized by immersion in 95% ethanol for 30 s, scarified using 95% sulfuric acid for 1, 60, and 120 min, respectively, and then washed several times with sterile water. Chickpea, soybean and cowpea seeds were surface-sterilized by adding 95% ethanol for 30 s, H2O2 15% for 8 min, and washed with sterile water. Finally, the seeds were placed in Petri dishes containing sterile water-agar (0.6%), and allowed to germinate in the dark at 28 °C for 2–4 d. The seedlings were then transferred to 200 × 20 mm Gibson tubes (1 seedling/tube) containing Jensen’s N-free mineral solution (Roughley 1984) and inoculated independently with the different bacterial suspensions (approximately 108 cells ml−1). Uninoculated plants were used as a control. A set of plants was grown in the same mineral solution supplemented with 0.05% KNO3. The plants were placed in a growth chamber at 26 °C under a 16.0/8.0 h light/dark photoperiod and checked for nodules appearance 8 weeks after inoculation. The Jensen solution was supplied just once, after 4 weeks culture.
2.4 Phenotypic characterization
Tests were performed using YEM medium inoculated with an exponentially growing liquid culture. The ability of the isolates to grow in acidic or basic media was determined on solid YEM whose pH had been adjusted and buffered to 5.5, 6.0, 7.0, 8.0, 9.0, 9.5 or 10.0, as described by Zerhari et al. (2000). The salt tolerance was tested at 0, 170, 340, 510, 680, or 850 mM of NaCl using solid YEM. Utilization of the amino acids lysine, methionine, histidine, glycine and glutamic acid at 1% as sole nitrogen source was investigated on a modified liquid YEM medium in which the yeast extract was replaced by the different amino acids. Carbohydrate assimilation was carried out on solid YEM containing 0.005% of yeast extract and 0.4% of glucose, fructose, sucrose, maltose, lactose or starch. The carbohydrates were sterilized separately by filtration. The intrinsic antibiotics resistance of the isolates was determined on solid YEM supplemented with the following antibiotics (μg/ml): kanamycin (20), ampicillin (20), bacitracin (20), tetracycline (10), vancomycin (20), streptomycin (20), nalidixic acid (20), and chloramphenicol (30). Hydrolysis of urea and nitrate reduction by the rhizobial isolates was investigated on solid YEM amended with 2% urea, 0.0012% phenol red and 0.1% KNO3, respectively, as described by Lindström and Lehtomäki (1988). The gelatinase activity was investigated as reported by Missbah El-Idrissi et al. (1996), and the catalase activity was determined by the method of Graham and Parker (1964).
2.5 PGPR characteristics
2.5.1 Phosphate solubilization
The ability of the isolates for phosphate solubilization was tested using PVK medium (Pikovskaya 1948) containing 2.5 g l−1 of (Ca3PO4)2. Aliquots (5 μl) of the bacterial suspension were plated onto PVK medium in Petri dishes and incubated at 28 °C for 7 d. The ability of the isolates to solubilize phosphate was estimated by measuring the size of the halos that appeared around the colonies and the size of the colonies.
2.5.2 Siderophore production
Siderophore production was detected according to Lakshmanan et al. (2015). Isolates synthesizing siderophores were identified by the development of orange halos around the colonies.
2.5.3 Indole acetic acid production
The ability of the isolates to produce indol acetic acid (IAA) and IAA-related compounds was determined on solid YEM medium supplemented with 0.5 g l−1 tryptophan. Drops (5 μl) of the bacterial suspension were placed on the medium and incubated at 28 °C for 24 to 48 h. The production of IAA was detected using the Salkowski reagent followed by the appearance of a pink halo around the colonies after incubation for 30 to 60 min at 28 °C.
3 Results
The sampling site soils are composed of clay (3.15%), fine silt (3.03%), coarse silt (3.30%), fine sand (30.36%), and coarse sand (60.23%). The soil at the sampling site is neutral to slightly alkaline with a mean pH value of 7.3 ± 0.1, and an electrical conductivity of 143.45 s cm−1; an average annual precipitation of 500 mm, and 18 °C as mean annual temperature (Benabid and Fennane 1994).
