Abstract
Pochonia chlamydosporia (Pc123) is a fungal parasite of nematode eggs which can colonize endophytically barley and tomato roots. In this paper we use culturing as well as quantitative PCR (qPCR) methods and a stable GFP transformant (Pc123gfp) to analyze the endophytic behavior of the fungus in tomato roots. We found no differences between virulence/root colonization of Pc123 and Pc123gfp on root-knot nematode Meloidogyne javanica eggs and tomato seedlings respectively. Confocal microscopy of Pc123gfp infecting M. javanica eggs revealed details of the process such as penetration hyphae in the egg shell or appressoria and associated post infection hyphae previously unseen. Pc123gfp colonization of tomato roots was low close to the root cap, but increased with the distance to form a patchy hyphal network. Pc123gfp colonized epidermal and cortex tomato root cells and induced plant defenses (papillae). qPCR unlike culturing revealed reduction in fungus root colonization (total and endophytic) with plant development. Pc123gfp was found by qPCR less rhizosphere competent than Pc123. Endophytic colonization by Pc123gfp promoted growth of both roots and shoots of tomato plants vs. uninoculated (control) plants. Tomato roots endophytically colonized by Pc123gfp and inoculated with M. javanica juveniles developed galls and egg masses which were colonized by the fungus. Our results suggest that endophytic colonization of tomato roots by P. chlamydosporia may be relevant for promoting plant growth and perhaps affect managing of root-knot nematode infestations.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Plant parasitic nematodes are responsible for global agricultural losses amounting to an estimated $157 billion annually (Abad et al. 2008). The root-knot nematodes (Meloidogyne spp.) seriously affect many economically important agricultural crops worldwide (Wesemael et al. 2010).
Currently, soil fumigants and nematicides are used to control nematodes. However, the need to reduce dependence on nematicides, imposed by legislation and consumers, requires the development of new management strategies. Biological control is an important tool for plant-parasitic nematode management (Sorribas et al. 2003).
Nematophagous fungi are a diverse group of antagonists of nematodes which infect, kill and digest their hosts (Nordbring-Hertz et al. 2006). Some nematophagous fungi have been detected in the rhizosphere of important crops (Persmark and Jansson 1997), yet others colonize plant roots endophytically (Lopez-Llorca et al. 2006). The advantage of endophytism is that the endophyte occurs in the same ecological niche as the endoparasitic nematodes but is not subject to competition from microorganisms in the soil (Stirling 2011).
Pochonia chlamydosporia (Goddard) Zare and Gams is a parasite of female nematodes and eggs has been studied as a biological control agent due to its worldwide distribution. P. chlamydosporia has been isolated from economically important species of plant pathogens such as root-knot (Meloidogyne spp.) (Hidalgo-Díaz et al. 2000) or cyst nematodes (Heterodera spp., Globodera spp.) (Kerry and Crump 1977). Rhizospheres of crops of economic interest, such as tomato and barley, are also colonized by P. chlamydosporia (Bordallo et al. 2002; Maciá-Vicente et al. 2009a).
Endophytic colonization of roots by P. chlamydosporia has a number of benefits to the host plant, such as growth promotion or protection against different pathogens, such as nematodes and fungi (Maciá-Vicente et al. 2009b; Monfort et al. 2005; Siddiqi and Akhtar 2008). Therefore, studying root colonization by P. chlamydosporia in crops such as tomato is essential to assess the possibility of their improved use as a biological control agent of root-knot nematodes. For instance, the effect of fungus development on roots in nematode invasion, further multiplication and damage to crops is yet unknown (Kerry and Bourne 2002). A main problem with biological control agents is their detection and quantification in soil. Real-time or quantitative PCR (qPCR) provides a tool to assess population dynamics of particular biological control agents in soil (Stirling 2011). Likewise, the use of GFP-like fluorescent proteins as living cell markers (Wiedenmann et al. 2009) has allowed the study in-vivo of the mode of action of the organisms transformed with these genes. This technology has been applied to entomopathogenic (Barelli et al. 2011; Inglis et al. 2000; Kurtti and Keyhani 2008) and nematophagous fungi (Zhang et al. 2008).
We have recently developed a P. chlamydosporia GFP-tagged transformant (Maciá-Vicente et al. 2009a) to visualize the endophytic colonization of barley roots inoculated with the fungus. Besides, the endophytic capacity could be an advantage for the fungus to establish in agricultural systems and thus help their antagonism against nematodes. In this study we have inoculated endophytically P. chlamydosporia GFP strain in tomato seedlings (Solanum lycopersicum Mill cv. durinta) evaluating its root endophytism using culturing, microscopical and molecular techniques. We have then evaluated the pathogenicity (as percentage nematode egg-infection) of P. chlamydosporia GFP vs. that of the corresponding parent strain. The effect of root endophytic inoculation of P. chlamydosporia GFP on root-knot nematode M. javanica second-stage juveniles (J2) invasion and subsequent development was evaluated. Finally, fungus colonization of nematode galls and egg masses was also assessed.
