Abstract
The greater bulk of soil nitrogen is immobilized in chitinous cell walls of fungi. Mycophagous soil mites participate in chitin decomposition and, hence, in the subsequent mobilization of nitrogen. The source of the chitinolytic enzymes was searched in this study. A multimethodical approach was designed for these studies. Histology, plating and identification of bacteria from mite homogenate and, finally, homogenate and bacterial treatment of the soil fungi were applied. Here the presence and activity of chitinolytic bacteria inside mycophagous mites are reported. These bacteria form an extraintestinal group within the mite’s body and pass their enzymes into the mite’s gut. Our results demonstrate that true mycophagous mites, defined by their ability to digest chitin (i.e. the fungal cell wall), achieve this through internal “cooperation” with chitinolytic bacteria that provide the necessary chitinolytic enzymes. The nitrogen from chitin is thus made available to other soil organisms and plants.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
The greater bulk of soil nitrogen is immobilized as chitin in the cell walls of numerous species of micromycetes. Nitrogen mobilization, therefore, must include mycolytic activity and, indeed, this appears to be a common phenomenon in soil. In grasslands of the U.S.A. and Canada, for example, the dry mass of mycelium has been estimated at from 12.6 to 184 g/m2 (Kjøller and Struwe 1982). As chitin comprises from 3% to 60% of the dry mass in the cell wall (Hudson 1986). it represents an important source of nitrogen in the soil and chitin decomposers play a substantial role in mobilizing nitrogen and, more generally, in the entire nutrient cycling process. Some soil organisms do appear to be chitinolytic (i.e. able to convert chitin through enzyme activity) and so can be termed as mycolytic (microorganisms) or mycophagous (animals), i.e. as feeding on fungi.
Production of chitinolytic enzymes has been confirmed in literature in some bacteria, including Bacillus (Watanabe et al. 1990; Citterio et al. 2001; Aktuganov et al. 2004), Pseudomonas (Lim et al. 1991; Wang et al. 1997; Citterio et al. 2001), Serratia (Kishore et al. 2005), Janthinobacterium (Molloy and Burke 1997), Brevundimonas (Jaspers and Overmann 2004) and Stenotrophomonas (Zhang et al. 2001; Kobayashi et al. 2002). Autochthonous chitinolytic enzymes are generally absent in all hitherto studied animals including the man, by contrast, except for a few employing pathogenous processes (Aktuganov et al. 2004). Earlier opinion held that saprophagous soil mites exhibit low feeding selectivity, although they transport the microorganisms (bacteria, fungal spores as well as fragments of mycelium) on their body surfaces or in their guts (Wallwork 1976). Several more recent observations, however, have revealed conspicuous food selectivity in some groups of these mites, especially moss mites (Oribatida) and mould mites (Acaridida) (Luxton 1972; Siepel and de Ruiter-Dijkman 1993; Smrž 2002a).
Several methods have been reported in the literature for determining mite food selection. Clearing of mites with lactic acid and then observing them under a light microscope can indicate a “swallowing” of fungi. But “swallowed” fungal propagula are also able to pass through the gut as intact particles. Other approaches have included laboratory feeding tests, based on the one-way or cafeteria test (Czajkowska 1970) and recording whether mites make contact with a fungal colony, whether they move their chelicerae, and whether faeces are produced. Again, even all of those phenomena do not together provide conclusive evidence for true mycophagy. Indeed, consumption of fungi might also proceed through piercing of the cell wall and sucking out of its contents, which are rich in trehalose and other substances (Wigglesworth 1974). This form of feeding, however, cannot be termed true mycophagy as the chitinous cell wall remains intact and nitrogen is not mobilized from the cell walls. Further, substances such as trehalose, a so-called fungal sugar, can occur in many organisms (e.g. insects) (Wigglesworth 1974). In contrast, testing of enzyme activity has proven a useful method for assessing mode of feeding, and it has resulted in the classification of mites into a number of feeding groups or guilds (Luxton 1972; Dinsdale 1974; Siepel and de Ruiter-Dijkman 1993; Smrž 1996). Using labelled isotopes of nitrogen also appears to be a very useful method (Schneider et al. 2004).