A total of 30 bacterial isolates were obtained from root nodules of R. sphaerocarpa grown wild in the Maamora forest, in the vicinity of Rabat (Morocco). The colonies were fast growing and slightly pinkish after 4 days growth on YEM medium supplemented with Congo Red. After ERIC-PCR, the 30 strains distributed into 2 different groups (Fig. S1), of which the strains RSM25 and RSM32 were randomly chosen as the representative strains of each I and II groups, respectively. Members of the group I were all similar; as were the isolates of group II, which shows the high clonality of the isolates. Based on results of the Enterobacterial repetitive intergenic consensus -polymerase chain reactions (ERIC-PCR) the strains RSM25 and RSM32 were used for further genotypic studies.
3.1 16S rRNA, dnak, and gyrB genes phylogenetic analysis
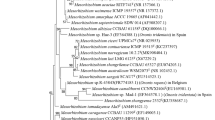
The analysis of the almost complete 16S rRNA gene sequence of the strains RSM25 and RSM32 showed that they were members of the genus Microvirga of the family Methylobacteriaceae of the Alphaproteobacteria, and the corresponding phylogenetic tree (Fig. 1) showed that the strains clustered with had “M. tunisiensis” LmiM8T with similarity values of 99.85 and 99.84%, respectively, and 98.33 and 98.20% with Microvirga makkahensis SV1470T, respectively. PCR amplification of the atpD and gyrB genes of the strains RSM25 and RSM32 gave DNA bands that were sequenced, and their concatenation confirmed they cluster with M. tunisiensis LmiM8T (Fig. 2), with which they shared 97.27 and 97.99% similarity, respectively. Phylogenetic trees based on individual atpD (Fig. 2S) and gyrB (Fig. 3S) gave similar results to those obtained for the concatenated tree, with no further insights into the affiliation of the isolates.
Neighbor-joining phylogenetic tree based on partial 16S rRNA gene sequences of the strains RSM25 and RSM32 (in bold) from nodules of R. sphaerocarpa and phylogenetically related type strains of the genus Microvirga. Bootstrap values are indicated as percentages derived from 1000 replications. Scale bar indicates substitutions per site. The tree is rooted with B. japonicum USDA6T
Neighbor-joining phylogenetic tree based on the concatenated partial atpD + gyrB gene sequences of the strains RSM25 and RSM32 (in bold) from nodules of R. sphaerocarpa and phylogenetically related Microvirga type species. Bootstrap values are indicated as percentages derived from 1000 replications. Scale bar indicates substitutions per site. The tree is rooted with B. japonicum LMG 6138T
3.2 Phylogenic analysis of the nodA and nodC genes
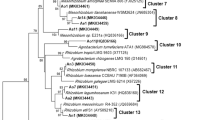
Utilization of the primer pair nodCF/nodCI resulted in amplification of the nodC gene from the strains RSM25 and RSM32. The phylogenetic tree (Fig. 3) built with the obtained sequences and those of the nodC genes from other Microvirga species as well as other Retama nodulating rhizobia such as B. retamae Ro19T, B. algeriense RST91T and E. aridi strains RMT3 and RMT46, revealed that RSM25 and RSM32 grouped with M. tunisiensis LmiM8T, with similarity values of 90.46 and 92.04%, respectively.
Neighbor-joining phylogenetic tree based on partial nodC sequences of the strains RSM25 and RSM32 (in bold) from nodules of R. sphaerocarpa and phylogenetically related species of the genus Microvirga. Bootstrap values are indicated as percentages derived from 1000 replications. Scale bar indicates substitutions per site. The tree is rooted with Cupriavidus taiwanensis STM6018
RSM25 and RSM32 have similarity percentages of 82.1 to 82.8% with Ensifer aridi strains RMT3 and RMT46; while the similarity percentages with B. retamae and B. algeriense vary between 83.4 and 83.8 (Table 3S).
Similar results were obtained with nodA gene, and the phylogenetic tree (Fig. 4S) revealed also that the two strains RSM25 and RSM32 grouped with M. tunisiensis LmiM8T, with similarity values of 95.20 and 95.10% respectively.
Based on the nodA, nodC sequences and their concatenation, Figs. 3, 4S and 5S also show that the strains isolated from R. sphaerocarpa belong to the symbiovar mediterranense. The type strains of B. retamae and B. algeriense clustered together with B. valentinum in the symbiovar retamae, while Ensifer aridi strains RM3 and RMT46 were very close to E. aridi strain JNVU TP6.