2 Materials and methods
2.1 Fungi, nematodes and plant materials
P. chlamydosporia used in this work was the wild type Pc123 (ATCC No. MYA-4875) (Olivares-Bernabeu and Lopez-Llorca 2002), and a GFP-tagged isolate (Pc123gfp) (Maciá-Vicente et al. 2009a). Root-knot nematodes (RKN) M. javanica were from a field population (Almeria, SE Spain) PCR identified as in Zijlstra et al. (2000). RKN populations were maintained in susceptible tomato plants (Solanum lycopersicum Mill. cv. durinta). Nematode egg masses were dissected from RKN-infested roots and kept at 4 ºC until used. Egg masses were hand-picked and surface-sterilized as in McClure et al. (1973) with slight modifications. M. javanica second-stage juveniles (J2) were hatched from surface-sterilized eggs at 28 ºC in the dark.
2.2 Parasitism of root-knot nematode eggs by Pochonia chlamydosporia
Surface-sterilized nematode eggs were placed in Petri dishes (90 mm diameter) containing 1 % water agar (Becton and Dickinson and Company, Le Pont de Claix, France) with 50 mg ml-1 Penicillin and 50 mg ml-1 Ampicillin (Sigma, St Louis, MO, USA). Each egg was inoculated with 10 μl of a 106 conidia ml-1 suspension of either Pc123 or Pc123gfp (Lopez-Llorca & Claugher, 1990). Plates were incubated at 25 ºC in the dark and fungal infection of individual eggs scored daily for a four-day period. Eggs developing fungal colonies were scored as infected and the percentage of egg-infection was then calculated. Control plates were made with eggs without conidia. The experiment was carried out twice.
Infection of eggs by Pc123gfp was monitored microscopically every day in ten randomly selected infected eggs. GFP fluorescence emission was visualized in a Leica TCS-SP2 laser-scanning confocal microscope. Samples were excited with a 488 nm laser, GFP fluorescence was detected at 505-530 nm and eggs autofluorescence was detected at 580-620 nm.
2.3 Inoculation of tomato seedlings with Pochonia chlamydosporia
Surface-sterilized tomato seeds were plated on germination medium and incubated at 25 ºC in dark for 7 days (Bordallo et al. 2002). Seedlings free from contaminants were inoculated by plating them on Petri dishes (5 per dish) with 21 day-old colonies of either Pc123 or Pc123gfp on corn meal agar (CMA), or without fungal colonies for controls. To determine the optimum inoculation time, inoculations were performed for 1, 2, 3, 5 or 7 days (Monfort et al. 2005) at 25 ºC under a photoperiod of 16:8 h (light:dark). The experiment consisted of 10 replicates/treatment in the first repetition and 20 replicates/treatment in the second one.
2.4 Distribution and quantification of Pochonia chlamydosporia in tomato roots
Root culturing was used to evaluate the inoculation of P. chlamydosporia (e.g. detecting the fungus) in tomato seedling roots. P. chlamydosporia total and root endophytic colonization were analyzed (Maciá-Vicente et al. 2009a).
The spatial pattern of root colonization by Pc123gfp was assessed with laser-scanning confocal microscopy. Ten fragments/root system (5 to 10 mm long) were examined in a Leica DM IRBE2 confocal microscope. GFP fluorescence and root cell autofluorescence were detected as described in Maciá-Vicente et al. 2009a.
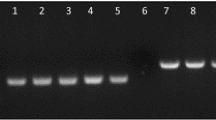
Further detection of P. chlamydosporia in roots was performed by PCR. For this purpose, DNA was extracted from roots of 10 seedlings/treatment (non-sterilized or surface-sterilized) as in Lopez-Llorca et al. (2010). Primers for PCR amplification of tomato PR1 gene (Gayoso et al. 2007), P. chlamydosporia VCP1 gene (Lopez-Llorca et al. 2010), and GFP gene (Lee et al. 2002) were used (Table 1). PCR reactions were carried out as in Lopez-Llorca et al. (2010). PCR products were visualized on 2 % agarose gels stained with GelRed (Biotium, Hayward, USA). These experiments were performed twice.
2.5 Effects of Pochonia chlamydosporia on development of tomato plants
Tomato seedlings inoculated for 3 days with Pc123gfp or uninoculated (20 each) were placed in cylindrical sterile plastic containers each containing 70 cm3 of sterilized sand and 23 ml of 1/10 Gamborg´s basal salt mixture (Sigma), and incubated at constant temperature (25 ºC) and relative humidity (65 %) under the photoperiod previously described. Controls were uninoculated seedlings. Soil moisture was kept constant by adding 1/10 Gamborg’s when necessary. After 10 and 20 days, 10 plants per treatment were sampled and fresh and dry aerial weight, maximum aerial length and fresh root weight per plant were scored. Roots were sampled for evaluating root colonization by culturing techniques, confocal laser microscopy and qPCR. Experiments were repeated twice. The experiment consisted of 10 replicates/treatment.
Total and endophytic root colonization by Pc123gfp were assessed by culturing techniques as described, except that root pieces were plated onto a semi-selective medium for P. chlamydosporia (Lopez-Llorca and Duncan 1986; Kerry and Bourne 2002). Whole roots or longitudinal 50-μm-thick root cryosections (obtained with a Leica CM1510 cryostat (Leica Microsystems, Wetzlar, Germany) were examined by confocal microscopy to analyze the dynamics of root colonization by GFP-expressing transformants.