Several authors observed bacteria inside the mites. Wolf and Rockett (1984) isolated several bacterial genera from two oribatids (Rhysotritia and Pergalumna). According to those authors, the effect of habitats was supposed to be important for the bacterial internal communities. The bacterial production of enzymes, however, was not assumed, although Bacillus or Pseudomonas were isolated. The participation of the gut bacteria was considered in the digestion processes by the Stefaniak and Seniczak (1976; 1981) and Seniczak and Stefaniak (1978), but without any identification of bacteria or specification of those processes. The type of food was assumed to be the substantial factor for the structure of the gut bacterial community. Again, the digestion of grazed food was not observed at the grazed food in gut.
To confirm whether mycophagy including chitinolytic activity is really occurring, a series of tests was undertaken in order to answer three questions: 1) are animals really able to digest chitin; if so, 2) how does the animal digest the chitin and utilize it for nutrition; and 3) are chitinolytic enzymes available to animals? 4) are we able to define the actual mycophagy?
2 Materials and methods
Tested mites and fungi
Several species of mites were examined, all previously considered as mycophagous: Damaeus onustus C.L.Koch, Damaeus riparius Nicolet, Damaeus auritus C. L. Koch, Metabelba pulverosa Strenzke, Belba pseudocorynopus Märkel & Mayer, or as occasional mycophagous - Scutovertex minutus (C.L.Koch) (all Oribatida sampled in forest and steppe biotopes) (see Pauly 1956; Luxton 1972; Smrž 1992, 2006). These mites were compared with three control oribatid species known to avoid fungi (non-mycophagous) sampled in forest and steppe soils: Platynothrus peltifer (C. L. Koch), Phthiracarus sp., and Trichoribates trimaculatus (C. L. Koch) (see Luxton 1972; Dinsdale 1974; Smrž 1992, 2006, 2007). The latter oribatids rejected any fungi, and therefore they were reared for the short time of the tests on algae covering tree bark (Protococcus sp.). The acaridid mycophagous mite Tyrophagus putrescentiae (Schrank) (Czajkowska 1970; Smrž and Čatská 1987; Smrž and Jungová 1989; Smrž 2003) was reared in the laboratory on the tested fungi. All experiments were performed in 220 cm3 glass boxes with a plaster of Paris/charcoal bottom.
The tested fungi were applied from our reference collection (Alternaria sp.—strain M 100, Fusarium oxysporum—strain M 26, Mucor racemosus—strain M 6, Penicillium claviforme—strain M 83, Penicillium griseofulvum—strain M 66, and Verticillium sp.—strain M 21). All those fungi were consumed by mites as sporulating ones on malt agar. Only the algal cover on the tree bark (Protococcus sp.) was invaded in rearing boxes by the sterile mycelium and the identification of those mycelium was not possible.
Histology
Mites (20 individuals per experimental group) were fixed in modified Bouin-Dubosque-Brasil fluid (Smrž 1989) and embedded in Paraplast (Polysciences). The samples were then sectioned on a Leica 2155 rotation microtome (5 μm thickness), stained using Masson’s triple stain, and observed under an AX-70 Provis (Olympus) light microscope, which included using a Nomarski DIC prism for unstained objects (Smrž 1989). Ten mites from each group were fixed in cacodylate-buffered glutaraldehyde (4%), embedded in Poly/Bed epoxy resin (Polysciences), and sectioned on a Leica 2155 rotation microtome by tungsten carbide knife (1 μm thickness).
Six microanatomical characteristics were examined (Smrž 1996, 2002a).
-
content of the gut (type of food consumed and stage of digestion),
-
activity of gut walls,
-
metabolite deposits (guanine crystals). Guanine, a metabolite deposit, is typically found following digestion of large amounts of nitrogenous food (e.g. chitin). Viewed under great magnification, these crystals are birefringent, conspicuously silver, and appear as concentric circles. They are whitish when seen in the mite’s body. The great bulk of those crystals is able to damage the internal mite organ (Smrž and Čatská 1987, 1989; Smrž et al. 1991).
-
nutrient deposits (glycogen granules),
-
conspicuous free cells (haemocytes) between the internal organs,
-
bacteria-containing bodies inside the intestinal lining.