3.3 Plant nodulation tests
The two strains RSM25 and RSM32 nodulated not only R. sphaerocarpa, but also R. monosperma, R. dasycarpa and L. luteus, and were unable to form nodules on P. vulgaris, C. arietinum, V. gummifera, G. max and V. unguiculata. Under the conditions used in this study, the inoculation of yellow retama with strains RSM25 and RSM32 produced a 1.22-, and 1.36-fold increase in the dry weight of the plants compared to those grown in the presence of 0.05% KNO3 (Table 2).
3.4 Phenotypic characterization
Phenotypic characteristics of the strains RSM25 and RSM32 are presented in Table 1S. The two strains were fast growing with an average generation time of 3 and 5 d to give colonies on solid YEM medium with a diameter higher than 2 mm, respectively. They grew well at 3% NaCl, 35 °C, and within a pH range from 6.5 to 9.0. The two Microvirga strains grow in YEM medium with pH values ranging from 6.5 to 9, and at 3% NaCl and produce catalase and urease. Out of the 14 C sources tested in this study, the strains RSM25 and RSM32 were unable to use arabinose, galactose, fructose, starch and glucose. They grew in the presence of arginine, tyrosine, leucine, aspartic acid, tryptophan, histidine, valine, lysine, and phenylalanine as the only N source, and no strain grew on glycine. They are resistant to ampicillin, kanamycin, bacitracin, tetracycline, streptomycin, vancomycin, and nalidixic acid.
3.5 PGPR activities
Neither siderophore production nor phosphate solubilization by strains RSM25 and RSM32 was observed; however, they were able to produce IAA (Table 1S).
4 Discussion
Although previous work has established that Bradyrhizobium is the dominant genus found in nodules of R. sphaerocarpa, rhizobial strains of Phyllobacterium and Ensifer have also been isolated (Ruiz-Díez et al. 2009; Guerrouj et al. 2013; Ahnia et al. 2018; Lamin et al. 2019). In this work we extend those findings by showing that M. tunisiensis is the most abundant microsymbiont in nodules of R. sphaerocarpa growing in soils of the Maamora forest, where, together with legumes of the genera Teline and Cytisus, and various species of the Poaceae, they are overgrazed by an abundant livestock. The sampling sites were located in the cool and temperate sub-humid bio-climate western part of this cork oak ecosystem whose climatic conditions contrast with those of the arid areas of eastern Morocco and the Middle and High-Atlas mountains where the yellow retama thrives spontaneously under hard weather conditions with low and random rainfall as well as very cold winters and hot summers.
In this study 30 bacterial strains were isolated from nodules of R. sphaerocarpa whose overall diversity was determined by rep-PCR using ERIC primers, a technique that has proven useful to avoid clonality as it discriminates among the molecular diversity of the isolates at the intra-species level (Versalovic et al. 1994). ERIC-PCR fingerprinting of the isolates allowed the selection of strains RSM25 and RSM32 that were chosen for further genetic identification.
The phylogenetic analysis of the 16S rDNA sequences and of the combined housekeeping genes atpD and gyrB revealed that the strains RS25 and RSM32 are related to M. tunisiensis isolated from L. micranthus grown in Northern Tunisia (Msaddak et al. 2017b). Members of Microvirga were also obtained from nodules of L. cosentinii from the Moroccan Maamora forest (Missbah El-Idrissi et al. 2019), and both R. sphaerocarpa and L. cosentinii can be found in the sub-humid part of the Maamora forest from which Microvirga strains have been isolated. Soil structure, pH, salinity, phosphate concentration, moisture and type of soil play a key role in the bacterial communities’ biogeography and selection of symbionts by legumes (Han et al. 2009; Rathi et al. 2018; Fan et al. 2018; Wang et al. 2019). The Maamora forest sandy soils are generally acidic under the cork oak trees, with pH values ranging from 4.8 to 6 (El Boukhari et al. 2016). However, in clear areas and under some legumes shrubs such as Retama and Teline, soils have neutral or slightly alkaline pH (El Boukhari et al. 2016; Belghazi and Mounir 2016; Gaddas 1976; Laouina et al. 1997). The presence of Microvirga in root nodules of plants growing on the slightly alkaline soils of the Maamora agrees with previous reports on Microvirga strains isolated from L. micranthus, L. luteus and L. angustifolius in alkalinized soils of Tunisia (Msaddak et al. 2017a, b; Rejili et al. 2019) and from L. texensis and L. subcarnosus grown in soils with pH varying from 7.9 to 8.4 in the United States (Beligala et al. 2017). The soils used in this study are slightly alkaline reddish sandy soils and it is therefore very likely that the pH played a role in the selection of Microvirga as microsymbionts of Retama instead of members of the genera Bradyrhizobium or Ensifer as reported previously (Guerrouj et al. 2013; Ahnia et al. 2018; Lamin et al. 2019). All these results support that the soil pH could be a key factor for determination of the biogeography of the Microvirga species.