Colonization of tomato roots by P. chlamydosporia was also measured using quantitative PCR, with LEPR1F/R and VCP1q1F/R primers (Table 1). Five μl root DNA extracts from each treatment were mixed with FastStart Universal SYBR Green Master (Roche, Barcelona, Spain) and 0.88 μl of each primer at 10 μM. Negative controls contained 5 μl sterile water instead of DNA. Thermal cycling conditions consisted of an initial denaturation step at 95 ºC for 2 min then 40 cycles at 95 ºC for 30 s and 60 ºC for 1 min. Reactions were run in triplicate in an ABI PRISM 7000 Sequence Detection System (Applied Biosystem, Foster City, CA, USA). Serial dilutions of Pc123 genomic DNA defined a calibration curve, using three independent calibrations for each DNA sample. After each run, a dissociation curve was acquired to check for amplification specificity. Results are pg P. chlamydosporia DNA/ng tomato DNA.
2.6 Effect of root colonization by Pochonia chlamydosporia on infection and multiplication of Meloidogyne javanica
Thirteen-day-old Pc123gfp-inoculated or uninoculated (control) tomato plants, growing on sterilized sand as described above, were inoculated with 100 M. javanica juveniles (J2) per plant. Seven days later, a subsample of 10 plants per treatment was used to measure growth parameters described above. Numbers of J2 per root were scored by root staining as in Byrd et al. (1983) and observed under an Olympus BH-2 light microscope. Roots from Pc123gfp-inoculated plants were imprinted onto semi-selective medium prior to staining, to test the presence of the fungus.
Remaining plants (5 per treatment) were kept until completion of one life cycle of M. javanica (about 50 days). At this time, plant growth parameters were measured and the numbers of galls and egg masses per plant scored. Egg-masses were plated on semi-selective medium for P. chlamydosporia (Lopez-Llorca and Duncan 1986; Kerry and Bourne 2002) to estimate the percentage of colonization by the fungus. These experiments were repeated twice.
2.7 Statistical analyses
For all data sets obtained in this work, normality and homoscedasticity were checked using the Shapiro-Wilk and Levene tests, respectively. Data following a normal distribution were compared using Student’s t or ANOVA tests for differences between treatments. Non-normal data were compared using either the Wilcoxon or Kruskal-Wallis tests. The level of significance in all cases was 95 %. All statistical analyses were performed with R version 2.11.1 (R Development Core Team, 2009).
3 Results
3.1 Parasitism of RKN eggs by Pochonia chlamydosporia
Pc123 and Pc123gfp did not infect M. javanica eggs 24 h after inoculation. Percentage of egg infection by the fungus increased from 9.7 ± 1.7 and 7.2 ± 1.6 (48 h after inoculation) to 73.4 ± 2.00 and 71.5 ± 2.6 At (96 h after inoculation) for Pc123 and Pc123gfp respectively. No differences (P < 0.05) were found between virulence of Pc123 and Pc123gfp on M. javanica eggs.
Egg-infection by Pc123gfp was analyzed with confocal laser microscopy (Fig. 1). Fine penetration hyphae through the egg-shell were observed 48 h after inoculation (Fig. 1a, arrows). The inside of most eggs did not show signs of fungus colonization at this time (Fig. 1a). Egg-shell penetration by Pc123gfp sometimes involved apressoria formation (Fig. 1b). With a hyphal-tip swelling and thin penetration hyphae (Fig. 1b, insert) in the egg-shell. This penetration hyphae gave rise in late infected eggs to trophic hyphae inside the egg (Fig. 1b) and in their surrounding (Fig. 1c). These colonies from infected eggs finally formed reproductive structures (e.g. conidiophores and conidia) (Fig. 1c, circle).
Main steps of the mode of parasitism of M. javanica (RKN) eggs by P. chlamydosporia (Pc123gfp). a Note fine Pc123gfp penetration hyphae (2 dai) (arrows) on RKN egg-shell. b RKN egg-shell penetration by Pc123gfp appressoria (insert) (3dai). c Late infection of a RKN egg by Pc123gfp (4 dai). Note heavy fungus growth inside and outside egg and conidiophore with conidia (circle). Abbreviation: dai, days after inoculation. Bar 10 μm
3.2 Inoculation of tomato seedlings with Pochonia chlamydosporia
We measured total and endophytic colonization 1, 2, 3, 5 and 7 days after inoculation. Tomato seedlings 3 days after of inoculation with the fungus were selected for the following experiments, since this was the minimum incubation period for achieving most root endophytic colonization. There were no significant differences between root colonization by Pc123 and Pc123gfp isolates (data not shown).
3.3 Detection of Pochonia chlamydosporia in roots of tomato seedlings
Three days after inoculation Pc123gfp colonization was low in the vicinity of the apical meristem close to the apex of tomato root seedlings (Fig. 2a). Biologically active Pc123gfp chlamydospores were sometimes formed in the root cap (Fig. 2b). An increase in fungus colonization was found in areas progressively more distant from the actively growing root apex.
Laser confocal microscopy of tomato roots colonization by P. chlamydosporia (Pc123gfp): a-b seedlings 3 days after inoculation (dai). Plants thirteen dai c-e. a General view of root apex showing Pc123gfp colonization of areas adjacent to meristematic zone (bar = 75 μm). b Close-up of root cap showing Pc123gfp chlamydospore (arrow for magnification) (bar = 75 μm). c Longitudinal section (LS) showing Pc123gfp colonization of epidermal cells (bar = 75 μm). d LS showing colonization Pc123gfp of cortex and outer vascular tissue (bar = 50 μm). e Plant defence induction (Papillae, arrows) associated to fungus colonization (bar = 20 μm). Note phialide forming conidia (asterisk)
Using PCR we could detect the presence of P. chlamydosporia in both unsterilized as well as surface sterilized tomato seedling roots using primers for P. chlamydosporia VcP1 gene (data not shown).