Mite faeces
Mite faeces were smeared, stained by acridine orange, and observed under an AX-70 fluorescence microscope (Olympus). The presence of fungal propagula and bacterial cells were ascertained through differential staining, i.e. dead fungal cells would show up red while living bacteria would show up luminous green (Smrž 2002b).
Transmission electron microscopy
Mites were fixed in cacodylate-buffered glutaraldehyde (4%), postfixed in 1% osmium tetroxide, embedded in Poly/Bed medium (Polysciences), and sectioned using a Reichert Ultracut E ultramicrotome. Sections were stained in lead citrate and uranyl acetate and observed under a Philips EM 300 transmission electron microscope.
Internal bacteria (Smrž 2000)
Living mites were rinsed twice in 96% ethyl alcohol followed by 10% JAR (Procter & Gamble) domestic detergent (to remove microorganisms from the body surface) and finally washed twice in distilled water. The mites were subsequently homogenized in Ringer’s solution. The internal bacteria from the mite homogenate (homogenate from 1 larger mite of Damaeus, or from 10 smaller individuals of the other mites) were plated on MPA (meat peptone agar) (pH = 7) and identified at the internationally certified microbiological institute of the Czech Collection of Microorganisms (Brno, Czech Republic). Bacteria were only determined as present or absent, no quantitative correlations were established. During bacteria plating, species dominance was very clear. The dominant bacterial species formed around 8 colony forming units (CFU) per dish. The maximum number of CFU was 12 per dish.
Enzyme activity (Smrž 2000)
Mite homogenate (for method, see previous paragraph) was tested on a thin layer of carboxymethyl-chitin on a microslide, with subsequent staining using basic fuchsin. The same test was performed on a bacterial suspension isolated from the homogenate that had been previously identified at the Czech Collection of Microorganisms
3 Results
In the real mycophagous mite species (Damaeus onustus, D. riparius, D. auritus, Metabelba pulverosa, Belba pseudocorynopus), the alimentary system was observed to be crowded with fungal propagula (Fig. 1) (spores and fragments of mycelium) in all three gut parts (mesenteron, colon, rectum). These corpuscles displayed increasingly high levels of destruction, especially as regards their cell walls, as they passed through the system. Cell damage was clearly visible in all three gut parts. The cell walls were nearly intact in the mesenteron (the first part of the entodermal gut), became thinner in the colon (the second gut section), and an amorphous mass with almost no wall structures, or with only scarce fragments of the walls, was visible at the third gut section (i.e. the rectum, including the anal atrium). “Swallowed” fungal propagula were scarce in non-mycophagous mites, and usually absent. When observed, they were never seen in more than one gut section at the same time and were passed through all three gut sections as intact corpuscles.
Alimentary tract: 1—Mesenteron of Belba pseudocorynopus crowded by fungal spores (Alternaria sp. dominated), 2—Apocrine secretion (red arrowheads) in the mesenteric caecum wall and the incorporated hemocytes (green arrows), a nymph of the mite Belba pseudocorynopus, 3—Scutovertex minutus, the mesenteric wall with guanine crystals (arrowheads), 4—Tyrophagus putrescentiae, guanine crystals within mesenchymous tissue between the internal organs. Masson´s trichrome (1), autofluorescence under a confocal microscope (2), Nomarski DIC prism (3), TEM, pseudorelief in PC (4). Scales: 0.1 mm (1,2), 0.02 mm (3), 0.003 mm
Cells in the gut wall of mycophages turned voluminous and contained dark enzyme granules and vacuoles. They exhibited apocrine secretion (Fig. 2). The gut walls of non-mycophages were thin, without any of the conspicuous micromorphological characteristics mentioned above.
Conspicuous crystals of guanine were observed within the gut walls (Fig. 3) of mycophages, as well as around the gut (Fig. 4). No guanine crystals were observed in non-mycophages.
Nutrient deposits, in the form of red-stained granules of glycogen (a polysaccharide without nitrogen), indicated a response to a high intake of palatable food and a high rate of digestion in mycophages. Red-stained glycogen granules were also deposited in non-mycophagous mites (Platynothrus peltifer, Trichoribates trimaculatus). Indeed, glycogen granules appeared to be deposited in the presence of any palatable food (in this case, plant litter) and, therefore, were not a good indicator for mycophagy.