Symbiovars are generally used to distinguish between different subgroups that are symbiotically distinct within a single legume-nodulating species (Rogel et al. 2011), and they are defined mainly on the basis of nod genes involved in legume nodulation.
Since Retama is nodulated by members of three different genera, this would suggest that a horizontal transfer of nodulation genes would have occurred between these bacteria. However, analyzes of the three genera nodC nodulation genes sequences and their phylogeny showed that strains RSM25 and RSM32 cluster together with Microvirga tunisiensis LmiM8T (Msaddak et al. 2019) and P. sophorae LmiT21 (Msaddak et al. 2018) in the mediterranense symbiovar, while B. retamae and B. algeriense clustered together with B. valentinum in the symbiovar retamae. The two strains of Ensifer aridi isolated from Retama monosperma in Morocco were closer to E. aridi strain JNVU TP6 isolated from Tephrosia purpurea in India.
These results show the aptitude of Retama to interact a wide range of rhizobia with different types of Nod factors. This ability contributes to the great resilience of the plant, which can develop in different agroecosystems and fix nitrogen under different conditions. It can thus establish a symbiosis with Ensifer aridi under metallic stress (Lamin et al. 2019), or with B. retamae (Guerrouj et al. 2013) or B. algeriense (Ahnia et al. 2018) under arid conditions, and it could associate with Microvirga under milder conditions of climate and soil pH.
To our knowledge, no strain of Microvirga has been previously reported to be a R. sphaerocarpa endosymbiont. Coincident with other Microvirga strains isolated from Lupinus (Ardley et al. 2012; Msaddak et al. 2017a, b; Rejili et al. 2019), the strains RSM25 and RSM32 also formed effective nodules on L. luteus.
Each strain RSM25 and RSM32 nodulated R. sphaerocarpa, R. monosperma, and R. dasycarpa. Moreover, the two strains established effective symbiosis with their original host, and the inoculation with any of them produced clear increases in the dry weight of the plants after growth for two months under axenic conditions (Table 1).
The strains RSM25, and RSM32 used a variety of carbohydrates as the sole C source and no strain used glycine for growth, a characteristic reported by Guerrouj et al. (2013) for all of the strains isolated from R. monosperma and R. sphaerocarpa.
Plant growth-promoting rhizobacteria (PGPR) have a favorable effect on plant growth and tolerance against biotic and abiotic stresses. In addition to supplying N to compatible host plants, legume-nodulating bacteria may also act as a crop enhancer and biofertilizer for increased non-legume production (Zaim et al. 2017; Patil et al. 2017; Vargas et al. 2017). However, the strains RSM25 and RSM32 were unable to produce siderophores or solubilize phosphate, but they produced IAA or IAA-related compounds derived from the metabolism of tryptophan.
The results in this study enlarge the scientific information about the diversity of bacteria associated with species of the genus Retama in Morocco by adding Microvirga to the previously reported species capable of nodulating yellow retama such as Bradyrhizobium, Ensifer, and Phyllobacterium. This, in turn, supports the idea that host plant promiscuity could be an adaptation strategy to grow in diverse ecosystems, with different types of soils and bioclimatic conditions and could aid the future development of bioinocula for use in sustainable agrosystems.