3.4 Evaluation of root colonization of tomato plants by P. chlamydosporia by culturing, microscopy and molecular techniques
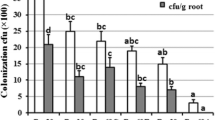
qPCR allowed an accurate, reliable and high-throughput quantification of target fungal DNA. The standard curves obtained revealed high precision and reproducibility between replications (Fig. 3a and b). Molecular quantification of tomato root colonization by P. chlamydosporia with qPCR detected significant differences between P. chlamydosporia strain and time after inoculation (Fig. 3c and d). Total root colonization (non-surface sterilized roots) measured by culturing techniques was 100 % for all treatments (Fig. 3c). On the contrary, differences in the endophytic colonization of tomato roots by Pc123 and Pc123gfp were found. Thirteen days after inoculation the endophytic colonization was 44.7 ± 4.7 % and 33.3 ± 8.8 % for Pc123 and Pc123gfp respectively. Twenty-three days after inoculation, colonization was 19.3 ± 11.6 % and 10 ± 5.8 % for Pc123 and Pc123gfp respectively (Fig. 3d). qPCR which was more sensitive than culturing techniques for detecting total and endophytic root colonization detected higher rhizosphere competence for Pc123 than for Pc123gfp. Colonization of tomato roots by Pc123 and Pc123gfp decreased with time (13d vs. 23 days after inoculation) for both total and endophytic root colonization. The only exception was endophytic colonization by Pc123gfp. This could be due to the low values of root colonization of this transformant strain.
Quantification of Pochonia chlamydosporia colonization of tomato roots. a-b Standard curves for real-time PCR of 10-fold serial dilutions of DNA from a Solanum lycopersicum; b P. chlamydosporia. Cycle thresholds (Ct) were plotted against the log of known concentrations of genomic DNA standards and linear regression equations were calculated for the quantification of the unknown samples by interpolation. c Quantification results for total colonization using real-time PCR (gray) and culture techniques (dark gray) at 13 and 23 days. d Quantification results for endophytic colonization using real-time PCR (gray) and culture techniques (dark gray) at 13 and 23 days. Each value (± SE) represents the mean of ten replicates (p-value <0.05)
Confocal microscopy showed details of root colonization by Pc123gfp 13 days after inoculation of seedlings. A widespread colonization of the rhizoplane and the first layers of the epidermis (Fig. 2c) was consistent with the high total rhizosphere colonization detected by culturing techniques. In some parts both epidermal cells and the first layers of the cortex also showed fungal colonization (Fig. 2d). Pc123gfp also induced root cell defences (cell wall papillae) as seen in Fig. 2e (arrows).
3.5 Effects of Pochonia chlamydosporia on tomato plants development
Thirteen days after inoculation with Pc123gfp tomato seedlings showed no differences (p < 0.05) in plant growth respect to uninoculated control plants, although at 23 days all parameters measured were higher than Pc123gfp inoculated plants (data not shown). Twenty days after inoculation Pc123gfp promoted aerial growth measured as fresh or dry shoot weight of tomato plants irrespective of J2 inoculation (Fig. 4a and b). Nematode inoculation reduced maximum shoot length of tomato plants, irrespective of fungus inoculation (Fig. 4c). Pc123gfp promoted root growth irrespective of J2 inoculation. Plants inoculated with both Pc123gfp and J2 had the largest root system of all treatments (Fig. 4d).
Effect of P. chlamydosporia on plant development a-d and infection/multiplication e-f of M. javanica in tomato roots. a Fresh shoot weight (FSW). b Dry shoot weight (DSW). c Maximum shoot length (MSL). d Fresh root weight (FRW). e M. javanica infection on tomato root per root system (gray) or per g of root (dark grey). f) M. javanica multiplication on tomato roots per root system (gray) or per g of root (dark grey). Each value (± SE) represents the mean of ten replicates (p-value <0.05). Abbreviations: (C) Control plant (uninoculated); (Pc123GFP) Plant inoculated with P. chlamydosporia 123gfp; (J2) Plants inoculated with M. javanica juveniles; (Pc123GFP+J2) Plants inoculated with P. chlamydosporia 123gfp and M. javanica juveniles; SE, Standard error
3.6 Effect of root colonization by Pochonia chlamydosporia on infection and multiplication of Meloidogyne javanica
Under our experimental conditions, endophytic development of P. chlamydosporia in tomato roots had no statistically significant effect on M. javanica J2 invasion in roots inoculated with the nematode vs. control roots (which had been nematode but not fungus inoculated) (Fig. 4e). When time was allowed for M. javanica to complete its life cycle, nematode multiplication was estimated as numbers of galls (Fig. 4f). M. javanica galls and egg masses were colonized by Pc123gfp (53.6 ± 14.8 and 32 ± 14 % respectively) when the fungus was applied endophytically in tomato seedlings.