Both the presence and number of free cells within mites feeding on fungal food were conspicuous. Haemocytes were observed penetrating into the gut walls between wall cells (Figs. 2, 5 and 6). In spite of the palatability of non-fungal food, free cells were very rarely observed in non-mycophagous mites.
Alimentary tract and mesenchymal tissue between the internal organs: 5—mesenteral caecum with incorporated hemocytes (arrows), arrowheads point to the wall cells, 6—the vacuolized hemocytes adjacent the mesenteral caecum, 7—a cluster of associated bacteria (arrow) near mesenteron inside oribatid Damaeus onustus, 8—bacteria associated with Tyrophagus putrescentiae. Masson´s trichrome (5), Nomarski DIC prism (6,7), TEM (8). Scales: 0.050 mm (5), 0.02 mm (6), 0.01 mm (7), 0.003 mm (8). Abbreviation used: me—mesenteron, mec—mesenteric caecum
Bacterial bodies were especially observed around the posterior part of the mesenteron and around the blind mesenteric caeca (a projection of the mesenteron) of mycophages (Fig. 7). The structure of the bacterial bodies (density of bacteria and size of body) varied according to the fungus consumed and digested. Their presence and shape were confirmed by transmission electron microscopy (TEM—Fig. 8). No such bodies were recorded in non-mycophages.
Mycophage faeces contained numerous digested fragments of fungal propagula. Many luminous green (living) bacterial cells were also present (Fig. 9). Mite digestion did not appear to be rigorous. No digested fungal fragments were observed in non-mycophage faeces. In addition, markedly fewer or no bacterial cells were observed there in comparison with the mycophagous mites (Fig. 10).
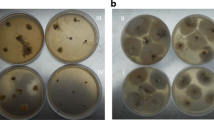
Consuption and digestion of food: 9—10 Faecal smears: 9—Damaeus riparius under fluorescent light—digested (dead) fungal mycelium (arrows) and living bacteria (green points), 10—Trichoribates trimaculatus , living algae (green spots); 11—12: Fungal colonies on the Petri dish with malt agar treated by bacterial suspension, restricted spots: 11—Alternaria sp., tretaed with Serratia liquefaciens—black spot (white arrow), intact fungus (black arrow), 12—Penicillium griseofulvum (white asterisk) and P.claviforme (black asterisk) tretaed by Pseudomonas fluorescens. Arrows point to treated spot, arrowheads to the control water drops. Acridine orange under fluorescent light (9, 10). Scales: 0.01 mm (9, 10)
Clear areas were formed in a basic fuchsin purple-blue layer when mycophage homogenate was applied to a carboxymethyl-chitin film on a microscopic slide, clearly indicating the digestion of chitin. The homogenate of non-mycophagous mites did not produce clear areas.The bacteria isolated and identified from the mite homogenate were found to be chitinolytic (Table 1).
Drops of a suspension of the chitinolytic bacteria isolated from the mycophagous mites digested the test fungi (Alternaria sp., Penicillium griseofulvum, Penicillium claviforme, Mucor racemosus and Fusarium oxysporum). Areas of digested fungus were very clearly delineated and visible, and they were surrounded by intact fungus (Figs. 11 and 12). The treatment of fungus by homogenate of mycophagous mites resulted in the same phenomenon. The population of Tyrophagus putrescentiae, chosen because of its rapid rate of reproduction, was introduced onto Alternaria sp. established on a malt agar Petri dish. After three weeks, an amorphous “mud” covered the dish, with no fungus visible. Mites introduced onto Fusarium oxysporum, and Penicillium claviforme produced the same results.
4 Discussion and conclusions
Many authors have reported the consumption of fungi by soil saprophagous mites (Coleman and McGinnis 1970; Pande and Berthet 1973; Mitchell and Parkinson 1976; Stefaniak and Seniczak 1981; Schneider et al. 2004). On that basis, these mites were classified into feeding guilds based on different forms of consumption (sucking, cell walls grazing) (Siepel and de Ruiter-Dijkman 1993). The digestion of the substrates, especially chitin in the fungal cell walls, however, has been neglected. Therefore, the autochthonous or allochthonous origin of the chitinases in mites was not defined or considered. Overgaard Nielsen (1962) assumed the occurence and origin of enzymes and he reported the bacterial production of chitinolytic enzymes in pulmonate snails. Shimano and Matsuo (2002) discussed the production of enzymes by the microorganisms with respect to their microanatomical results. The biochemical and immunological definition of enzyme activities, classified as allergens Der p 15 and Der p 18, were reported in Dermatophagoides pteronyssinus (O'Neil et al. 2006). They were considered as potentially important allergens for humans as well as dogs. The source of two allergens, hovewer, were not identified.