References
Aafi A, Achhal EL Kadmiri A, Benabid A, Rouchdi M (2005) Richesse et diversité floristique de la subéraie de la Mamora (Maroc). Acta Bot Mal 30:127–138. https://doi.org/10.24310/abm.v30i0.7187
Ahnia H, Bourebaba Y, Durán D, Boulila F, Palacios JM, Rey L, Ruiz-Argüeso T, Boulila A, Imperial J (2018) Bradyrhizobium algeriense sp. nov., a novel species isolated from effective nodules of Retama sphaerocarpa from northeastern Algeria. Syst Appl Microbiol 41(4):333–339. https://doi.org/10.1016/j.syapm.2018.03.004
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215:403–410
Ardley JK, Parker MA, De Meyer SE, Trengove RD, O'Hara GW, Reeve WG, Yates RJ, Dilworth MJ, Willems A, Howieson JG (2012) Microvirga lupini sp. nov., Microvirga lotononidis sp. nov. and Microvirga zambiensis sp. nov. are alphaproteobacterial root nodule bacteria that specifically nodulate and fix nitrogen with geographically and taxonomically separate legume hosts. Int J Syst Evol Microbiol 62:2579–2588. https://doi.org/10.1099/ijs.0.035097-0
Armas C, Rodríguez-Echeverría S, Pugnaire FI (2011) A field test of the stress-gradient hypothesis along an aridity gradient. J Veg Sci 22:818–827
Belghazi B, Mounir F (2016) Analyse de la vulnérabilité au changement climatique du couvert forestier. Forêt de la Maâmora (Maroc). Edit. FAO, rapport technique, 124 p
Beligala DH, Michaels HJ, Devries M, Phuntumart V (2017) Multilocus sequence analysis of root nodule Bacteria associated with Lupinus spp. and Glycine max. Adv Microbiol 7(11):790–812. https://doi.org/10.4236/aim.2017.711063
Benabid A, Fennane M (1994) Connaissances sur la végétation du Maroc. Phytogéographie, phytosociologie et séries de végétation. Lazaroa 14:21–97
Boulila F, Depret G, Boulila A, Belhadi D, Benallaoua S, Laguerre G (2009) Retama species growing in different ecological-climatic areas of northeastern Algeria have a narrow range of rhizobia that form a novel phylogenetic clade within the Bradyrhizobium genus. Syst Appl Microbiol 32:245–255. https://doi.org/10.1016/j.syapm.2009.01.005
Brenner DJ, Mcwhorter AC, Leete Knutson JK, Steigerwalt AG (1982) Escherichia vulneris a new species of Enterobacteriaceae associated with human wounds. J Clin Microbiol 15:1133–1140
de Lajudie P, Andrews M, Ardley J, Eardly B, Jumas-Bilak E, Kuzmanović N, Lassalle F, Lindström K, Mhamdi R, Martínez-Romero E, Moulin L, Abdollah Mousavi S, Nesme X, Peix A, Puławska J, Steenkamp E, Stępkowski T, Tian CF, Vinuesa P, Wei G, Willems A, Zilli J, Young P (2019) Minimal standards for the description of new genera and species of rhizobia and agrobacteria. Int J Syst Evol Microbiol 69:1852–1863. https://doi.org/10.1099/ijsem.0.003426
El Boukhari EM, Brhadda N, Gmira N (2016) Contribution à l’étude de la régénération artificielle du chêne liège (Quercus suber L.) vis-à-vis du contenu minéral des feuilles et des paramètres physicochimiques des sols de la Maâmora (Maroc). Nature Technol 14:26–23
Estrada-de los Santos P, Palmer M, Steenkamp ET, Maluk M, Beukes C, Hirsch AM, James EK, Venter SN (2018) Trinickia dabaoshanensis sp. nov., a new name for a lost species. Arch Microbiol 201(9):1313–1316
Fan K, Weisenhorn P, Gilbert JA, Shi Y, Bai Y, Chu H (2018) Soil pH correlates with the co-occurrence and assemblage process of diazotrophic communities in rhizosphere and bulk soils of wheat fields. Soil Biol Biochem 121:185–192. https://doi.org/10.1016/j.soilbio.2018.03.017
Gaddas RR (1976) Aménagement et Amélioration des Parcours, Royaume du Maroc. Les Sols de la Mamora et du Rif Occidental, (zones caractéristiques d'intervention). Document de Travail