4 Discussion
In this paper we have evaluated the effects on plant growth and root-knot nematode (M. javanica) invasion of an endophytic GFP transformant of the nematophagous fungus P. chlamydosporia (Pc123gfp). Pc123gfp was as virulent to nematode eggs as the corresponding wild type strain. This would indicate that the transformation for obtaining this strain did not affect its pathogenicity to nematode eggs. Pc123gfp infecting M. javanica eggs analyzed with confocal laser microscopy revealed details of the infection process not found in previous studies. We showed Pc123gfp forming early appressoria and penetration hyphae on the egg shell which later developed into trophic hyphae inside the infected egg. Previously SEM (Lopez-Llorca and Duncan 1987) and Field Emission SEM studies mostly showed images on the egg-shell surface of this process (Lopez-Llorca and Claugher 1990). Ultrastructural studies of fungal parasites of nematode eggs were only limited to advanced stages of infection because of the impermeability of egg-shells to TEM embedding mixtures and resins (Lopez-Llorca and Robertson 1992). Pc123gfp could be used in future studies to investigate the infection process in plant parasitic nematodes other than root-knot (e.g. cyst nematodes) with different egg-shell characteristics (Bird and Bird 1991).
P. chlamydosporia is a facultative parasite of eggs and females of plant parasitic nematodes, which also colonizes endophytically plant roots (Lopez-Llorca et al. 2002; Bordallo et al. 2002). Pc123gfp was found to colonize endophytically barley roots under axenic conditions (Maciá-Vicente et al. 2009a). In this study we have measured colonization of tomato roots by P. chlamydosporia using culturing techniques, molecular markers and confocal laser microscopy. Using cultural techniques and surface sterilization we distinguished total vs. endophytic (internal) root colonization. The latter being lower than the former. We found no differences in the endophytic colonization of tomato seedlings by either Pc123 or Pc123gfp. In previous studies using culturing techniques (Bourne et al. 1996) 17 plant species including tomato were found to differ in their ability to support P. chlamydosporia in their rhizospheres (Bourne et al. 1996). Tomato was, depending on the cultivar, a moderate or poor host of the fungus. A recent study (Manzanilla-López et al. 2011) has shown the ability of break crops (oilseed rape, sugarbeet and wheat) in a potato rotation to support P. chlamydosporia in their rhizospheres. In these studies only total root colonization was estimated since no surface sterilization was performed. However, culturing techniques for estimating rhizosphere colonization by fungi are biased and therefore more sensitive PCR-based methods are presently being used.
We found qPCR more accurate and sensitive than culturing methods to detect differences in tomato rhizosphere colonization by P. chlamydosporia. Total root colonization, was 100 %, for both Pc123 and Pc123gfp irrespective of time after inoculation measured by culturing methods. However, we have detected by qPCR higher rhizosphere competence (total and endophytic) in tomato for Pc123 than for Pc123gfp. Estimated by qPCR, total as well as endophytic root colonization by both fungi mostly decreased with time. Our strategy for estimating root colonization by P. chlamydosporia involving amplification of single gene sequences from plant and fungus origins was previously used to quantify Verticillium dahliae in Solanaceae cultivars (Gayoso et al. 2007). Previous studies for qPCR quantification of P. chlamydosporia in roots used fungus genes only. (Atkins et al. 2009; Ciancio et al. 2005; Maciá-Vicente et al. 2009a). The quantification by qPCR of fungi/plant DNA in roots would allow in future studies to analyze rhizosphere competence of P. chlamydosporia isolates in diverse plant cultivars (e. g. tomato) or different crop species. Besides, root samples from soil contain, other rhizosphere microorganisms that would not be amplified with fungus and plant species primers instead of total DNA as with one gene target (fungus) based methods. The main drawback of standard qPCR for fungus quantification is that the method does not distinguish viable from dead propagules. Reverse transcription-qPCR has been applied for detection an enumeration of yeast with a low detection limit and higher accuracy than qPCR because dead cells were not quantified with this RNA-based technique (Hierro et al. 2006).
Using confocal laser microscopy we analyzed the pattern of colonization by Pc123gfp in tomato roots. Fungus colonization was low in the vicinity of the apical meristem close to the root apex and increased in areas progressively more distant from the actively growing root apex. This would explain the reduction in rhizosphere competence of Pc123/Pc123gfp with time found in this study. To this respect, reduction in barley rhizosphere colonization by P. chlamydosporia implied that free root niches were colonized by other fungi (Maciá-Vicente et al. 2009b). Pc123gfp colonized epidermal and cortex cells and induced defences (papillae) in tomato roots, similarly to that found for barley (Maciá-Vicente et al. 2009a). Papillae and other plant defences responses have also been observed in Pc123 colonizing endophytically barley and tomato roots (Bordallo et al. 2002). Papillae have also been found for other P. chlamydosporia isolates colonizing endophytically wheat roots (Manzanilla-López et al. 2011).
Pc123gfp promoted root and shoot growth of tomato plants 20 days after inoculation (dai) irrespective of M. javanica J2 inoculation. In previous studies, P. chlamydosporia and Pseudomonas aeruginosa applied together promoted growth of tomato plants compared with either antagonist alone or untreated controls (Siddiqui and Ehteshamul-Haque 2000; Siddiqui and Shaukat 2003). Manzanilla-López et al. (2011) found variable growth promotion (shoot and root weight) depending on the crop and P. chlamydosporia isolate combinations. P. chlamydosporia also promoted root and shoot weight of wheat in pots (ca. 20 dai) irrespective of infection by the fungus root pathogen Gaeumannomyces graminis var. tritici (Ggt) (Monfort et al. 2005). Plant growth promotion by Pc123 in barley was found to increase with time and was maximum c.a. 60 dai (Maciá-Vicente et al. 2009b). These authors found higher growth promotion by two P. chlamydosporia isolates (including Pc123) for roots than for shoots respect to uninoculated control plants. This agrees with our findings for Pc123gfp in tomato.