The multimethodical approach used to examine chitinolysis and mycophagy in soil organisms (i.e. bacteria and mites) indicated that while many soil mites consume parts of fungi only some can be considered as truly mycophagous, i.e. consuming all parts of the fungus cell, including the chitinous cell wall (Smrž 2007).
Microanatomical methods clearly showed not only “swallowing” of fungi fragments but also progressive digestion of fungal cells through the guts of mites considered as mycophages, while their faeces contained a large proportion of dead fungal fragments and many live bacteria. In the 1990 s, several chitinolytic bacteria (Bacillus badius, B. cereus, B. megatherium, Janthinobacterium lividum, Serratia marcescens, S. liquefaciens and Pseudomonas fluorescens) had been isolated (Smrž et al. 1991; Smrž and Trelová 1995) from mites known to be mycophagous (Damaeus riparius, D. auritus, Achipteria coleoptrata, Chamobates voigtsii, and Tyrophagus putrescentiae) (Smrž et al. 1991; Smrž and Trelová 1995). A multimethodical approach, especially enzyme activity and homogenate tests, had not been applied in those studies, however, and it had been possible only to hypothesize a process of “cooperation” between the isolated bacteria and mites for chitinolysis resulting in mycophagy.
The tested mycophagous mites were able to digest the whole fungal propagula, i.e. including the chitinous cell walls, through “cooperation” with internal extraintestinal bacteria that produced chitinolytic enzymes. These passed into the gut lumen and participated in chitin digestion. Chitinolysis by some bacterial species isolated from these (Serratia marcescens, S. liquefaciens, S.rubidea and Pseudomonas fluorescens, P.stutzeri, Brevundimonas vesicularis, Stenotrophomonas maltofillia ) was confirmed on thin carboxymethyl-chitin film and on a test fungus sample (Smrž and Čatská 1987; Smrž et al. 1991; Smrž 2003; Smrž and Norton 2004; Smrž and Soukalová 2008).
The fungal digestion differed whether bacteria were in suspension or were “cooperating” inside the mycophagous mites. The only bacterial digestion without mites resulted in restricted spots or area in fugal cover. The association of chitinolytic bacteria inside mites resulted in the destruction of complete fungal colonies in the Petri dish due to the mite’s ability to move and graze (Smrž and Čatská 1987; Smrž et al. 1991; Smrž and Norton 2004; Smrž and Soukalová 2008).
References
Aktuganov GE, Melent'ev AI, Kuz'mina LJu, Galimzyanova NF, Shirokov AV (2004) The Chitinolytic Activity of Bacillus Cohn Bacteria Antagonistic to Phytopathogenic Fungi. Microbiology 72:313–317
Citterio B, Malatesta M, Battistelli S, Marcheggiani F, Baffone W, Saltarelli R, Stocchi V, Gazzanelli G (2001) Possible involvement of Pseudomonas fluorescens and Bacillaceae in structural modifications of Tuber borchii fruit bodies. Can J Microbiol 47:264–268
Coleman D, McGinnis JT (1970) Quantification of fungus—small arthropod food chains in the soil. Oikos 21:134–137
Czajkowska B (1970) Rozwój rozkruszków na niektórych gatunkach grzybów. Zesz Problemowe Postepów Nauk Rolniczych 109:219–227
Dinsdale D (1974) The digestive activity of a phthiracarid mite mesenteron. J Zool Soc Lond 174:15–21
Hudson HJ (1986) Fungal biology. Edward Arnold, London, 298 pp
Jaspers E, Overmann J (2004) Ecological significance of microdiversity:identical 16 S rRNA gene sequence can be found in bacteria with highly divergent genomes and ecophysiologies. Appl Environ Microbiol 70:4831–4839
Kjøller A, Struwe S (1982) Microfungi in ecosystems: fungal occurrence and activity in litter and soil. In: A comparative analysis of soil fauna populations and their role in decomposition processes. Petersen, H. and Luxton, M., eds. Oikos 39 (Suppl.) pp 391–422