Graham PH, Parker CA (1964) Diagnostic features in the characterization of the root nodule bacteria of legumes. Plant Soil 20:338–396
Guerrouj K, Ruíz-Díez B, Chahboune R, Ramírez-Bahena MH, Abdelmoumen H, Quiñones M, Missbah El Idrissi M, Velázquez E, Fernández-Pascual M, Bedmar EJ, Peix A (2013) Definition of a novel symbiovar (sv. retamae) within Bradyrhizobium retamae sp. nov., nodulating Retama sphaerocarpa and Retama monosperma. Syst Appl Microbiol 36:218–223. https://doi.org/10.1016/j.syapm.2013.03.001
Han LL, Wang ET, Han TX, Liu J, Sui XH, Chen WF, Chen WX (2009) Unique community structure and biogeography of soybean rhizobia in the saline-alkaline soils of Xinjiang, China. Plant Soil 324:291–305. https://doi.org/10.1007/s11104-009-9956-6
Hannane FZ, Kacem M, Kaid-Harche M (2014) Retama-rhizobia symbiosis studies in some countries of the Mediterranean Basin. Ecol Medit 40:5–18
Kimura M (1980) A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 16(2):111–120. https://doi.org/10.1007/BF01731581
Kumar S, Stecher G, Tamura K (2016) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33(7):1870–1874. https://doi.org/10.1093/molbev/msw054
Laguerre G, Nour SM, Macheret V, Sanjuan J, Drouin P, Amarger N (2001) Classification of rhizobia based on nodC and nifH gene analysis reveals a close phylogenetic relationship among Phaseolus vulgaris symbionts. Microbiol 147:981–993
Lakshmanan V, Shantharaj D, Li G, Seyfferth AM, Sherrier DJ, Bais HP (2015) A natural rice rhizospheric bacterium abates arsenic accumulation in rice (Oryza sativa L.). Planta 242:1037–1050. https://doi.org/10.1007/s00425-015-2340-2
Lamin H, Alami S, Bouhnik O, ElFaik S, Abdelmoumen H, Bedmar EJ, Missbah-El Idrissi M (2019) Nodulation of Retama monosperma by Ensifer aridi in an Abandonned Lead mine soils in eastern Morocco. Front Microbiol 10:1456. https://doi.org/10.3389/fmicb.2019.01456
Laouina A, Nafaa R, Wtfeh A (1997) Occupation des sols et dégradation des terres, le cas de la Mamora et de ses bordures. Méditerranée, vol. 86, n° 1–2 ("Impact anthropique en milieu méditerranéen"). p. 45–51. https://doi.org/10.3406/medit.1997.2989
León-González AJ, Navarro I, Acero N, Muñoz Mingarro D, Martín-Cordero C (2018) Genus Retama: a review on traditional uses, phytochemistry, and pharmacological activities. Phytochem Rev 17:701–731. https://doi.org/10.1007/s11101-018-9555-3
Lindström K, Lehtomäki S (1988) Metabolic properties, maximum growth temperature and phage sensitivity of Rhizobium sp. (Galega) compared with other fast-growing rhizobia. FEMS Microbiol Let 50:277–287
Martens M, Delaere M, Coopman R, De Vos P, Gillis M, Willems A (2007) Multilocus sequence analysis of Ensifer and related taxa. Int J Syst Evol Microbiol 57:489–503
Menna P, Barcellos FG, Hungria M (2009) Phylogeny and taxonomy of a diverse collection of Bradyrhizobium strains based on multilocus sequence analysis of the 16S rRNA gene, ITS region and glnII, recA, atpD and dnaK genes. Int J Syst Evol Microbiol 59:2934–2950
Missbah El-Idrissi M, Aujjar N, Belabed A, Dessaux Y, Filali-Maltouf A (1996) Characterization of rhizobia isolated from carob tree (Ceratonia siliqua). J Appl Bacteriol 80(2):165–173. https://doi.org/10.1111/j.1365-2672.1996.tb03205.x
Missbah El-Idrissi M, Lamin H, ElFaik S, Tortosa G, Peix A, Bedmar EJ, Abdelmoumen H (2019) Microvirga sp. symbiovar mediterranense nodulates Lupinus cosentinii grown wild in Morocco. J Appl Microbiol 128(4):1109–1118. https://doi.org/10.1111/jam.14526