Tomato roots endophytically colonized by Pc123gfp showed less M. javanica juveniles per g of root than fungus uninoculated plants also infected with the nematode. However, these differences were not statistically significant (p > 0.05). Previous studies, which did not involve root but soil inoculation by P. chlamydosporia, concluded that the presence of the fungus in the rhizosphere did not affect the invasion of tomato roots by infective juveniles of M. incognita (Bailey et al. 2008; Tobin et al. 2008). The reduction in nematode invasion may be due to the production by Pochonia spp. of secondary metabolites with antifugal and nematicidal activity (Hellwig et al. 2003; Khambay et al. 2000; Lopez-Llorca and Boag 1993; Niu et al. 2010) or proteases (e.g. VcP1) with nematicidal activity. To this respect, VcP1 is a serine protease involved in the pathogenesis of nematode eggs (Segers et al. 1996) and in the root endophytic colonization by P. chlamydosporia (Lopez-Llorca et al. 2010). However, the growth promotion of roots induced by P. chlamydosporia could also explain our results. P. chlamydosporia colonizing endophytically wheat also reduced root colonization by Ggt (Monfort et al. 2005).
In our study, P. chlamydosporia applied only endophytically in tomato roots then post-inoculated with RKN could colonize RKN galls and egg masses. In previous studies P. chlamydosporia was inoculated by applying chlamydospores of the fungus to the plant substrate (Bourne and Kerry 1999; Kerry and Hidalgo-Díaz 2004; Manzanilla-López et al. 2011; Sorribas et al. 2003; Tzortzakakis and Petsas 2003; Van Damme et al. 2005; Verdejo-Lucas et al. 2003). Our semi-axenic experimental system did not allow proper root development. This may explain a larger number of galls in plants colonized with Pc123gfp than in controls. Therefore, future studies (in progress) should involve tomato growth in pots for allowing completion of plant and plant-parasitic nematode development.
Endophytic nematophagous fungi such as P. chlamydosporia share the niche with endoparasitic nematodes but are less subject to competition from soil microorganisms (Stirling 2011). Therefore, rhizosphere colonization (endophytism) by biocontrol agents such as Pochonia chlamydosporia is a promising strategy for implementing biocontrol/IPM of root pathogens such plant-parasitic nematodes. From the perspective of biocontrol of nematodes, endophytes should be relatively easy to apply as inoculants to seed or seedlings and could therefore be established in the root system before nematodes are attracted to root and begin to feed (Stirling 2011). Although mutualist endophytic fungi have been used for biocontrol of plant parasitic nematodes (Sikora et al. 2008), the use of nematophagous fungi as endophytes has not been fully exploited yet.
This study shows that P. chlamydosporia growing endophytically can promote root and shoot growth and colonizes egg masses of root-knot nematodes. However, future research is needed to increase the amount of fungus in the root, since this became reduced with plant development.
References
Abad P, Gouzy J, Aury JM, Castagnone-Sereno P, Danchin EGJ, Deleury E, Perfus-Barbeoch L, Anthouard V, Artiguenave F, Blok VC (2008) Genome sequence of the metazoan plant-parasitic nematode Meloidogyne incognita. Nat biotechnol 26(8):909–915
Atkins SD, Peteira B, Clark IM, Kerry BR, Hirsch PR (2009) Use of real-time quantitative PCR to investigate root and gall colonisation by co-inoculated isolates of the nematophagous fungus Pochonia chlamydosporia. Ann Appl Biol 155(1):143–152
Bailey DJ, Biran GL, Kerry BR, Gilligan CA (2008) Pathozone dynamics of Meloidogyne incognita in the rhizosphere of tomato plants in the presence and absence of the nematophagous fungus, Pochonia chlamydosporia. Plant Pathol 57(2):354–362
Barelli L, Padilla-Guerrero IE, Bidochka MJ (2011) Differential expression of insect and plant specific adhesin genes, Mad1 and Mad2, in Metarhizium robertsii. Fungal Biol 115(11):1174–1185
Bird AF, Bird J (1991) The structure of nematodes. The egg. Academic, San Diego, pp 7–43
Bordallo JJ, Lopez-Llorca LV, Jansson HB, Salinas J, Persmark L, Asensio L (2002) Colonization of plant roots by egg-parasitic and nematode-trapping fungi. New Phytol 154(2):491–499
Bourne JM, Kerry BR (1999) Effect of the host plant on the efficacy of Verticillium chlamydosporium as a biological control agent of root-knot nematodes at different nematode densities and fungal application rates. Soil Biol and Biochem 31:75–84
Bourne JM, Kerry BR, De Leig FAAM (1996) The importance of the host plant on the interaction between root-knot nematodes Meloidogyne spp. and the nematophagous fungus, Verticillium chlamydosporium goddard. Biocontrol Sci Tech 6(4):539–548