Kishore GK, Pande S, Podile AR (2005) Chitin-supplemented Foliar Application of Serratia marcescens GPS 5 Improves Control of Late Leaf Spot Disease of Groundnut by Activating Defence-related Enzymes. J Phytopathol 153:169–173
Kobayashi DY, Reedy RM, Bick JA, Oudemans PV (2002) Characterization of chitinase gene from Stenotrophomonas maltophilia Strain 34 S1 and its involvement in biological control. Appl Environ Microbiol 68:1047–1054
Lim HS, Kim YS, Kim SD (1991) YPL-1 Genetic Transformation and Antifungal Mechanism against Fusarium solani, an Agent of Plant Root Rot. Appl Environ Microbiol 57:510–516
Luxton M (1972) Studies on the oribatid mites of a Danish beech wood soil. I. Nutritional biology. Pedobiologia 2:434–463
Mitchell MJ, Parkinson D (1976) Fugal feeding of oribatid mites (Acari: Cryptostigmata) in an aspen woodland soil. Ecology 57:302–312
Molloy C, Burke B (1997) Expression and secretion of Janthinobacterium lividum chitinase in Saccharomyces cerevisiae. Biotechnol Lett 19:1161–1164
O'Neil SE, Heinrich TK, Hales BJ, Hazell LA, Holt DC, Fischer K, Thomas WR (2006) The chitinase allergens Der p15 and Der p18 from Dermatophagoides pteronyssinus. Clin Exp Allergy 36:831–839
Overgaard Nielsen C (1962) Carbohydrates in soil and litter invertebrates. Oikos 13:200–215
Pande YD, Berthet P (1973) Studies on the food and feeding habits of soil Oribatei in a black pine plantation. Oecologia Berl 12:413–426
Pauly F (1956) Zur Biologie einiger Belbiden (Oribatei, Moosmilben) und zur Funktion ihrer pseudostigmatischen Organe. Zoologische Jahrbücher Systematik 84:275–328
Schneider K, Migge S, Norton RA, Scheu S, Langel R, Reineking A, Maraun M (2004) Trophic niche differentiation in soil microarthropods (Oribatida, Acari: evidence from stable isotope ratios (15 N/14 N). Soil Biol Biochem 36:1769–1774
Seniczak S, Stefaniak O (1978) The microflora of the alimentary canal of Oppia nitens (Acarina, Oribatei). Pedobiologia 18:110–119
Siepel H, de Ruiter-Dijkman EM (1993) Feeding guilds of oribatid mites based on their carbohydrase activities. Soil Biol Biochem 25:1491–1497
Shimano S, Matsuo T (2002) Morphological studies on the digestive tract of Scheloribates azumaensis (Acari: Oribatida). J Acarological Soc Jpn 11:37–40
Smrž J (1989) Internal anatomy of Hypochthonius rufulus (Acari, Oribatida). J Morphol 200:215–230
Smrž J (1992) Some adaptive features in the microanatomy of moss-dwelling oribatid mites (Acari: Oribatida) with respect to their ontogenical development. Pedobiologia 36:306–320
Smrž J (1996) Some aspects of the life strategy of oribatid mites. In Acarology IX., Mitchell, Horn, R. D. J., Needham, G. R. and Welbourn, W. C., eds. Ohio Biol. Survey, Columbus, Ohio, USA. pp 553–555
Smrž J (2000) A modified test for chitinase and cellulase activity in soil mites. Pedobiologia 44:186–189
Smrž J (2002a) Nutritional biology: the basic step in the autecological studies (multi-methodical approach). Eur J Soil Biol 38:35–38
Smrž J (2002b) The excrement analysis—the useful tool for the biological and autecological studies in soil zoology. In: Studies on Soil Fauna in Central Europe. Tajovský, K., Balík, V. and Pižl, V. eds. Biology Centre of the Academy of Sciences of the Czech Republic, České Budějovice. pp.185–189
Smrž J (2003) Microanatomical and biological aspects of bacterial associations in Tyrophagus putrescentiae (Acari: Acaridida). Exp Appl Acarol 31:105–113
Smrž J (2006) Microhabitat selection in the simple oribatid community dwelling in epilithic moss cover (Acari: Oribatida). Naturwisseschaften 93:570–576