Msaddak A, Rejili M, Durán D, Rey L, Imperial J, Palacios J, Ruiz-Argüeso T, Mars M (2017a) Members of Microvirga and Bradyrhizobium genera are native endosymbiotic bacteria nodulating Lupinus luteus in northern Tunisian soils. FEMS Microbiol Ecol 93. https://doi.org/10.1093/femsec/fix068
Msaddak A, Durán D, Rejili M, Mars M, Ruiz-Argüeso T, Imperial J, Palacios JM, Rey L (2017b) Diverse Bacteria affiliated with the genera Microvirga, Phyllobacterium, and Bradyrhizobium Nodulate Lupinus micranthus growing in soils of northern Tunisia. Appl Environ Microbiol 83(6). https://doi.org/10.1128/AEM.02820-16
Msaddak A, Rejili M, Durán D, Rey L, Palacios JM, Imperial J, Ruiz-Argüeso T, Mars M (2018) Definition of two new symbiovars, sv. lupini and sv. mediterranense, within the genera Bradyrhizobium and Phyllobacterium efficiently nodulating Lupinus micranthus in Tunisia. Syst Appl Microbiol 41(5):487–493
Msaddak A, Rejili M, Durán D, Mars M, Palacios JM, Ruiz-Argüeso T, Imperial J (2019) Microvirga tunisiensis sp. nov., a root nodule symbiotic bacterium isolated from Lupinus micranthus and L. luteus grown in northern Tunisia. Syst Appl Microbiol 42(6):126015. https://doi.org/10.1016/j.syapm.2019.126015
Patil A, Kale A, Ajane G, Sheikh R, Patil S (2017) Plant growth-promoting rhizobium: mechanisms and biotechnological prospective. In: Hansen A, Choudhary D, Agrawal P, Varma A (eds) Rhizobium Biology and Biotechnology. Soil Biology, vol 50. Springer, Cham. https://doi.org/10.1007/978-3-319-64982-5_7
Pikovskaya RI (1948) Mobilization of phosphorus in soil in connection with the vital activity of some microbial species. Mikrobiologiya 17:362–370
Prieto I, Kikvidze Z, Pugnaire FI (2010) Hydraulic lift: soil processes and transpiration in the Mediterranean leguminous shrub Retama sphaerocarpa (L.) Boiss. Plant Soil 329:447–456. https://doi.org/10.1007/s11104-009-0170-3
Rathi S, Tak N, Bissa G, Chouhan B, Ojha A, Adhikari D, Barik SK, Satyawada RR, Sprent JI, James EK, Gehlot HS (2018) Selection of Bradyrhizobium or Ensifer symbionts by the native Indian caesalpinioid legume Chamaecrista pumila depends on soil pH and other edaphic and climatic factors. FEMS Microbiol Ecol 94(11):fiy180. https://doi.org/10.1093/femsec/fiy180
Rejili M, Msaddak A, Filali I, Benabderrahim MA, Mars M, Marín M (2019) New chromosomal lineages within Microvirga and Bradyrhizobium genera nodulate Lupinus angustifolius growing on different Tunisian soils. FEMS Microbial Ecol 95(9):fiz118. https://doi.org/10.1093/femsec/fiz118
Rodríguez-Echeverría S, Pérez-Fernández MA (2003) Soil fertility and herb facilitation mediated by Retama sphaerocarpa. J Veg Sci 14(6):807–814. https://doi.org/10.1111/j.1654-1103.2003.tb02213.x
Rodríguez-Echeverría S, Pérez-Fernández MA (2005) Potential use of Iberian shrubby legumes and rhizobia inoculation in revegetation projects under acidic soil conditions. Appl Soil Ecol 29(2):203–208. https://doi.org/10.1016/j.apsoil.2004.11.004
Rogel MA, Ormeño-Orrillo E, Martinez Romero E (2011) Symbiovars in rhizobia reflect bacterial adaptation to legumes. Syst Appl Microbiol 34:96–104
Roughley RJ (1984) Effect of Soil Environmental Factor on Rhizobia. In R. Shiblies (ed.) Proceedings of World Soybean Research Conference III. August 12-17 1984. Westview Press London. 903-910
Ruiz-Díez B, Fajardo S, Puertas-Mejía MA, del Rosario de Felipe M, Fernández-Pascual M (2009) Stress tolerance, genetic analysis and symbiotic properties of root-nodulating bacteria isolated from Mediterranean leguminous shrubs in Central Spain. Arch Microbiol 191(1):35–46. https://doi.org/10.1007/s00203-008-0426-y