Byrd DW Jr, Kirkpatrick T, Barker KR (1983) An improved technique for clearing and staining plant tissues for detection of nematodes. J Nematol 15(1):142
Ciancio A, Loffredo A, Paradies F, Turturo C, Sialer MF (2005) Detection of Meloidogyne incognita and Pochonia chlamydosporia by fluorogenic molecular probes*. EPPO Bull 35(1):157–164
Gayoso C, Ilárduya OM, Pomar F, Merino de Caceres F (2007) Assessment of real-time PCR as a method for determining the presence of Verticillium dahliae in different Solanaceae cultivars. Eur J Plant Pathol 118(3):199–209
Hellwig V, Mayer-Bartschmid A, Müller H, Greif G, Kleymann G, Zitzmann W, Tichy HV, Stadler M (2003) Pochonins A-F, New Antiviral and Antiparasitic Resorcylic Acid Lactones from Pochonia chlamydosporia var. catenulata. J Nat Prod 66:829–837
Hidalgo-Díaz L, Bourne JM, Kerry BK, Rodríguez MG (2000) Nematophagous Verticillium spp. in soils infested with Meloidogyne spp. in Cuba: Isolation and screening. Int J Pest Manage 46(4):277–284
Hierro N, Esteve-Zarzoso B, González A, Mas A, Guillamón JM (2006) Real-time quantitative PCR (QPCR) and reverse transcription-QPCR for detection and enumeration of total yeasts in wine. Appl Environ Microl 72(11):7148–7145
Inglis PW, Aragao FJL, Frazao H, Magalhaes BP, Valadares-Inglis MC (2000) Biolistic co-transformation of Metarhizium anisopliae var. acridum strain CG423 with green fluorescent protein and resistance to glufosinate ammonium. FEMS Microbiol Lett 191:249–254
Kerry B, Hidalgo-Diaz L (2004). Application of Pochonia chlamydosporia in the integrated control of root-knot nematodes on organically grown vegetable crops in Cuba. Bulletin OILB/SROP 27:123–126
Kerry BR, Bourne JM (2002). A manual for Research on Verticillium chlamdosporium, as Potential Biological Control Agent for Root-Knot Nematodes. Ed WPRS/SROP
Kerry BR, Crump DH (1977) Observations on fungal parasites of females and eggs of the cereal cyst-nematode, Heterodera avenae, and other cyst nematodes. Nematologica 23:193–201
Khambay B, Bourne JM, Cameron S, Kerry B, Zaki M (2000) A nematicidal metabolite from Verticillium chlamydosporium. Pest Manag Sci 56:1098–1099
Kurtti TJ, Keyhani NO (2008) Intracellular infection of tick cell lines by the entomopathogenic fungus Metarhizium anisopliae. Microbiology 154(6):1700–1709
Lee S, Kim SH, Breuil C (2002) The use of the green fluorescent protein as a biomarker for sapstain fungi. Forest Pathol 32(3):153–161
Lopez-Llorca LV, Boag B (1993) Biological properties of a red pigment produced by the nematophagous fungus Verticillium suchlasporium. Nematol medit 21:143–149
Lopez-Llorca LV, Claugher D (1990) Appressoria of the nematophagous fungus Verticillium suchlasporium. Micron Microscoc Acta 21(3):125–130
Lopez-Llorca LV, Duncan JM (1986) New media for the estimation of fungal infection in eggs of the cereal cyst nematode. Nematologica 32:486–490
Lopez-Llorca LV, Duncan GH (1987) A study of fungal endoparasitism of the cereal cyst nematode Heterodera avenae by scanning electron microscopy. Can J Microbiol 34:613–619
Lopez-Llorca LV, Robertson WM (1992) Ultrastructure of infection of cyst nematode eggs by the nematophagous fungus Verticillium Suchlasporium. Nematologica 39(4):65–74
Lopez-Llorca LV, Bordallo JJ, Salinas J, Monfort E, Lopez-Serna ML (2002) Use of Light and scanning electron microscopy to examine colonisation of barley rhizosfere by the nematophagous fungus Verticillium chlamydosporium. Micron 33:61–67
Lopez-Llorca LV, Jansson HB, Maciá-Vicente JG, Salinas J (2006) Nematophagous fungi as root endophytes. In: Microbial root endophytes, Springer–Verlag, pp 191–206
Lopez-Llorca LV, Gomez-Vidal S, Monfort E, Larriba E, Casado-Vela J, Elortza F, Jansson HB, Salinas J, Martín-Nieto J (2010) Expression of serine proteases in egg-parasitic nematophagous fungi during barley root colonization. Fungal Genet Biol 47(4):342–351
Maciá-Vicente JG, Jansson HB, Talbot NJ, Lopez-Llorca LV (2009a) Real-time PCR quantification and live-cell imaging of endophytic colonization of barley (Hordeum vulgare) roots by Fusarium equiseti and Pochonia chlamydosporia. New Phytol 182(1):213–228
Maciá-Vicente JG, Rosso LC, Ciancio A, Jansson HB, Lopez-Llorca LV (2009b) Colonisation of barley roots by endophytic Fusarium equiseti and Pochonia chlamydosporia: Effects on plant growth and disease. Ann Appl Biol 155(3):391–401
Manzanilla-López RH, Esteves I, Powers SJ, Kerry BR (2011) Effects of crop plants on abundance of Pochonia chlamydosporia and other fungal parasites of root-knot and potato cyst nematodes. Ann Appl Biol 159(1):118–129
McClure MA, Kruk TH, Misaghi I (1973) A method for obtaining quantities of clean Meloidogyne eggs. J Nematol 5(3):230
Monfort E, Lopez-Llorca LV, Jansson HB, Salinas J, Park JO, Sivasithamparam K (2005) Colonisation of seminal roots of wheat and barley by egg-parasitic nematophagous fungi and their effects on Gaeumannomyces graminis var. tritici and development of root-rot. Soil Biol Biochem 37(7):1229–1235