Smrž J (2007) Nutritional biology in the oribatid mites (Acari: Oribatida) communities in the different, closely neighbouring microhabitats in the steppe biotope—preliminary report. In: Tajovský K, Schlaghamerský J, Pižl V (eds) Contributions to Soil Zoology in Central Europe II. Biology Centre of the Academy of Sciences of the Czech Republic, České Budějovice, pp 153–160
Smrž J, Čatská V (1987) Food selection of the field population of Tyrophagus putrescentiae (Schrank) (Acari, Acaridida). Z Angewadte Entomologie 104:329–335
Smrž J, Čatská V (1989) The effect of the consumption of some soil fungi on the internal microanatomy of the mite Tyrophagus putrescentiae (Schrank)(Acari, Acaridida). Acta Univ Carol Biol 33:81–93
Smrž J, Jungová E (1989) The ecology of a field population of Tyrophagus putrescentiae (Acari, Acaridida). Pedobiologia 33:183–192
Smrž J, Norton RA (2004) Food selection and internal processing in Archegozetes longisetosus (Acari: Oribatida). Pedobiologia 48:111–120
Smrž J, Trelová M (1995) The associations of bacteria and some soil mites (Acari: Oribatida and Acaridida). Acta Zool Fennica 196:120–123
Smrž J, Soukalová H (2008) Mycophagous mites (Acari: Oribatida and Acaridida) and their cooperation with chitinolytic bacteria. In: Bertrand M. Kreiter, S. McCoy, K.D. Migeon A.,Navajas M., Tixier M.-S. Vial L. eds.: Integrative acarology. EURAAC. pp 359–362
Smrž J, Svobodová J, Čatská V (1991) Synergetic participation of Tyrophagus putrescentiae (Schrank) (Acari, Acaridida) and its associated bacteria on the destruction of some soil micromycetes. J Appl Entomol 111:206–210
Stefaniak O, Seniczak S (1976) The microflora of the alimentary canal of Achipteria coleoptrata (Acarina, Oribatei). Pedobiologia 16:185–194
Stefaniak O, Seniczak S (1981) The effect of fungal diet on the development of Oppia nitens (Acari, Oribatei) and on the miocroflora of its alimentary tract. Pedobiologia 21:202–210
Wallwork JA (1976) The distribution and diversity of soil fauna. Academic, London, 353 pp
Wang SL, Chiou SH, Chang WT (1997) Production of chitinase from shellfish waste by Pseudomonas aeruginosa K-187. Proc Natl Sci Counc Repub China 21:71–78
Watanabe T, Oyanagi W, Suzuki K, Tanaka H (1990) Chitinase system of Bacillus circulans WL-12 and importance of chitinase A1 in chitin degradation. J Bacteriol 172:4017–4022
Wigglesworth VB (1974) The Principles of Insect Physiology, 7th edn. Chapman and Hall, London, 827 pp
Wolf MM, Rockett CL (1984) Habitat changes affecting bacterial composition in the alimentary canal of oribatid mites (Acari: Oribatida). Int J Acarol 10:209–215
Zhang Z, Yuen GY, Sarath G, Penheiter AR (2001) Chitinases from the plant disease biocontrol agent, Stenotrophomonas maltophilia C3. Phytopathology 91:204–211
Acknowledgements
The fieldwork and histological work for this study were supported by Grant No. 526/07/0393 from the Grant Agency of the Czech Republic. The other laboratory work was supported by grant MSM 0021620828 from the Ministry of Education of the Czech Republic. We wish to thank ing.M.Doubek, State Health Institute, Prague, for his very kind help with the TEM observations, prof.dr.J.Vávra, Department of Parasitology, Charles University, Prague, who rendered possible to make the ultramicrotome sections and mgr.T.Erban for his information about Dermatophagoides chitinases. We wish to thank also Mrs. Milada Řeháková for her technical help.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Smrž, J., Čatská, V. Mycophagous mites and their internal associated bacteria cooperate to digest chitin in soil. Symbiosis 52, 33–40 (2010). https://doi.org/10.1007/s13199-010-0099-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13199-010-0099-6