Ruiz-Díez B, Quiñones MA, Fajardo S, López MA, Higueras P, Fernández-Pascual M (2012) Mercury-resistant rhizobial bacteria isolated from nodules of leguminous plants growing in high Hg-contaminated soils. Appl Microbiol Biotechnol 96:543–554
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Vargas LK, Volpiano CG, Lisboa BB, Giongo A, Beneduzi A, Passaglia LMP (2017) Potential of Rhizobia as Plant Growth-Promoting Rhizobacteria. In A. Zaidi et al. (eds.), Microbes for Legume Improvement, Springer International Publishing AG 2017. https://doi.org/10.1007/978-3-319-59174-2_7
Versalovic J, Koeuth T, Lupski R (1991) Distribution of repetitive DNA sequences in eubacteria and application to fingerprinting of bacterial genomes. Nucleic Acids Res 19:6823–6831
Versalovic J, Schneider M, de Bruijn FJ, Lupski JR (1994) Genomic fingerprinting of bacteria using repetitive sequence-based Polymerase Chain Reaction. Methods Mol Cell Biol 5:25–40
Vincent JM (1970) A manual for the practical study of the root-nodule Bacteria. Blackwell Scientific Publications, Oxford
Vinuesa P, Silva C, Werner D, Martínez-Romero E (2005) Population genetics and phylogenetic inference in bacterial molecular systematics: the roles of migration and recombination in Bradyrhizobium species cohesion and delineation. Mol Phylogenet Evol 34:29–54. https://doi.org/10.1016/j.ympev.2004.08.020
Wang C, Zhou X, Guo D, Zhao JH, Yan L, Feng GZ, Gao Q, Yu H, Zhao LP (2019) Soil pH is the primary factor driving the distribution and function of microorganisms in farmland soils in northeastern China. Ann Microbiol 69:1461–1473. https://doi.org/10.1007/s13213-019-01529-9
Weisburg WG, Barns SM, Pelletier DA, Lane DJ (1991) 16S ribosomal DNA amplification for phylogenetic study. J Bacteriol 173(2):697–703
Yoon S-H, Ha S-M, Kwon S, Lim J, Kim Y, Seo H, Chun J (2017) Introducing EzBioCloud: a taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int J Syst Evol Microbiol 67(5):1613–1617
Zaim S, Bekkar AA, Belabid L (2017) Rhizobium as a Crop Enhancer and Biofertilizer for Increased Non-legume Production. A.P. Hansen et al. (eds.), Rhizobium Biology and Biotechnology, Soil Biology 50, Springer International Publishing AG 2017. ISBN978-3-319-64981-8. https://doi.org/10.1007/978-3-319-64982-5_4
Zerhari K, Aurag J, Khbaya B, Kharchaf D, Filali-Maltouf A (2000) Phenotypic characteristics of rhizobia isolates nodulating Acacia species in the arid and Saharan regions of Morocco. Lett Appl Microbiol 30(5):351–357. https://doi.org/10.1046/j.1472-765x.2000.00730.x
Zohary M (1959) A revision of the genus Retama ( Boiss). Bull Res Counc Isr 7(D):1–2
Acknowledgements
The authors want to thank the Hassan II Academy of Sciences for funding this research in the frame of the BIOMICS project. Financial support was also obtained from the ERDF-cofinanced project AGL2017–85676R from Ministerio de Economía, Industria y Competitividad (Spain).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare there is no conflict of interest.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
ESM 1
(DOCX 184 kb)
Rights and permissions
About this article
Cite this article
Mouad, L., Hanane, L., Omar, B. et al. Nodulation of Retama species by members of the genus Microvirga in Morocco. Symbiosis 82, 249–258 (2020). https://doi.org/10.1007/s13199-020-00725-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13199-020-00725-5