Niu XM, Wang YL, Chu YS, Xue HX, Li N, Wei LX, Mo MH, Zhang KQ (2010) Nematodetoxic aurovertin-type metabolites from a rook-knot nematode parasitic fungus Pochonia chlamydosporia. J Agric Food Chem 58:828–834
Nordbring-Hertz B, Jansson HB, Tunlid A (2006) Encyclopedia of life sciences. pp 1–11
Olivares-Bernabeu CM, Lopez-Llorca LV (2002) Fungal egg-parasites of plant-parasitic nematodes from Spanish soils. Rev Iberoam Micol 19(2):104–110
Persmark L, Jansson HB (1997) Nematophagous fungi in the rhizosphere of agricultural crops. FEMS Microbiol Ecol 22(4):303–312
Segers R, Butt MT, Kerry BR, Beckett A, Peberdy JF (1996) The role of the proteinase VCP1 produced by the nematophagous Verticillium chlamydosporium in the infection process of nematode eggs. Mycol Res 100:421–428
Siddiqi ZA, Akhtar MS (2008) Synergistic effects of antagonistic fungi and a plant growth promoting rhizobacterium, an arbuscular mycorrhizal fungus, or composted cow manure on populations of Meloydogyne incognita and growth of tomato. Biocontrol Sci Techn 18(3):207–290
Siddiqui IA, Ehteshamul-Haque S (2000) Effect of Verticillium chlamydosporium and Pseudomonas aeruginosa in the control of Meloidogyne javanica in tomato. Nematol Medit 28:193–196
Siddiqui IA, Shaukat SS (2003) Combination of Pseudomonas aeruginosa and Pochonia chlamydosporia for Control of Root-Infecting Fungi in Tomato. J Phytopathology 151:215–222
Sikora RA, Pocasangre L, Felde A, Niere B, Vu TT, Dababat AA (2008) Mutualistic endophytic fungi and in-planta suppressiveness to plant parasitic nematodes. Biol Control 46:15–23
Sorribas FJ, Ornat C, Galeano M, Verdejo-Lucas S (2003) Evaluation of a Native and Introduced Isolate of Pochonia chlamydosporia against Meloidogyne javanica. Biocontrol Sci Techn 13(8):707–714
Stirling GR (2011) Biological control of plant-parasitic nematodes: an ecological perspective, a review of progress and opportunities for further research. Biological Control of Plant-Parasitic Nematodes. pp 1–38
Tobin JD, Haydock PPJ, Hare MC, Woods SR, Crump DH (2008) Effect of the fungus Pochonia chlamydosporia and fosthiazate on the multiplication rate of potato cyst nematodes (Globodera pallida and G. rostochiensis) in potato crops grown under UK field conditions. Biol Control 46(2):194–201
Tzortzakakis EA, Petsas SE (2003) Investigation of alternatives to methyl bromide for management of Meloidogyne javanica on greenhouse grown tomato. Pest Manag Sci 59:1311–1320
Van Damme V, Hoedekie A, Viaene N (2005) Long-term efficacy of Pochonia chlamydosporia for management of Meloidogyne javanica in glasshouse crops. Nematology 7:741–745
Verdejo-Lucas S, Sorribas FJ, Galeno M (2003) Evaluating Pochonia chlamydosporia in a double-cropping system of lettuce and tomato in plastic houses infested with Meloidogyne javanica. Plant Pathol 52:521–528
Wesemael WML, Viaene N, Moens M (2010) Root-knot nematodes (Meloidogyne spp.) in Europe. Nematology 13(1):3–16
Wiedenmann J, Oswald F, Nienhaus GU (2009) Fluorescent proteins for live cell imaging: Opportunities, limitations, and challenges. IUBMB Life 61(11):1029–1042
Zhang L, Yang J, Niu Q, Zhao X, Ye F, Liang L, Zhang KQ (2008) Investigation on the infection mechanism of the fungus Clonostachys rosea against nematodes using the green fluorescent protein. Appl Microbiol Biot 78(6):983–990
Zijlstra C, Donkers-Venne D, Fargette M (2000) Identification of Meloidogyne incognita, M. javanica and M. arenaria using sequence characterised amplifed region (SCAR) based PCR assays. Nematology 2(8):847–853
Acknowledgements
This research was funded by the Spanish Ministry of Science and Innovation Grants AGL 2008-00716/AGR and AGL 2011-29297. We thank Mr. E. Larriba (University of Alicante, Spain) for designing qPCR primers for VcP1 gene and Dr. F. J. Sorribas (Universitat Politècnica de Catalunya) for his help in the establishment of the nematode population. We also thank Professor Hans-Börje Jansson (University of Alicante) for critical comments on the manuscript.
In Memoriam
This paper is dedicated to the memory of the late Prof. Brian Kerry (Rothamsted Research, UK). He made key discoveries in the field of nematophagous fungi finding that soil suppression to plant parasitic nematodes was due to fungal parasites of nematode eggs. His work and scientific thought inspired the research of biologists all over the world including us. Many thanks Brian!
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Escudero, N., Lopez-Llorca, L.V. Effects on plant growth and root-knot nematode infection of an endophytic GFP transformant of the nematophagous fungus Pochonia chlamydosporia . Symbiosis 57, 33–42 (2012). https://doi.org/10.1007/s13199-012-0173-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13199-012-0173-3